Abstract
Zinc finger E-box binding (ZEB) proteins ZEB1 and ZEB2 are transcription factors essential in transforming growth factor (TGF)-β-mediated senescence, epithelial to mesenchymal transition (EMT) and cancer stem cell function. ZEBs are negatively regulated by members of the miR-200 microRNA family, but precisely how tumor cells expressing ZEBs emerge during invasive growth remains unknown. Here we report that NOTCH3-mediated signaling prevents expansion of a unique subset of ZEB-expressing cells. ZEB expression was associated with the lack of cellular capability of undergoing NOTCH3-mediated squamous differentiation in human esophageal cells. Genetic inhibition of the Notch-mediated transcriptional activity by dominant-negative Mastermind-like1 (DNMAML1) prevented squamous differentiation and induction of Notch target genes including NOTCH3. Moreover, DNMAML1 enriched EMT competent cells exhibited robust upregulation of ZEBs, downregulation of the miR-200 family, and enhanced anchorage independent growth and tumor formation in nude mice. RNA interference (RNAi) experiments suggested the involvement of ZEBs in anchorage independent colony formation, invasion and TGF-β-mediated EMT. Invasive growth and impaired squamous differentiation was recapitulated upon Notch inhibition by DNMAML1 in organotypic 3D culture, a form of human tissue engineering. Together, our findings indicate that NOTCH3 is a key factor limiting the expansion of ZEB-expressing cells, providing novel mechanistic insights into the role of Notch signaling in the cell fate regulation and disease progression of squamous esophageal cancers.
Keywords: Notch, EMT, squamous cell differentiation, ZEB1, miR-200
Introduction
The stratified squamous epithelium of the esophagus is regulated at an exquisite level. Exiting from the cell cycle, basal keratinocytes migrate towards the luminal surface. They undergo terminal differentiation in the suprabasal layer, expressing Involucrin (IVL) and cytokeratins such as CK13, and eventually desquamated into the lumen to complete epithelial renewal. Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive forms of squamous cell carcinomas (SCCs)(1) and is a paradigm for the investigation for all types of SCCs. Squamous differentiation contributes to tumor heterogeneity in SCCs. An individual tumor often consists of both well-differentiated cell nests with central keratinization (i.e. keratin pearl) and poorly-differentiated cell nests.
The Notch pathway regulates cell fate and differentiation through cell-cell communication. Ligand binding triggers a series of enzymatic cleavages of one of four Notch receptor paralogs (NOTCH1-4), resulting in nuclear translocation of the intracellular domain of Notch (ICN). ICN forms a transcriptional activation complex containing a common transcription factor CSL [CBF-1/RBP-jκ, Su(H), Lag-1] and the coactivator Mastermind-like (MAML)(2). Notch target genes include the HES/HEY family of transcription factors. CSL-dependent canonical Notch signaling regulates squamous differentiation and epidermal barrier functions (3). Loss of Notch receptors or γ-secretase, a key Notch processing enzyme, or inhibition of the CSL-dependent activity by dominant negative MAML1 (DNMAML1) impairs squamous differentiation and causes epidermal barrier defects, promoting SCC (4–9). We have demonstrated recently that NOTCH1 regulates esophageal epithelial homeostasis through NOTCH3, which is activated in a CSL-dependent manner at the onset of squamous differentiation and required for induction of IVL and CK13 (10). However, it remains to be elucidated as to how Notch signaling may contribute to esophageal cancer initiation and progression.
Epithelial-to-mesenchymal transition (EMT) is marked by loss of epithelial characteristics (e.g. cell polarity and cell-cell junctions) and gain of mesenchymal characteristics (e.g. fibroblastic spindle-shaped morphology and an increased motility). EMT occurs during cancer cell invasion and metastasis (11–13). In a mouse xenograft model using viral-oncogene transformed human esophageal cells, we have documented EMT in vivo (14). In primary ESCC, EMT markers are upregulated at the invasive front (15–19). TGF-β is a potent EMT inducer in the tumor microenvironment (20) and expressed by both tumor and stromal cells in ESCC (21). EMT occurs also during the early stages of carcinogenesis to bypass oncogene-induced senescence (19, 22). We have found recently that malignant transformation of human esophageal cells by EGFR oncogene causes enrichment of EMT-competent cells negating oncogene-induced senescence through transcriptional repression of the INK4 locus by zinc finger E-box binding (ZEB) proteins ZEB1 and ZEB2 (23), transcription factors essential in TGF-β-mediated EMT, senescence and maintenance of cancer stem cells (24–25). ZEBs are subjected to negative regulation by the microRNA (miR)-200 family members (26). However, neither the status of ZEBs nor their regulation in ESCC is known to date.
Herein, we demonstrate that ZEB1 is induced in ESCC at the invasive front undergoing EMT-like dedifferentiation. Loss of the NOTCH3-mediated CSL-dependent transcriptional activity allows expansion of EMT-competent cells expressing ZEBs, providing a novel mechanistic link between the Notch pathway and cell fate regulatory transcription factors during cancer progression.
Materials and Methods
Tissue samples
Paraffin blocks containing primary ESCC and adjacent normal tissues were procured via surgery as described previously (n=31)(27) and at the Hospital of the University of Fukui (n=20). All of the clinical materials were obtained from informed-consent patients in accordance with Institutional Review Board standards and guidelines.
Cell lines, treatment and organotypic 3D culture
HCE7 and other (TE series) ESCC cell lines were described previously (28). EPC2-hTERT and derivatives transformed by either SV40 Large T antigen and Ha-RasV12 (T-TeRAS) or EGFR, p53R175H and cyclin D1 (EPC2-T) were described (27, 29–30). EPC2-hTERT derivatives stably expressing short hairpin RNA (shRNA) directed against NOTCH3 (two cell lines Notch3-A and Notch3-B expressing independent shRNA sequences V2LHS_229748 and N3-B, V2LHS_93017, respectively) or a non-silencing control sequence (Open Biosystems) was described previously (10). Cells were treated with 0.6 mM calcium chloride (Ca2+), Compound E, a γ-secretase inhibitor (GSI) or 5 ng/ml TGF-β1 as described (10, 14, 23). Phase contrast images were acquired to score spindle-shaped cells by counting at least 100 cells per high-power field (n=6) as described (14, 23). Organotypic 3D culture was done as described previously (10, 14, 31).
Retrovirus and lentivirus-mediated gene transfer and RNA interference (RNAi)
Retroviruses expressing ICN1 or DNMAML1 (10) and tetracycline-inducible lentiviruses (Open Biosystems, Huntsville, AL) expressing short hairpin RNA (shRNA) directed against ZEB1 (clones V2THS_116663 and V2THS_116659), ZEB2 (clones V2THS_95420 and V3THS_373827) or a non-silencing control sequence (clone RHS4743) were produced and transduced as described (10, 14, 23). Cells were labeled with tdTomato for xenograft transplantation experiments as described (32). Cells transduced with GFP (for DNMAML1) or tdTomato were selected for the brightest level of fluorescence (top 20%) by flow sorting. Small interfering RNA (siRNA) directed against NOTCH1 (two independent sequences Notch1-A, HSS181550 and Notch1-B, HSS107249), or a non-silencing scramble control sequence (12935-300)(Invitrogen) was transfected transiently using the Lipofectamine™ RNAiMAX reagent (Invitrogen) as described previously (10).
