Abstract
Microscale hydrogels have been shown to be beneficial for various applications such as tissue engineering and drug delivery. A key aspect in these applications is the spatial organization of biological entities or chemical compounds within hydrogel microstructures. For this purpose, sequentially patterned microgels can be utilized to spatially organize either living materials to mimic biological complexity or to encapsulate multiple chemicals to design functional microparticles for drug delivery. Photolithographic methods are the most common way to pattern microscale hydrogels, but are limited to photocrosslinkable polymers. So far, conventional micromolding approaches use static molds to fabricate structures, which limits the resulting shapes that can be generated. In this study, we describe a dynamic micromolding technique to fabricate sequentially patterned hydrogel microstructures by exploiting the thermo-responsiveness of poly(N-isopropylacrylamide) based micromolds. These responsive micromolds exhibited shape changes under temperature variations, facilitating the sequential molding of microgels at two different temperatures. We fabricated multi-compartmental striped, cylindrical, and cubic microgels that encapsulated fluorescent polymer microspheres or different cell types. These responsive micromolds can be used to immobilize living materials or chemicals into sequentially patterned hydrogel microstructures which may potentially be useful for a range of applications at the interface of chemistry, biology, materials science and engineering.
In this communication, we introduce responsive micromolds to sequentially pattern hydrogel microstructures. Hydrogels are readily engineered with tuned biodegradability, high permeability to oxygen or other soluble factors, and mechanical stability1,2 and are therefore ideal for tissue engineering3 and drug delivery applications.2 Micro- and nano-engineering methods have been used to fabricate shape-controlled micro- and nano-scale hydrogels to encapsulate living materials for tissue engineering and bioprocess applications4-6 or chemicals for controlled drug delivery.7,8 Cell-laden hydrogels have been sequentially photopatterned to generate biomimetic microtissues.9,10 Multi-compartment hydrogels were also used to encapsulate and deliver drugs in a controlled manner.11,12 Traditionally, sequential patterning of hydrogels has relied on photolithographic methods, which are not applicable to a wide variety of polymers such as those that require thermal or ionic crosslinking.6 Herein, we describe a simple method to sequentially pattern hydrogel microstructures by utilizing the temperature dependent shape change properties of poly(N-isopropylacrylamide) (PNIPAAm) based micromolds.
The shape of microgels can control the loading and release of the drugs13 and the design of constructs for tissue engineering applications.4-6,14 Drugs can be encapsulated in multi-compartment microgels to control the release of drugs sequentially from different compartments.11,12 Sequentially patterned hydrogels have also been used to fabricate tissue constructs that mimic native tissue architecture.9,10 Microfluidic methods have been used to generate microparticles15 or pattern microgels,16,17 though it is challenging to use these methods to fabricate sequentially patterned hydrogels with different shapes and the associated apparatus is generally complex and requires multiple fabrication steps, limiting a high-throughput production. Therefore, a simple method to fabricate sequentially patterned microgels could be useful.