Soft agar colony formation assays
Soft agar colony formation assays were done as described previously (33). In brief, 2.5×104 cells were suspended in 0.67% agarose containing media and overlaid on top of a 1% agarose containing the medium per well, and grown for two weeks.
RNA isolation, cDNA synthesis, real-time reverse-transcription polymerase chain reactions (RT-PCR) and microarray analysis
RNA isolation, cDNA synthesis and real-time RT-PCR were done as described (10, 23) using TaqMan® Gene Expression and MicroRNA® Assays (Applied Biosystems) (Supplementary Table S1). Gene array experiments were done using an Affymetrix gene chip (U133+v2.0)(Affymetrix, Santa Clara, CA) as described previously (27, 34). Data was deposited at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo)(accession # GSE27424).
Western blot analysis
Western blotting was done as described (10, 23). Supplementary Table S2 lists primary antibodies and the titers used for Western blotting.
Flow cytometry
Flow cytometry was done using the FACSCalibur (BD Biosciences). In brief, cells were fixed and permeabilized in cold acetone at −20°C for 10 min, and washed twice, followed by incubation on ice for 30 min with primary rabbit monoclonal anti-ZEB1 antibody (1:200)(Cell Signaling, #3396) or rabbit IgG as a control, and then on ice for 30 min with Alexa Fluor 633 dye-conjugated secondary anti-rabbit IgG (1:200)(Invitrogen).
Immunofluorescence (IF) and immunohistochemistry (IHC)
IF and IHC were done as described previously (10, 23, 27, 35). Supplementary Table S2 summarizes antibodies, titers and specific conditions. Stained objects were examined with a Nikon Microphot microscope and imaged with a digital camera. The immunohistochemical staining was assessed independently (SN, HI and AKS) and the intensity was expressed as negative (−), weakly positive (+) or moderately positive (++).
Xenograft transplantation experiments and in vivo fluorescence imaging
Xenograft transplantation experiments were done as described previously. In brief, 3 × 106 cells were suspended in 50% Matrigel and implanted subcutaneously into the dorsal skin of athymic nu/nu mice (4–6 weeks old)(Charles RiverBreeding Laboratories). Tumor growth was monitored by in vivo fluorescence imaging of tdTomato using Maestro instrumentation (Cambridge Research and Instrumentation, Woburn, MA) with DsRed filter settings. Anesthetized mice were scanned (550 to 700 nm) using a 5-nm step and background fluorescence was eliminated by employing the spectral unmixing capability. Tumor volumes were also measured. All experiments were done under approved protocols from the University of Pennsylvania Institutional Animal Care and Use Committee and NIH guidelines.
Statistical analyses
Data from triplicate and hexaduplicate experiments in real-time RT-PCR, luciferase assays, soft agar colony formation assays and xenograft transplantation were presented as mean ± SE and analyzed by two-tailed Student’s t test. P <0.05 was considered to be statistically significant.
Results
ZEB1 is localized to the invasive fronts exhibiting downregulation of the miR-200 family
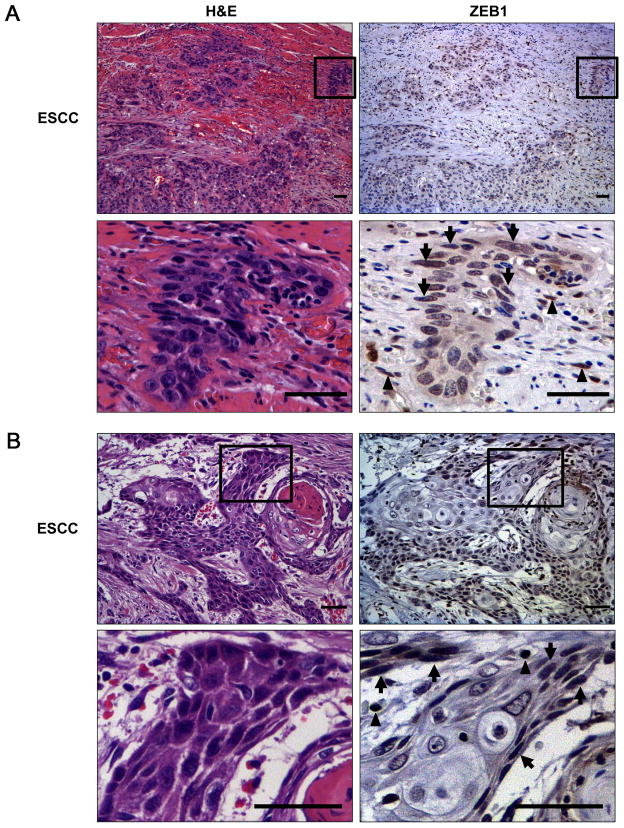
The expression status of ZEB1 in primary ESCC remains unknown. IHC revealed nuclear expression of ZEB1 in ESCC tumor cells in 17 out of 51 cases (33%). ZEB1 positive tumor cells were found focally either within a cord-like invasive small nest (Fig. 1A and supplementary Fig. S1) or at the periphery, but not the center, of well-differentiated tumor nests forming a keratin pearl (Fig. 1B). ZEB1 positive tumor cells were of basaloid type or less differentiated (dedifferentiated) and often displaying spindle-cell shape, reminiscent of EMT. ZEB1 was not detected in the normal adult esophageal epithelia. However, it was detectable in early lesions such as carcinoma in situ (Supplementary Fig. S1), albeit infrequently (16%, 3 out of 19 informative cases). Quantitative RT-PCR coupled with laser capture microdissection documented downregulation of the miR-200 family, a chief negative regulator of ZEBs at the invasive fronts of tumors showing ZEB1 upregulation (Supplementary Fig. S2, and not shown). ZEB1 was not detected in well-differentiated tumor nests expressing IVL along with NOTCH1 and NOTCH3 (Supplementary Fig. S3). Based upon these observations, we hypothesized that ZEB1 is induced in dedifferentiated ESCC cells at the interface of the microenvironment and that EMT may occur upon loss of the commitment toward Notch-mediated squamous differentiation.
Fig. 1. ZEB1 upregulation at the invasive front of ESCC.
Representative images for H&E and corresponding IHC for ZEB1 in two primary ESCC cases featuring poorly differentiated invasive tumor nests (A) and tumor cells surrounding a well-differentiated lesion with keratin pearl formation (B). The selected areas were enlarged in the respective lower panels. Note that ZEB1 positive tumor cells tend to show spindle-cell differentiation (arrows). Stromal inflammatory cells and fibroblasts (arrow heads) are also positive for ZEB1. Scale bar, 100 μm.
Upregulation of ZEBs and reciprocal downregulation of the miR-200 family are associated with suppression of Notch signaling and squamous differentiation
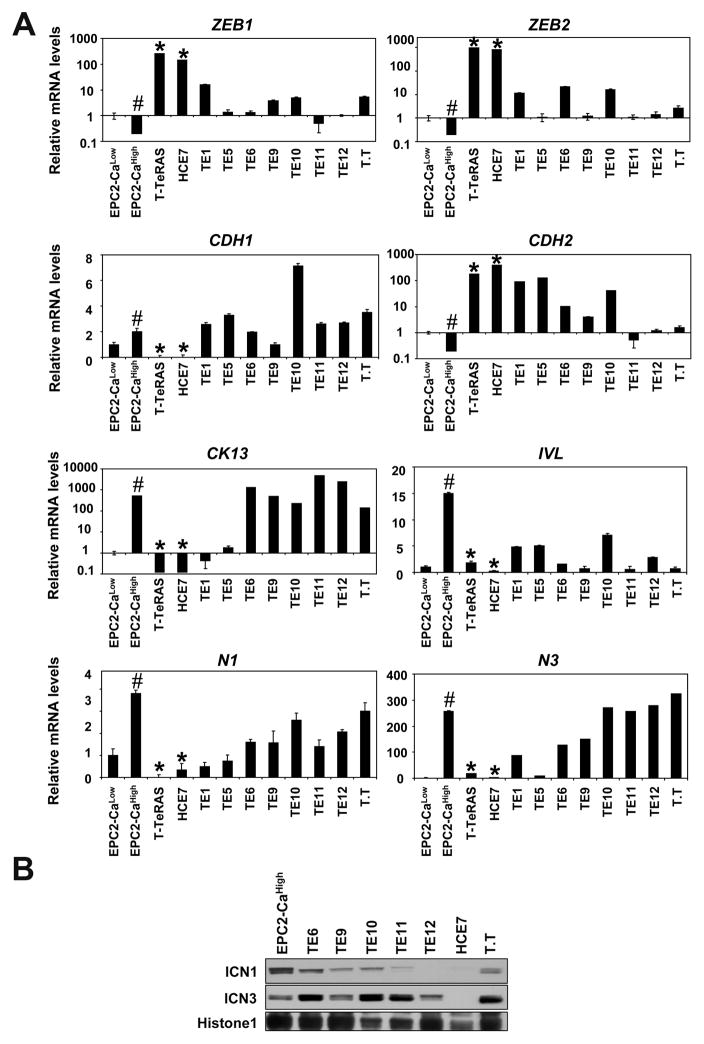
Next, we determined ZEBs and other markers for EMT as well as squamous differentiation in ESCC cell lines. As controls, we used immortalized human esophageal cell line EPC2-hTERT and its transformed derivative T-TeRAS. EPC2-hTERT cells express low levels of ZEBs consistent with its minimal capability of undergoing EMT (14, 23). By contrast, ZEBs were upregulated in T-TeRAS and HCE7 cells, exhibiting a fibroblastic morphology and changes in EMT indicators such as loss of CDH1 (E-cadherin) and upregulation of CDH2 (N-cadherin)(Fig. 2A and Supplementary Fig. S4). Reciprocally, the miR-200 family members were suppressed sharply in these cell lines (Supplementary Fig. S5). Calcium induces squamous differentiation in keratinocytes. EPC2-hTERT cells are maintained in medium containing low Ca2+ concentration (0.09 mM). Addition of calcium (0.6 mM) induced mRNA for CK13 and IVL, markers for squamous differentiation (Fig. 2A). Calcium induced concurrently mRNA and ICN for NOTCH1 and NOTCH3, indicating activation of Notch signaling (Fig. 2) as described previously (10). Interestingly, ZEBs were suppressed in EPC2-hTERT cells upon calcium-induced squamous differentiation (Fig. 2A). Unlike EPC2-hTERT, T-TeRAS and ESCC cell lines grow in the presence of high concentrations of Ca2+ (0.95 mM and 1.8 mM, respectively). Nonetheless, they expressed relatively low levels of IVL (Fig. 2A), implying restricted capability of undergoing squamous differentiation. In T-TeRAS and HCE7, there were inverse relationships between the level of ZEBs and that of NOTCH1, NOTCH3 and CK13 (Fig. 2A). Given low levels of ICN1 and ICN3 in HCE7 and T-TeRAS (Fig. 2B and data not shown), we hypothesized that restriction in Notch-mediated squamous differentiation may alter ESCC cell fate through induction of ZEBs.
Fig. 2. Upregulation of ZEBs in ESCC cells expressing markers for EMT and impaired squamous differentiation associated with loss of Notch activities.
Indicated cell lines were subjected to real-time RT-PCR in (A) and Western blotting in (B). EPC2-hTERT cells were treated with 0.6 mM Ca2+ (EPC2-CaHigh) for 72 hrs to induce Notch-dependent terminal differentiation or left untreated (EPC2-CaLow).
A. Relative mRNA levels for indicated genes were determined. β-actin served as an internal control. CDH1, E-cadherin; CDH2, N-cadherin; N1, NOTCH1; N3, NOTCH3; #, P< 0.01 vs. EPC2-CaLow; *, P< 0.01 vs. EPC2-CaHigh (n=3).
B. Nuclear extracts were used to determine the Notch activation status. Histone H1 served as a loading control. ICN1 and ICN3 represent the active form of NOTCH1 and NOTCH3, respectively.