While photolithographic methods have been used to sequentially pattern multilayer hydrogel microstructures,9,10 they cannot be used with non-photocrosslinkable hydrogels. Furthermore, photoinitiators may cause cytotoxicity for applications where cells are encapsulated.9 Micromolding is an alternative approach for forming hydrogel microstructures,5,6,14 though the static nature of conventional micromolding templates inhibits sequential molding. Therefore, micromolding hydrogel microstructures with two or more spatially organized gel portions remains a challenge. This is because as one material gels there is no additional free space for subsequent gels. Herein, we exploited the thermoresponsiveness of PNIPAAm to create dynamic micromolds to overcome the static nature of conventional micromolding techniques. PNIPAAm is a temperature responsive polymer which exhibits a lower critical solution temperature (LCST) of ~32 °C.18 PNIPAAm swells when the temperature is below its LCST and shrinks when the temperature is raised above. Recently, soft lithographically fabricated PNIPAAm-based microwells demonstrated shape changes in response to variations in temperature.19
By using a soft lithographic method,19 we fabricated PNIPAAm micromolds with various patterns, such as microgrooves, as well as circular and square microwells (see Supporting Information). We subjected these structures to different temperatures (4 °C, 24 °C and 37 °C) to test the responsiveness. After 15 min of incubation at 4 °C, microstructured PNIPAAm stripes swelled significantly (Figure 1a). After raising the temperature from 4 °C to 24 °C, the space within the PNIPAAm stripes significantly increased within 15 min as the gels shrank (Figure 1a,d). The space within the PNIPAAm stripes further expanded (Figure 1a,d) within 30 min after changing the temperature from 24 °C to 37 °C. We also analyzed the temperature dependent shape change of circular microwells. When circular microwells were subjected to the same experimental procedure, swelling of PNIPAAm caused circular microwells to lose their circularity at 4 °C (Figure 1b). Within 15 min after changing the temperature from 4 °C to 24 °C, microwells had returned to their original circular shapes with significantly increased surface area (Figure 1b,e). Similarly, microwells retained their circularity while increasing their surface area significantly (Figure 1b,e) within 30 min following a temperature increase from 24 °C to 37 °C. Square microwell arrays were also tested in a similar manner, exhibiting non-square microwell shapes at 4 °C with reduced surface area (Figure 1c). The microwells began to return to their square shapes with a significant change in surface area (Figure 1c,f) within 15 min after changing the temperature from 4 °C to 24 °C. Microwell size increased further (Figure 1c,f) within 30 min following a temperature increase from 24 °C to 37 °C. Taken together these results show that PNIPAAm micromolds exhibited significant changes in the patterned areas at different temperatures. Therefore, these micromolds posses sufficient shape changing behavior for sequential patterning of hydrogels at different temperatures.
Figure 1.
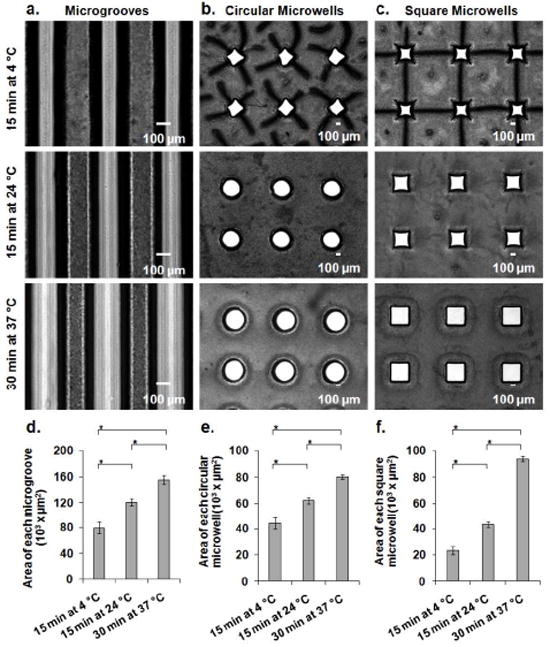
Shape responsiveness of PNIPAAm micromolds at three different temperatures. Time lapse images for a) microgrooves, b) circular microwells, and c) square microwells. Corresponding responsiveness plot based on top-viewed surface areas for d) microgrooves, e) circular microwells, and f) square microwells. * indicates a statistically significant difference in variance (p < 0.05).
We demonstrated the capability of responsive micromolds for fabrication of multi-compartment hydrogels by generating sequentially patterned microstructures of agarose. Agarose is a thermoresponsive polysaccharide20 which can be tailored to be gelled or melted at different temperatures.21 The mechanical stiffness of agarose can be altered by varying the gelling temperature, prepolymer concentration, or curing time.21,22 Agarose has previously been used in various applications such as cell encapsulation,20,23 hyrogel microfluidics,21 and drug delivery.24,25 Agarose is also biocompatible for in vivo applications.26 We therefore fabricated sequentially patterned agarose microstructures for potential use in biotechnological applications. To generate multi-compartment agarose microstructures, we molded agarose at two different temperatures by exploiting the temperature dependent shape changing properties of PNIPAAm based micromolds.