Inhibition of Notch-mediated squamous differentiation increases malignant potential of transformed human esophageal cells
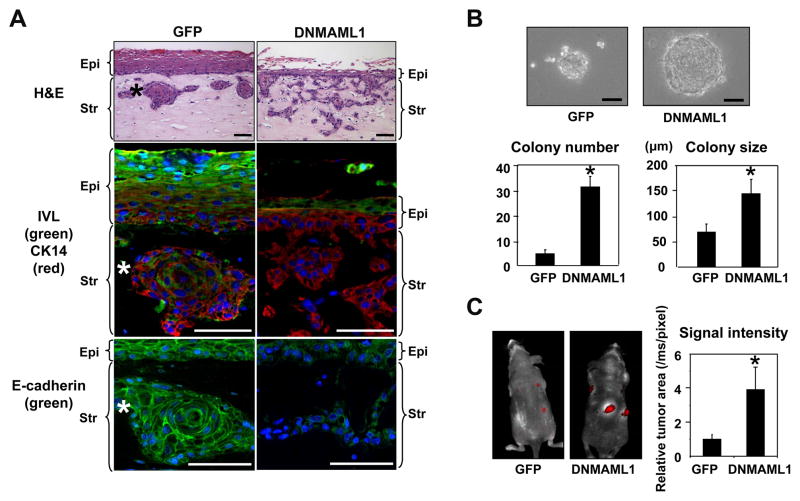
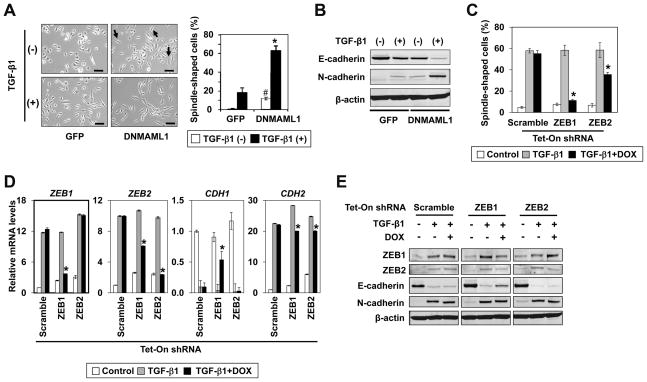
To determine the roles of Notch signaling in cell fate switch in transformed esophageal epithelial cells, we transduced EPC2-T cells stably with DNMAML1, a genetic pan-Notch inhibitor (Supplementary Fig. S6A). We used EPC2-T since it maintains epithelial characteristics unlike T-TeRAS and HCE7 cells. DNMAML1 prevented ICN1, an active form of NOTCH1, from activating a CSL-reporter and Notch target genes in EPC2-T cells (Supplementary Fig. S6, B and C). DNMAML1 alone suppressed the basal level of CK13 and IVL (Supplementary Fig. S6D), suggesting an inhibited squamous differentiation status. Pharmacological inhibition of Notch signaling by GSI induced similar inhibitory effects upon these Notch target genes (data not shown) as observed in the parental EPC2-hTERT cells (10). We assessed the biological impact of Notch inhibition in EPC2-T cells by carrying out organotypic 3D culture, a form of human tissue engineering. As observed with other transformed human esophageal cells (14, 31, 36), control EPC2-T cells not only formed a dysplastic stratified squamous epithelium but exhibited downward invasive growth into the matrix compartment to form cell nests resembling keratin pearls, a hall mark of well-differentiated SCC (Fig. 3A, left). By contrast, DNMAML1 impaired epithelial stratification sharply as observed in non-transformed human esophageal cells (10). Interestingly, DNMAML1 stimulated massive invasion of EPC2-T cells with loss of IVL expression and mislocalization of E-cadherin (Fig. 3A, right). DNMAML1 also enhanced cell migration and invasion in Boyden chamber assays (Supplementary Fig. S7). These findings indicate that the functional consequences of Notch inhibition in transformed cells may be not merely suppression of CSL-dependent squamous differentiation, but activation of alternative cell fates such as EMT, thereby enhancing the malignant potential of cells. In agreement with such a premise, DNMAML1 increased anchorage independent growth and tumorigenicity in immunodeficient mice (Fig. 3, B and C, and supplementary Fig. S8). Interestingly, ZEB1 was upregulated also in the invasive cells expressing DNMAML1 in organotypic 3D culture (Fig. 4A). Such findings prompted us to determine the roles of ZEBs in EPC2-T cells with altered Notch signaling by DNMAML1.
Fig. 3. DNMAML1 inhibits Notch signaling to suppress squamous differentiation and enhance EMT and malignant potentials in esophageal cells.
EPC2-T cells expressing either DNMAML1 or GFP (control) were compared in organotypic 3D culture (A), soft agar colony formation assays (B) and a xenograft transplantation model (C).
A. Stratified squamous epithelia were reconstituted in organotypic 3D culture. Serial sections from the paraffin embedded end products were analyzed morphologically. Note that DNMAML1 suppressed epithelial thickness sharply and stimulated cell invasion into the stroma. IVL and E-cadherin were lost or mislocalized in CK14 positive invasive cells expressing DNMAML1. *, Note that keratin pearl-like invasive cell nests, formed by the GFP control cells, express IVL and E-cad on the cell membrane, and thus consistent with squamous differentiation. Blue, DAPI; Epi, epithelium; Str, stroma; Scale bar, 50 μm.
B. Cells were grown in soft agar for 2 weeks and photomicrographed. Colony number and size were determined per low-power field under light microscopy. *, P< 0.01 vs. GFP, n=6.
C. Tumor growth in nude mice was monitored for 8 weeks by in vivo tdTomato red fluorescent protein imaging. Histogram represents tumor size. *, P< 0.01 vs. GFP, n=3.
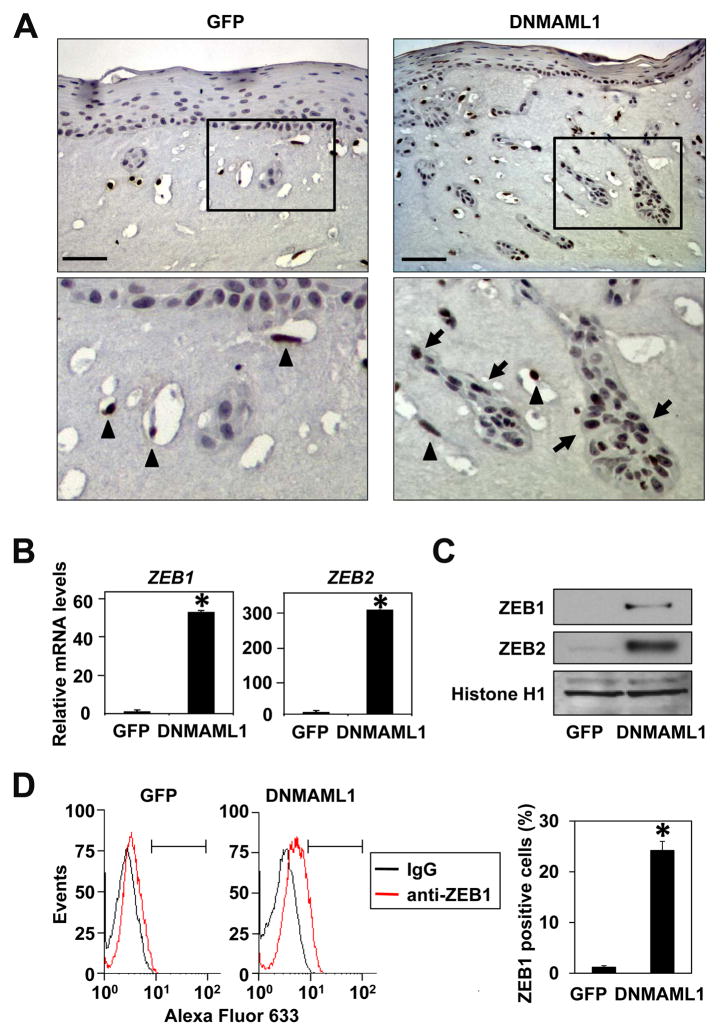
Fig. 4. DNMAML1 induces ZEBs in transformed human esophageal cells.
Expression of ZEBs was determined in EPC2-T cells in the presence or absence of DNMAML1.
A. IHC was done for ZEB1 in the cells grown in organotypic 3D culture. The selected areas were enlarged in the respective lower panels. Note that light nuclear staining was observed in the epithelial compartment in both GFP control and DNMAML1 cells, which was occasionally observed in carcinoma in situ, but not normal human esophageal mucosa (Supplementary Fig. S1). Nonetheless, more intense nuclear ZEB1 (arrow) was detected in the presence of DNMAML1. Stromal fibroblasts (arrow heads) are also positive for ZEB1. Scale bar, 100 μm.
B. Quantitative real-time RT-PCR determined relative mRNA levels for ZEB1 and ZEB2. β-actin served as an internal control. *, P< 0.01 vs. GFP (n=3).
C. Western blotting determined ZEB1 and ZEB2 in the nuclear extracts. Histone H1 served as a loading control.
D. Flow cytometry determined ZEB1 positive cells as indicated by a segmented line and presented as a histogram (P< 0.01 vs. GFP, n=3).
Inhibition of Notch signaling promotes TGF-β-mediated EMT through ZEBs
ZEBs were upregulated at the mRNA and protein levels in the presence of DNMAML1 (Fig. 4, B and C). Moreover, the miR-200 family was downregulated reciprocally (Supplementary Fig. S9). We hypothesized that DNMAML1 may enrich a unique subset of cells expressing ZEBs with increased malignant potential. Flow cytometry revealed a significantly increased ZEB1 positive cells in the presence of DNMAML1 (Fig. 4D). However, a small number of ZEB1 positive cells were present in the control cells (Fig. 4D), suggesting that DNMAML may allow expansion of a preexisting ZEB1 positive cell population. Interestingly, spindle-shaped cells were spontaneously induced in the presence of DNMAML1, which were augmented further by TGF-β stimulation (Fig. 5A). At least 60% of cells showed spindle-shaped cell morphology after TGF-β stimulation for two weeks although a subset of cells remained unchanged (Fig. 5A), implying cell heterogeneity. EMT was validated by an E-cadherin to N-cadherin class switch (Fig. 5B and supplementary Fig. S10). TGF-β induced both ZEB1 and ZEB2, reinforcing their roles in EMT. To better define the specific functions of ZEBs, we conducted RNAi experiments targeting either ZEB1 or ZEB2 using the tetracycline-inducible system. ZEB1 knockdown prevented TGF-β from inducing spindle-shaped cells and suppressing CDH1 (E-cadherin) more efficiently than ZEB2 knockdown (Fig. 5, C–E). However, knockdown of either ZEB1 or ZEB2 had a limited impact upon TGF-β-induced N-cadherin expression (Fig. 5, D and E), implying both ZEBs and/or other factors such as SNAI1 and TWIST1 in EMT (supplementary Fig. S10). Interestingly, there was also a significant RNAi effect upon anchorage independent growth of EPC2-T cells, where knockdown of ZEB1, but not ZEB2 reduced colony formation in soft agar by 35% (supplementary Fig. S11). In addition, knockdown of ZEB1, but not ZEB2 restored the expression of miR-205 and miR-200b, yet only to a limited extent (supplementary Fig. S12), consistent with the above described incomplete RNAi effect upon TGF-β-mediated EMT.
Fig. 5. ZEBs mediate EMT in the transformed human esophageal cells expressing DNMAML1.
EPC2-T cells expressing either DNMAML1 or GFP (control) were stimulated with TGF-β1 for 2 weeks in (A)-(E). In (C)-(E), cells expressing tetracycline-inducible (Tet-On) shRNA directed against either ZEB1, ZEB2 or a non-silence control sequence (Scramble) were subjected to TGF-β1 stimulation along with or without 1 μg/ml of doxycycline (DOX) for 2 weeks. Representative data are shown with comparable results using two independent shRNA sequences.
A and C. Phase-contrast images were taken to score spindle-shaped cells (arrows) as shown in the histogram. Scale bar, 50 μm. In A, *, P < 0.01 vs. GFP plus TGF-β1 (+); #, P < 0.05 vs. GFP plus TGF-β1 (−)(n = 6). In C, *, P < 0.01 vs. TGF-β1 only (n = 6).
B and E. Western blotting determined indicated molecules with β-actin as a loading control.
D. Real-time RT-PCR determined mRNA for indicated genes with β-actin as an internal control. CDH1, E-cadherin; CDH2, N-cadherin; *, P < 0.01 vs. TGF-β1 only (n=3).
To confirm these findings in an independent cell line, we knocked down ZEBs in HCE7 cells. The RNAi effect upon E-cadherin and other EMT markers appeared to be similar to those observed in EPC2-T cells expressing DNMAML1 (supplementary Fig. S13A). HCE7 cells remained mesenchymal morphologically despite highly efficient shRNA induction (supplementary Fig. S13B). In addition, knockdown of ZEBs did not affect transcription factors such as SNAI1 and the miR-200 family (supplementary Fig. S13C and data not shown) Nonetheless, knockdown of both ZEBs suppressed anchorage independent growth and invasion in HCE7 cells (supplementary Fig. S13D and S14).
In sum, inhibition of Notch-mediated squamous differentiation may divert the esophageal epithelial cell fate towards that of mesenchymal and raise cellular malignant potential in concert with ZEBs.
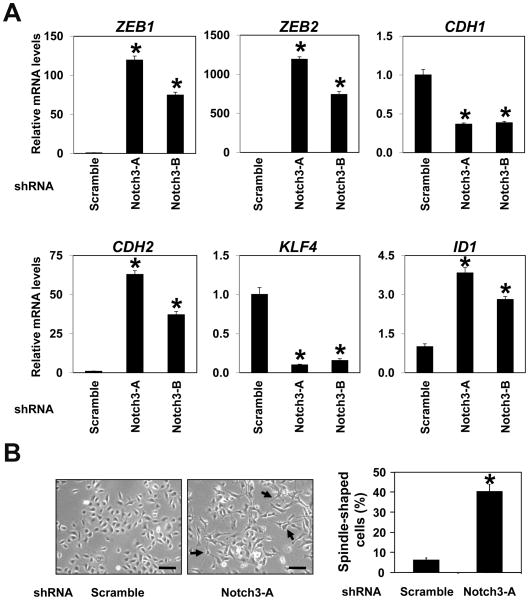
NOTCH3 contributes to esophageal cell fate decisions
Suppression of NOTCH1 and NOTCH3 was associated with lack of squamous differentiation markers and induction of ZEBs (Fig. 2). We have demonstrated recently that NOTCH1 regulates squamous differentiation through NOTCH3 in normal esophageal epithelial cells (10). NOTCH1 knockdown did not have an immediate impact upon the basal expression of NOTCH3 (10) and ZEBs (Supplementary Fig. S15). DNMAML1 not only prevented ICN1 from inducing NOTCH3 mRNA, but suppressed the NOTCH3 basal level in EPC2-T cells (Supplementary Fig. S6). Searching for NOTCH3-regulated genes, gene expression profiling was done using EPC2-hTERT cells stably expressing shRNA directed against NOTCH3 or a non-silencing control shRNA (Supplementary Table S4). Interestingly, ZEBs were amongst the most highly upregulated genes along with N-cadherin, whereas E-cadherin was downregulated as validated by quantitative RT-PCR (Fig. 6A). Corroborating such observations was spontaneous emergence of spindle-shaped cells compatible with EMT in NOTCH3 knockdown cells (Fig. 6B). Furthermore, knockdown of NOTCH3 led to downregulation of the miR-200 family (Supplementary Fig. S16). Interestingly, there were changes in expression of transcription factors implicated in squamous epithelial biology and carcinogenesis. Amongst them were downregulation of KLF4 (required for squamous cell maturation)(37) and upregulation of ID1 (inhibitor of differentiation)(38)(Fig. 6A). These findings suggest that NOTCH3 may contribute to esophageal cell fate decision by promoting squamous cell differentiation while preventing dedifferentiation to mesenchymal cell lineages expressing ZEBs.