The schematic for sequential patterning of hydrogel microstructures is shown in Figure 2. After fabrication, responsive micromolds were first kept at 24 °C for 15 min to reach their ambient temperature shape. In our process, the prepolymer of the first gel was then placed onto a responsive micromold and molded with a flat poly(dimethylsiloxane) (PDMS) substrate at 24 °C. The molded gel was then placed at 4 °C for 15 min to induce rapid gelling. Then, the responsive micromold containing the first crosslinked gel was placed at 37 °C for 30 min to allow the micromold to expand, resulting in more free space. The second gel solution was then placed on the micromold and immediately pressed with a new PDMS slab at 37 °C and crosslinked for 30 min. As the micromolds have already maximized their free spaces during the first incubation at 37 °C due to PNIPAAm shrinking, the second incubation at 37 °C did not cause a further significant expansion in patterned areas of the micromolds. Three different sequentially patterned hydrogel microstructures were obtained with this process, particularly stripe microgels from microgroove molds, cylindrical microgels from circular microwell molds, and cubic microgels from square micromolds.
Figure 2.
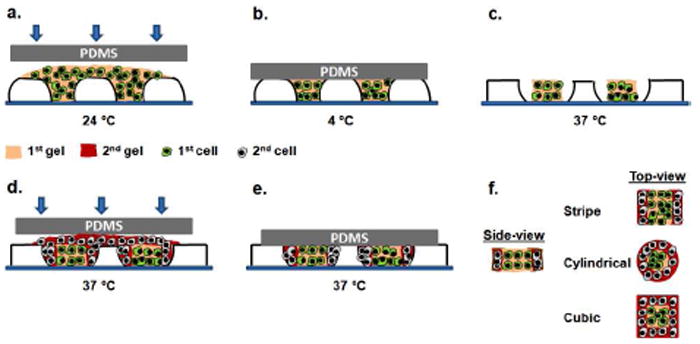
Schematic diagram of sequential patterning of hydrogel microstructures with responsive micromolds. Either fluorescent microbeads or cells (3T3 fibroblasts, HepG2 cells and HUVECs) were encapsulated within agarose microgels during fabrication process. a) The first gel precursor with encapsulated cells or microbeads was put on a responsive micromold and molded with a PDMS slab at 24 °C. b) 15 min incubation at 4 °C for crosslinking the first gel. c) 30 min incubation at 37 °C to allow responsive micromolds to shrink. d) The second gel precursor with encapsulated cells or microbeads was put on a responsive micromold and molded with a PDMS slab at 37 °C. e) 30 min incubation at 37 °C for crosslinking the second gel. f) Side and top views of resulting hydrogel microstructures.
To model the encapsulation of organic materials, we encapsulated green fluorescent microbeads (1.2 μm) within the first gel and red fluorescent microbeads (2 μm) within the second gel. The first crosslinked gel roughly possessed the shape of the microgroove patterns at 24 °C and the second gel was spatially aligned parallel to the first gel by filling the free space that resulted from PNIPAAm contraction at 37 °C (Figure 3a). The resulting microstructures within the microgrooves were multi-compartment stripe microgels. For circular microwells, the first crosslinked gel had the similar circular shape of microwells at 24 °C and the second gel was molded around the first gel by keeping the free space that resulted from PNIPAAm shrinking at 37 °C (Figure 3a). Multi-compartment cylindrical microgels were fabricated by using the circular microwell molds. For square microwells, the first molded gel had a similar square-like shape of the microwells at 24 °C and second gel was spatially immobilized around the first gel pattern by obtaining a square shape at 37 °C (Figure 3a). Interestingly, the shapes of the first gels were similar to the shapes of all micromold patterns at 24 °C, suggesting that 15 min incubation at 4 °C to crosslink the first gels did not significantly affect their shape.
Figure 3.
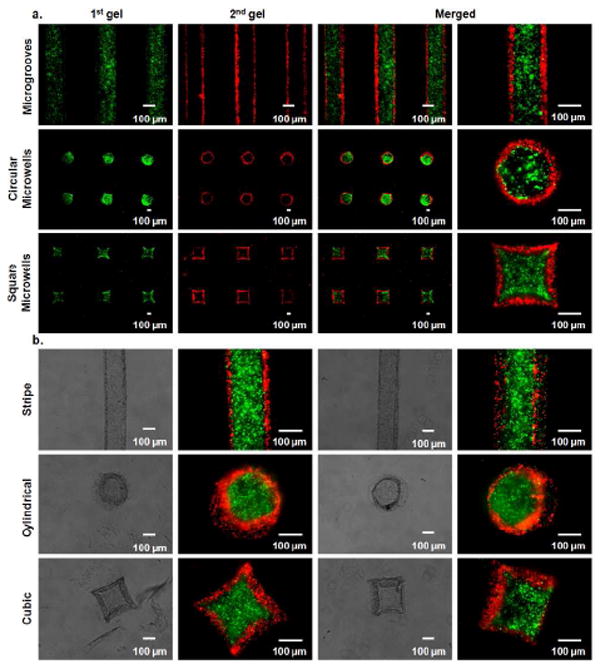
a) Fluorescent images of sequentially molded microgels within responsive microgrooves, circular microwells, and square microwells. Green microbeads were encapsulated within the first gels and red microbeads were encapsulated within the second gels. b) Hydrogel microstructures were recovered from responsive micromolds. Phase contrast and fluorescent images for stripe microgels, cylindrical microgels, and cubic microgels.
Resulting hydrogel microstructures were recovered from micromolds by gently flowing PBS over the micromold surfaces with a pipette. As shown in Figure 3b, stripe microgels were recovered from microgrooves and conserved their shapes and multi-compartmental structures. Multi-compartmental cylindrical microgels were also harvested from circular microwells and conserved their cylindrical geometry (Figure 3b). Harvested microgels from square microwells exhibited cubic geometry with two layers (Figure 3b). For all hydrogel microstructures, the second layer was patterned around the first gel. Therefore, this sequential micromolding process may be useful for encapsulating drugs or other organic functional materials within microgels to generate multi-compartment drug carriers or functional multilayer microstructures.
Native tissues contain multiple spatially organized cell types within a three dimensional (3D) microenvironment.6,17,27 Heterotypic cell-cell interactions and cell-matrix interactions play an important role in fabricating functional tissue constructs.14,17,28 3D microenvironments can be engineered with biocompatible or biodegradable hydrogels. The presented sequential micromolding technique can replicate the biological complexity of native tissues by arranging the orientation of different cell types within hydrogel microstructures. To showcase this method for tissue engineering applications, cells were encapsulated within hydrogel microstructures (20 × 106 cells/ml) (Figure 4a). 3T3 fibroblasts were used as one model cell type and human umbilical vein endothelial cells (HUVECs) were chosen as the second cell type due to their relevance in engineered vascularized tissues. 3T3 fibroblasts (red) were encapsulated in agarose molded as the first gel within microgrooves. Subsequently, green fluorescent protein (GFP) labeled HUVECs (green) were separately mixed with agarose which was then molded as the second gel. The second gel was finely patterned around the first gel which resulted in a stripe tissue construct containing two spatially organized cell types (Figure 4a). These cell-laden stripe microgels could be useful as models for cardiac, skeletal muscle, myoblast, and neural tissues.
Figure 4.
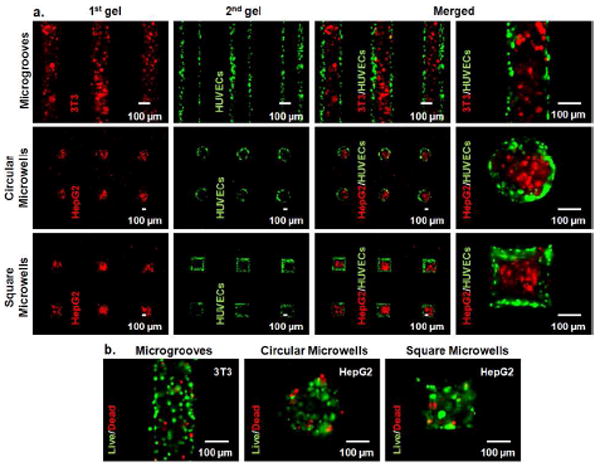
Living materials were encapsulated in agarose microgels. a) Cell encapsulated microgels were sequentially patterned with responsive micromolds. For microgrooves, agarose precursor containing 3T3 fibroblasts (red) was patterned as the first gel and gel precursor containing GFP-HUVECs was patterned as the second gel. For circular and square microwells, agarose prepolymer containing HepG2 cells (red) was patterned as the first gel and the second gel contained GFP-HUVECs. b) Fluorescent microscopy images for live/dead experiment. Microgels containing 3T3 fibroblasts were encapsulated within microgrooves while HepG2-laden microgels were within circular and square microwells.