Fig. 6. Notch3 knockdown promotes EMT with upregulation of ZEBs and impairs squamous differentiation mechanisms in human esophageal cells.
EPC2-hTERT derivatives expressing two independent shRNA sequences directed against NOTCH3 (Notch3-A and Notch3-B) or a non-silence control sequence (Scramble) were analyzed to validate the microarray data.
A. Real-time RT-PCR determined mRNA for indicated genes with β-actin as an internal control. CDH1, E-cadherin; CDH2, N-cadherin; *, P < 0.01 vs. Scramble (n=3).
B. Phase-contrast images were taken to score spindle-shaped cells (arrows) as represented in the histogram. *, P < 0.01 vs. Scramble (n=6). Scale bar, 50 μm. Representative data (Notch3-A) are shown with comparable results using Notch3-A and Notch3-B.
In aggregate, our data indicate that ZEB1 expression is associated with invasive growth of primary ESCC. Suppression of Notch-mediated squamous cell differentiation may be associated with induction of ZEBs as implicated in ESCC cell lines. Inhibition of the CSL-dependent canonical Notch activities abrogates the squamous differentiation program, allowing expansion of cells expressing ZEBs with enhanced malignant potential. ZEBs contribute to EMT in response to TGF-β stimulation. NOTCH3 limits EMT competence and may have a role in cell fate decisions. Thus, we provide a novel mechanistic link between ZEBs and the Notch pathway in esophageal carcinogenesis and disease progression.
Discussion
ZEBs and the miR-200 family in esophageal carcinogenesis and disease progression
ZEBs and the miR-200 family have been linked to EMT, invasion, metastasis, chemotherapeutic drug resistance and poor clinical outcomes in several cancers (26, 39). This is the first report about aberrant expression of ZEBs and the miR-200 family in SCCs including ESCC. More importantly, however, we provide a novel link between ZEBs and Notch signaling which control a cell fate switch from squamous differentiation to EMT during tumor progression. Notch-mediated squamous differentiation may be suppressed at the tumor invasive front where ZEBs facilitate a dedifferentiation process involving EMT in response to various stimuli such as TGF-β from the microenvironment as proposed in our model (Fig. 7). TGF-β, hypoxia and inflammatory cytokines are amongst many factors known to induce ZEBs and EMT (26).
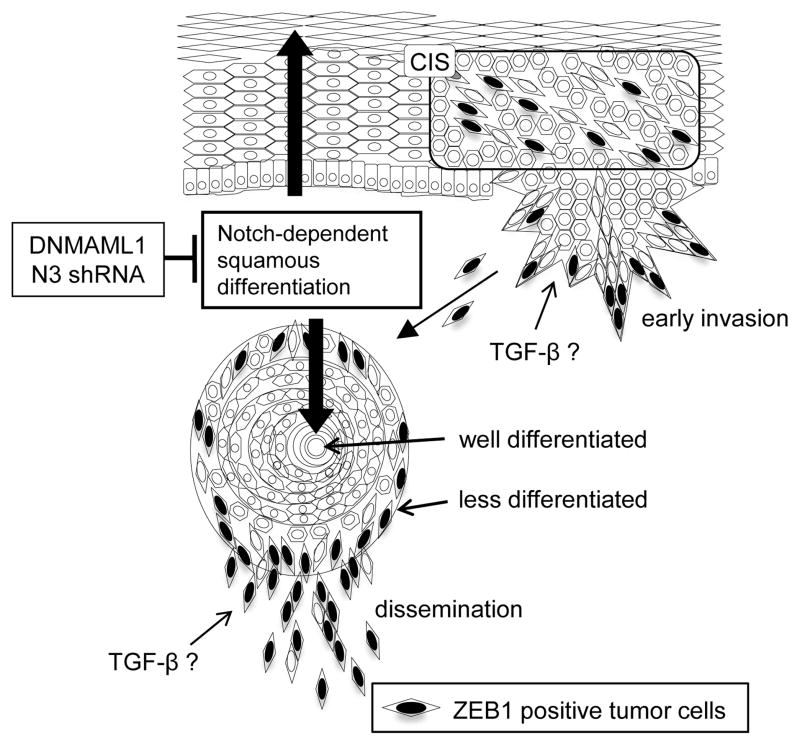
Fig. 7. Model.
Notch signaling regulates squamous differentiation in the normal esophageal epithelium. Notch signaling also contributes to keratin pearl formation in well-differentiated tumor nests. ZEB1 is expressed at the invasive fronts where tumor cells do not undergo squamous differentiation. In early lesions such as carcinoma in situ, ZEBs may be induced in response to oncogene activation (e.g. EGFR) to negate oncogene-induced senescence as a cellular fail safe mechanism (23). In advanced ESCC, NOTCH3 may be downregulated, resulting in dedifferentiated status where microenvironmental cues (e.g. TGF-β and hypoxia) may promote EMT in concert with ZEBs, leading to invasive growth and tumor cell dissemination. In this study, either DNMAML1 or shRNA directed against NOTCH3 (N3) was used to suppress canonical Notch-mediated squamous differentiation to facilitate ZEB-mediated EMT.
ZEB1 is focally expressed in invasive ESCC (Fig. 1 and supplementary Fig. S1) as observed in breast, colorectal and pancreatic carcinomas (25, 40). In addition, we detected ZEB1 in early lesions comprising spindle-shaped tumor cells (supplementary Fig. S1), implying ZEBs in early stages of carcinogenesis. ZEB1 plays a critical role in senescence as well as EMT. ZEB1−/− murine embryonic fibroblasts undergo premature senescence and ectopic E-cadherin induction (24). ZEBs are induced during malignant transformation of esophageal cells to negate EGFR oncogene-induced senescence in concert with mutant p53 (23). Thus, our data reinforce the roles of EMT as a fail-safe mechanism against oncogene-induced senescence at early stages of carcinogenesis (19). In addition, transformed human esophageal cells undergo EMT at the invasive front in organotypic 3D culture (14, 41). Importantly, EMT has been implicated in generation of migratory cancer stem cells (42), which may be subjected to regulation by the ZEB1-miR-200 feedback loop (25). Since the miR-200 family regulates tumor cell plasticity, invasiveness and metastasis (43), downregulation of the miR-200 family in invasive primary ESCC tumors (supplementary Fig. S2) and cell culture (Fig. 2 and supplementary Fig. S5) implicate their roles in the pathogenesis of ESCC.