For circular and square microwells, we used human hepatoblastoma (HepG2) cells as the first cell type and HUVECs as the second cell type. Co-culturing hepatocytes with endothelial cells resulted in an improved hepatic functionality.28 An agarose solution containing HepG2 cells (red) was first molded within both circular and square microwells. The second gels containing GFP-HUVECs (green) were subsequently molded around the first gels. The second gels containing HUVECs were patterned around the first gels which produced circular and square shaped tissue constructs containing two spatially oriented cell types (Figure 4a). After 3 days of incubation, high cell-viability levels were observed, qualitatively suggesting that the hydrogel patterning process, the agarose precursor, and the PNIPAAm micromolds did not cause an adverse effect on cell viability (Figure 4b). These cell-laden hydrogel microstructures could be used as models of hepatic tissues. Other cell types could be substituted for 3T3 fibroblasts, HepG2 cells, and HUVECs to obtain different tissue models. These cell-laden modular tissues can further be assembled to obtain larger tissue constructs retaining the controlled microarchitecture and cell placement by using modular tissue engineering methods.29 Furthermore, mixing agarose with proteins such as collagen can induce cell spreading,30,31 which could be potentially useful for fabricating vascularized tissues with our micromolding method.
For microbead or cell experiments, we observed that some excess prepolymer solutions were adsorbed within the hydrogel structure of micromolds. We also observed that wrapping of the second gel over the first gel may occur with non-flat PDMS slabs or by using the second gel precursor over desired volume (60 μl). It should also be noted that our micromolding method may require further refinements to fabricate smaller hydrogel microstructures. For example, different PNIPAAm concentrations may be required to control the swelling/deswelling behavior of the micromold.
We have described a simple method, called responsive micromolding, to sequentially pattern hydrogel microstructures by using the thermo-responsive PNIPAAm-based micromolds. The patterned surface areas of responsive micromolds increased with increased temperature. This feature was exploited to sequentially mold microgels containing two distinct layers. This technique can encapsulate different organic materials into different layers of microgels for drug delivery or functional microparticle applications. Furthermore, this method can encapsulate living materials to create biomimetic modular tissue constructs with spatially organized different cell types in a single hydrogel microstructure. In summary, these responsive micromolds could be a versatile tool in several fields, including tissue engineering, drug delivery, diagnostics, drug discovery, and 3D cell culture systems.
Supplementary Material
Acknowledgments
This research was financially supported by the U.S. Army Research Office through the Institute for Soldier Nanotechnologies at MIT under the project DAAD-19-02-D-002, the NIH (DE013023 and DE016516 (R.L.), HL092836, DE019024, EB012597, AR057837, DE021468, and HL099073 (A.K.)), and the Office of Naval Research.
Footnotes
Detailed information for fabrication and experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Langer R, Peppas NA. AIChE Journal. 2003;49:2990. [Google Scholar]
- 2.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Advanced Materials. 2006;18:1345. [Google Scholar]
- 3.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 4.McGuigan AP, Sefton MV. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11461. doi: 10.1073/pnas.0602740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franzesi GT, Ni B, Ling YB, Khademhosseini A. Journal of the American Chemical Society. 2006;128:15064. doi: 10.1021/ja065867x. [DOI] [PubMed] [Google Scholar]
- 6.Tang MD, Golden AP, Tien J. Journal of the American Chemical Society. 2003;125:12988. doi: 10.1021/ja037677h. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman AS. Advanced Drug Delivery Reviews. 2002;54:3. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 8.Kelly JY, DeSimone JM. Journal of the American Chemical Society. 2008;130:5438. doi: 10.1021/ja8014428. [DOI] [PubMed] [Google Scholar]
- 9.Liu VA, Bhatia SN. Biomedical Microdevices. 2002;4:257. [Google Scholar]
- 10.Du YA, Ghodousi M, Qi H, Haas N, Xiao WQ, Khademhosseini A. Biotechnology and Bioengineering. 2011;108:1693. doi: 10.1002/bit.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An YJ, Hubbell JA. Journal of Controlled Release. 2000;64:205. doi: 10.1016/s0168-3659(99)00143-1. [DOI] [PubMed] [Google Scholar]
- 12.Kiser PF, Wilson G, Needham D. Journal of Controlled Release. 2000;68:9. doi: 10.1016/s0168-3659(00)00236-4. [DOI] [PubMed] [Google Scholar]
- 13.Edwards DA, Hanes J, Caponetti G, Hrkach J, BenJebria A, Eskew ML, Mintzes J, Deaver D, Lotan N, Langer R. Science. 1997;276:1868. doi: 10.1126/science.276.5320.1868. [DOI] [PubMed] [Google Scholar]
- 14.Yeh J, Ling YB, Karp JM, Gantz J, Chandawarkar A, Eng G, Blumling J, Langer R, Khademhosseini A. Biomaterials. 2006;27:5391. doi: 10.1016/j.biomaterials.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Jeong WJ, Kim JY, Choo J, Lee EK, Han CS, Beebe DJ, Seong GH, Lee SH. Langmuir. 2005;21:3738. doi: 10.1021/la050105l. [DOI] [PubMed] [Google Scholar]
- 16.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Lab on a Chip. 2009;9:269. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 17.Huang CP, Lu J, Seon H, Lee AP, Flanagan LA, Kim HY, Putnam AJ, Jeon NL. Lab on a Chip. 2009;9:1740. doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alarcon CDH, Pennadam S, Alexander C. Chemical Society Reviews. 2005;34:276. doi: 10.1039/b406727d. [DOI] [PubMed] [Google Scholar]
- 19.Tekin H, Anaya M, Brigham MD, Nauman C, Langer R, Khademhosseini A. Lab on a Chip. 2010;10:2411. doi: 10.1039/c004732e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uludag H, De Vos P, Tresco PA. Advanced Drug Delivery Reviews. 2000;42:29. doi: 10.1016/s0169-409x(00)00053-3. [DOI] [PubMed] [Google Scholar]
- 21.Ling Y, Rubin J, Deng Y, Huang C, Demirci U, Karp JM, Khademhosseini A. Lab on a Chip. 2007;7:756. doi: 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]
- 22.Aymard P, Martin DR, Plucknett K, Foster TJ, Clark AH, Norton IT. Biopolymers. 2001;59:131. doi: 10.1002/1097-0282(200109)59:3<131::AID-BIP1013>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Jones KS, Sefton MV, Gorczynski RM. Transplantation. 2004;78:1454. doi: 10.1097/01.tp.0000142094.63083.fb. [DOI] [PubMed] [Google Scholar]
- 24.Haglund BO, Upadrashta SM, Neau SH, Cutrera MA. Drug Development and Industrial Pharmacy. 1994;20:947. [Google Scholar]
- 25.Wang JY, Wang ZH, Gao J, Wang L, Yang ZY, Kong DL, Yang ZM. Journal of Materials Chemistry. 2009;19:7892. [Google Scholar]
- 26.Rahfoth B, Weisser J, Sternkopf F, Aigner T, von der Mark K, Brauer R. Osteoarthritis and Cartilage. 1998;6:50. doi: 10.1053/joca.1997.0092. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg CB, Bell E. Science. 1986;231:397. doi: 10.1126/science.2934816. [DOI] [PubMed] [Google Scholar]
- 28.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Faseb Journal. 1999;13:1883. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 29.Nichol JW, Khademhosseini A. Soft Matter. 2009;5:1312. doi: 10.1039/b814285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batorsky A, Liao JH, Lund AW, Plopper GE, Stegemann JP. Biotechnology and Bioengineering. 2005;92:492. doi: 10.1002/bit.20614. [DOI] [PubMed] [Google Scholar]
- 31.Ulrich TA, Jain A, Tanner K, MacKay JL, Kumar S. Biomaterials. 2010;31:1875. doi: 10.1016/j.biomaterials.2009.10.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