Although ZEB2 was detectable in culture (Fig. 2, 4, 5 and 6), IHC failed to localize ZEB2 in ESCC with primary antibodies tested. Both ZEBs repress E-cadherin (39). Our RNAi data suggest that ZEB1 may have a predominant role over ZEB2 in EMT, especially in E-cadherin repression (Fig. 5 and supplementary Fig. S13). However, both ZEBs had inhibitory effects upon colony formation and invasion in HCE7 cells (supplementary Fig. S13 and S14). ZEB1 knockdown had indirect effects upon ZEB2 expression as described previously (23, 44–45). Thus, it is plausible that both ZEBs contribute to EMT and other biological processes. In addition, ZEB1 knockdown had limited RNAi effects. In particular, the failure of full restoration of the miR-200 family may suggest involvement of both ZEBs and/or other transcription factors induced by TGF-β (supplementary Fig. S10). Recent studies show that TWIST1 and SNAI1 may repress the miR-200 family directly (13, 46). Further studies are required to address their roles by RNAi designed to target simultaneously both ZEBs and/or other factors. Moreover, a subset of cells did not express detectable ZEB1 (Fig. 4D) and failed to undergo EMT in response to TGF-β stimulation (Fig. 5A). Thus, they may be refractory to RNAi directed against ZEBs. We are currently characterizing such cell populations.
Induction of EMT as a novel consequence of Notch inhibition in ESCC
Cancer invasion may involve a dedifferentiation process which was implicated in ESCC cells exhibiting mesenchymal characteristics with concomitant downregulation of NOTCH receptor paralogs (Fig. 2). Inhibition of canonical Notch signaling impaired squamous differentiation in transformed human esophageal cells (Supplementary Fig. S6). However, we observed additional changes in transformed cells.
First and foremost, Notch inhibition resulted in upregulation of ZEBs and enhancement of malignant potentials implicated by EMT, invasion, anchorage-independent growth and tumor formation (Fig. 3–5, supplementary Fig. S7 and S8). One may argue this as an off-target effect of DNMAML1. Transcription factors other than CSL may recruit Mastermind-like as a co-activator (2). However, knockdown of NOTCH3 also induced ZEBs and EMT (Fig. 6), implying Notch signaling more specifically in restriction of the EMT-competent cells. Although TGF-β and Notch signaling may cooperate to promote EMT during development and cancer progression (20), TGF-β robustly induced EMT in the presence of DNMAML1, suggesting that canonical Notch signaling may be dispensable. In addition, TGF-β suppressed CK13 and IVL (supplementary Fig. S10). Therefore, concurrent Notch inhibition and TGF-β stimulation may drive cancer cell fate towards a migratory mesenchymal cell lineage while suppressing squamous differentiation (Fig. 7).
Second, our data extends the current view of Notch as a tumor suppressor in skin SCCs. DNMAML1 promotes SCC in the mouse skin (8) and transforms epidermal keratinocytes in concert with oncogenic Ras. Moreover, DNMAML1 may enhance epidermal keratinocyte stem cell populations (5). Increased colony formation efficiency in soft agar and tumorigenesis by DNMAML1 (Fig. 3) may be indicative of potential cancer stem cells where ZEBs and the miR-200 family may have a critical role. Such a possibility is currently under investigation. Interestingly, the growing list of validated targets for the miR-200 family includes Notch signaling components such as Jag1, Maml2 and Maml3 (26). In fact, MAML2 and MAML3 were found upregulated with reciprocal downregulation of the miR-200 family in the presence of DNMAML1 (Supplementary Fig. S6 and S9). Although Notch signaling may contribute to ZEB1 induction in pancreatic cancer cells (47), it is unlikely that MAML2 and MAML3 have a role in ZEB1 induction in our system since DNMAML1 interferes with all MAML family members (48). Consistent with such a notion, Notch was not activated in HCE7 cells showing upregulation of ZEBs and downregulation of the miR-200 family (Fig. 2B, Supplementary Fig. S4 and S5). Therefore, ZEBs and the miR-200 family may influence Notch signaling in a cell-type or context dependent manner.
Finally, this study revealed NOTCH3 as a critical determinant in epithelial vs. mesenchymal specification. ICN3 was upregulated in most ESCC cells. Thus, NOTCH3 may maintain commitment of ESCC cells toward squamous cell differentiation with its potential downstream effectors such as KLF4 and ID1. In turn, acquisition of mesenchymal traits as observed in HCE7 cells may require inactivation of NOTCH3. Our data imply NOTCH3 in regulation of ZEBs and the miR-200 family (Fig. 6A and Supplementary Fig. S16). Moreover, NOTCH3 inhibition implicated many genes encoding essential cytokines, growth factors and enzymes such as IL6, FGF2 and PTGS2 (i.e. cyclooxygenase-2)(supplementary Table S4). Interestingly, NOTCH3 activity contributes to non-small cell lung cancer stem cells (49) and promotes tissue site-specific transformation of glial precursor cells (50). Therefore, it is imperative to understand the biological roles and regulation of NOTCH3 in the context of the tumor microenvironment as well as the molecular basis of NOTCH3-mediated regulation of ZEBs and the miR-200 family in cancer stem cells.
Supplementary Material
Acknowledgments
This study was supported in part by NIH Grants R01DK077005 (to MN, SO, HN), P01-CA-098101 (Mechanisms of Esophageal Carcinogenesis to HN, SO, MN, SK, AJPK, MH, JAD), R01DE01923 and P30ES014443 (to DD), P30-DK050306 (to RK and MN), University of Pennsylvania University Research Foundation Award (to HN), University of Pennsylvania, Abramson Cancer Center Pilot Project Grant (to HN), Japan Society for the Promotion of Science Grant-in-Aid for Young Scientists B-21790382 (to SN), American Gastroenterological Association Foundation Student Research Fellowship Award (to Momo Nakagawa), and the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) and its core facilities (Molecular Pathology and Imaging, Cell Culture, Molecular Biology/Gene Expression). We thank Dr. Warren S. Pear (Penn) for reagents and discussion. We thank Ms. Rachel Hammond, Dr. Don Baldwin and Dr. John Tobias (Penn Microarray Facility Bioinformatics Group), Dr. Charles H. Pletcher (Flow Cytometry & Cell Sorting Facility), Dr. Gary P. Swain and Ms. Daniela Budo (Morphology Core), Dr. Gary D. Wu and Dr. Sue Keilbaugh (Molecular Biology/Gene Expression Core) for assistance in data collection and analysis. We are appreciative of discussions with the lab of Dr. Anil K Rustgi and his editorial help.
Abbreviations
- CK
cytokeratin
- CSL
CBF-1/RBP-jκ, Su(H), Lag-1
- DNMAML1
dominant negative mastermind-like1
- DOX
doxycycline
- EMT
epithelial to mesenchymal transition
- ESCC
esophageal squamous cell carcinoma
- GFP
green fluorescent protein
- GSI
γ-secretase inhibitor
- ICN
intracellular domain of Notch
- IVL
Involucrin
- miR
microRNA
- MAML
Mastermind-like
- N1
NOTCH1
- N3
NOTCH3
- RT-PCR
reverse-transcription polymerase chain reaction
- RNAi
RNA interference
- SCC
squamous cell carcinomas
- Tet
tetracycline
- TGF-β
transforming growth factor-β
- ZEB
Zinc finger E-box binding
Footnotes
Disclosure: The authors disclose no conflicts.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–52. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.McElhinny AS, Li JL, Wu L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 2008;27:5138–47. doi: 10.1038/onc.2008.228. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–35. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 5.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–77. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Y, Lin MH, Tian X, Cheng HT, Gridley T, Shen J, et al. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–43. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Wen H, Brayton C, Das P, Smithson LA, Fauq A, et al. Epidermal growth factor receptor and notch pathways participate in the tumor suppressor function of gamma-secretase. J Biol Chem. 2007;282:32264–73. doi: 10.1074/jbc.M703649200. [DOI] [PubMed] [Google Scholar]
- 8.Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, et al. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006;66:7438–44. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- 9.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16:55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohashi S, Natsuizaka M, Yashiro-Ohtani Y, Kalman RA, Nakagawa M, Wu L, et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology. 2010;139:2113–23. doi: 10.1053/j.gastro.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 13.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Natsuizaka M, Ohashi S, Wong GS, Ahmadi A, Kalman RA, Budo D, et al. Insulin-like growth factor-binding protein-3 promotes transforming growth factor-{beta}1-mediated epithelial-to-mesenchymal transition and motility in transformed human esophageal cells. Carcinogenesis. 2010;31:1344–53. doi: 10.1093/carcin/bgq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuen HF, Chan YP, Wong ML, Kwok WK, Chan KK, Lee PY, et al. Upregulation of Twist in oesophageal squamous cell carcinoma is associated with neoplastic transformation and distant metastasis. J Clin Pathol. 2007;60:510–4. doi: 10.1136/jcp.2006.039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usami Y, Satake S, Nakayama F, Matsumoto M, Ohnuma K, Komori T, et al. Snail-associated epithelial-mesenchymal transition promotes oesophageal squamous cell carcinoma motility and progression. J Pathol. 2008;215:330–9. doi: 10.1002/path.2365. [DOI] [PubMed] [Google Scholar]
- 17.Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S, et al. Slug Expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1174–80. [PubMed] [Google Scholar]
- 18.Natsugoe S, Uchikado Y, Okumura H, Matsumoto M, Setoyama T, Tamotsu K, et al. Snail plays a key role in E-cadherin-preserved esophageal squamous cell carcinoma. Oncol Rep. 2007;17:517–23. [PubMed] [Google Scholar]
- 19.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 21.Natsugoe S, Xiangming C, Matsumoto M, Okumura H, Nakashima S, Sakita H, et al. Smad4 and transforming growth factor beta1 expression in patients with squamous cell carcinoma of the esophagus. Clin Cancer Res. 2002;8:1838–42. [PubMed] [Google Scholar]
- 22.Ansieau S, Hinkal G, Thomas C, Bastid J, Puisieux A. Early origin of cancer metastases: dissemination and evolution of premalignant cells. Cell Cycle. 2008;7:3659–63. doi: 10.4161/cc.7.23.7049. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–84. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–88. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 26.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–7. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JJ, Natsuizaka M, Ohashi S, Wong GS, Takaoka M, Michaylira CZ, et al. Hypoxia activates the cyclooxygenase-2-prostaglandin E synthase axis. Carcinogenesis. 2010;31:427–34. doi: 10.1093/carcin/bgp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okano J, Gaslightwala I, Birnbaum MJ, Rustgi AK, Nakagawa H. Akt/protein kinase B isoforms are differentially regulated by epidermal growth factor stimulation. J Biol Chem. 2000;275:30934–42. doi: 10.1074/jbc.M004112200. [DOI] [PubMed] [Google Scholar]
- 29.Harada H, Nakagawa H, Oyama K, Takaoka M, Andl CD, Jacobmeier B, et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–38. [PubMed] [Google Scholar]
- 30.Kim SH, Nakagawa H, Navaraj A, Naomoto Y, Klein-Szanto AJ, Rustgi AK, et al. Tumorigenic conversion of primary human esophageal epithelial cells using oncogene combinations in the absence of exogenous Ras. Cancer Res. 2006;66:10415–24. doi: 10.1158/0008-5472.CAN-06-2104. [DOI] [PubMed] [Google Scholar]
- 31.Okawa T, Michaylira CZ, Kalabis J, Stairs DB, Nakagawa H, Andl CD, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnstone CN, Mongroo PS, Rich AS, Schupp M, Bowser MJ, Delemos AS, et al. Parvin-beta inhibits breast cancer tumorigenicity and promotes CDK9-mediated peroxisome proliferator-activated receptor gamma 1 phosphorylation. Mol Cell Biol. 2008;28:687–704. doi: 10.1128/MCB.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takaoka M, Kim SH, Okawa T, Michaylira CZ, Stairs DB, Johnstone CN, et al. IGFBP-3 Regulates Esophageal Tumor Growth Through IGF-Dependent and Independent Mechanisms. Cancer Biol Ther. 2007:6. doi: 10.4161/cbt.6.4.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takaoka M, Harada H, Andl CD, Oyama K, Naomoto Y, Dempsey KL, et al. Epidermal growth factor receptor regulates aberrant expression of insulin-like growth factor-binding protein 3. Cancer Res. 2004;64:7711–23. doi: 10.1158/0008-5472.CAN-04-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darling DS, Stearman RP, Qi Y, Qiu MS, Feller JP. Expression of Zfhep/deltaEF1 protein in palate, neural progenitors, and differentiated neurons. Gene Expr Patterns. 2003;3:709–17. doi: 10.1016/s1567-133x(03)00147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, et al. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc Natl Acad Sci U S A. 2010;107:11026–31. doi: 10.1073/pnas.0914295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tetreault MP, Wang ML, Yang Y, Travis J, Yu QC, Klein-Szanto AJ, et al. Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology. 2010;139:2124–34. e9. doi: 10.1053/j.gastro.2010.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Tsao SW, Li YY, Wang X, Ling MT, Wong YC, et al. Id-1 promotes tumorigenicity and metastasis of human esophageal cancer cells through activation of PI3K/AKT signaling pathway. Int J Cancer. 2009;125:2576–85. doi: 10.1002/ijc.24675. [DOI] [PubMed] [Google Scholar]
- 39.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–88. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michaylira CZ, Wong GS, Miller CG, Gutierrez CM, Nakagawa H, Hammond R, et al. Periostin, a cell adhesion molecule, facilitates invasion in the tumor microenvironment and annotates a novel tumor-invasive signature in esophageal cancer. Cancer Res. 2010;70:5281–92. doi: 10.1158/0008-5472.CAN-10-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Gene Dev. 2009;23:2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–24. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 45.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327–34. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–7. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937–48. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierfelice TJ, Schreck KC, Dang L, Asnaghi L, Gaiano N, Eberhart CG. Notch3 activation promotes invasive glioma formation in a tissue site-specific manner. Cancer Res. 2011;71:1115–25. doi: 10.1158/0008-5472.CAN-10-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.