Abstract
Inflammation-induced activation of proto-oncogenic NF-κB/REL and dysfunction of tumor suppressor TP53/p63/p73 family transcription factors are key events in cancer progression. How inflammatory signaling coordinates dysregulation of these two transcription factor families during oncogenesis remains incompletely understood. Here, we observed that oncoprotein c-REL and tumor suppressor TAp73 are co-expressed and complex with ΔNp63α in the nucleus of a subset of head and neck squamous cell carcinoma (HNSCC) cell lines with mutant (mt)TP53. TNF-α a pro-inflammatory cytokine, promoted nuclear translocation and c-REL/ΔNp63α interaction and dissociation of TAp73 from nuclear ΔNp63α to the cytoplasm, while c-REL siRNA depletion attenuated this effect. Overexpression of c-REL or a c-REL κB-site DNA binding mutant enhanced protein interaction withΔNp63α and TAp73 dissociation, implicating c-REL/ΔNp63α-specific interactions in these effects. We discovered TNF-α- or genetic alteration of c-REL expression inversely modulatesΔNp63α/TAp73 interactions on distinct p63 DNA binding sites, including those for key growth arrest and apoptotic genes p21WAF1, NOXA, and PUMA. Functionally, c-REL repressed these genes and the anti-proliferative effects of TNF-α or TAp73. Conversely, c-REL siRNA depletion enhanced TAp73 promoter interaction, and expression of genes mediating growth arrest and apoptosis. Similar to TNF-α treated HNSCC lines, human HNSCC tumors and hyperplastic squamous epithelia of transgenic mice overexpressing ΔNp63α that exhibit inflammation, also demonstrate increased nuclear c-REL/ΔNp63α and cytoplasmic TAp73 localization. These findings unveil a novel and reversible dynamic mechanism whereby pro-inflammatory cytokine TNF-α-induced c-REL/ΔNp63α interactions inactivate tumor suppressor TAp73 function, promoting TNF-α resistance and cell survival in cancers with mtTP53.
Keywords: c-REL, ΔNp63, TAp73, TNF-α, squamous cell carcinoma
Introduction
TP53, a tumor suppressor that mediates growth arrest and apoptosis of critically damaged cells, is the most frequently mutated gene in cancer, including head and neck squamous cell carcinomas (HNSCC) (1, 2). The TP53 family also includes p63 and p73, which have overlapping functions in cell growth and apoptosis, as well as development of squamous epithelia of the skin and mucosa (3, 4). When TP53 is mutated or inactivated, transactivating (TA) isoforms TAp63 and TAp73 can potentially replace the tumor suppressor function of TP53. TAp63 and TAp73 have full length N-terminal domains which share homology, transactivating, and tumor suppressor function with TP53. While TAp63 and TAp73 are rarely mutated, expression of alternatively transcribed ΔN isoforms can differentially affect their transactivation and tetramerization, respectively. Recent evidence suggests that the ΔNp63α isoform is predominantly overexpressed together with TAp73 in major subsets of HNSCC, breast, and other cancers (5). Nuclear interactions between ΔNp63α and TAp73 proteins on regulatory promoters have been implicated in repression of genes mediating growth arrest and apoptosis (4, 5). However, what factor(s) regulate these interactions between overexpressed ΔNp63α and TAp73, and how their interaction leads to inactivation of TAp73 in cancers with mtTP53, remains unclear.
Among candidate regulatory factors, Tumor Necrosis Factor (TNF)-α is a cytotoxic cytokine which is expressed by cancer and infiltrating inflammatory cells in the tumor microenvironment of many cancers, including HNSCC (6, 7). Interestingly, we found that HNSCC are paradoxically resistant to TNF-α-mediated growth arrest and apoptosis, and that such resistance involves aberrant activation of NF-κB/REL transcription factors (8). Among these, TNF-α has been shown to signal via a canonical pathway to promote nuclear translocation and transactivation of RELA (p65) and c-REL (9). RELA (p65) promotes expression of genes that enhance cell proliferation and survival of cancers, including HNSCC (10–12). However, inhibition of RELA and these prosurvival genes by siRNA in vitro or by proteasome inhibitor in a phase I clinical trial in vivo demonstrated limited cytotoxic and clinical activity (11–13). Amplification and nuclear localization of c-REL has also been previously detected, and to be relatively unaffected by proteasome inhibition in HNSCC (13, 14). These observations raised the question whether TNF-α regulated or overexpressed c-REL may contribute to inhibition of growth arrest and apoptosis of HNSCC by a distinct mechanism.
We recently discovered that overexpressed ΔNp63α co-localizes and forms novel nuclear complexes with c-Rel in murine keratinocytes or orthologue c-REL in human HNSCC (15). Herein, we explored the hypothesis that TNF-α and c-REL activation is linked to modulation of the aforementioned ΔNp63α/TAp73 interactions and dysregulation of growth arrest and apoptosis in cancer. We reveal a dynamic and reversible mechanism whereby TNF-α promotes c-REL nuclear translocation and interaction with ΔNp63α, dissociation of TAp73 from distinct p63 promoter sites of key growth arrest and apoptotic genes, and TAp73 translocation to the cytoplasm. These results can help explain how inflammatory factor TNF-α and c-REL, throughΔNp63α, inhibit the compensatory ability of TAp73 to activate genes that mediate growth arrest and apoptosis in HNSCC with mutant TP53.
Materials and Methods
Cell Lines
The characteristics, TP53 genotype and culture of HNSCC (UMSCC) cell lines obtained from the University of Michigan were previously described (16, 17). UMSCC lines used herein were obtained in 2008. Authentication was performed at the University of Michigan by DNA genotyping of alleles for 9 loci, D3S1358, D5S818, D7S820, D8S1179, D13S317, D18S51, D21S11, FGA, vWA and amelogenin locus in 2008 and 2010, as recently described (18). Cell line stocks were preserved at −80 degrees C and cultured for fewer than 6 months before use.
Western Blot
Western blot analysis was performed as previously (17). Primary antibodies are listed in Supplemental Methods.
Real time RT-PCR
RNA isolation and cDNA synthesis were performed as previously (19). Real time PCR primers and probes for ΔNp63 and TAp63 validated previously (20) were synthesized by Applied Biosystems. Primers to amplify TAp73 and ΔNp73 are listed in Supplemental Methods.
Plasmid, siRNA Transfection and Reporter gene assays
These experiments were performed as described previously (19). ΔNp63 specific siRNA (21) was synthesized by Integrated DNA Technologies (IDT). TAp63 specific siRNA was purchased from Dharmacon. The 2.6kb p21-luc reporter was provided by Dr. Bert Vogelstein’s laboratory (22). Each sample was assayed in triplicate and data were presented as mean ± SD.
Coimmunoprecipitation analysis (IP)
Co-IP was performed as described previously (15). Antibodies used are listed in Supplemental Methods
Electrophoretic mobility shift assay
Oligonucleotides covering the p21WAF1 promoter p63-binding site (−2283bp, 5′-TGGCCGTCAGGAACATGTCCCAACATGTTGAGCTCTGGCA-3′, Fig. S3A) were [γ-32P]-end-labeled with 10 U of T4 polynucleotide kinase (New England Biolabs), and EMSA was performed as previously described (23).
Chromatin Immunoprecipitation Analyses (ChIP)
ChIP assays were performed using the Magna ChIP™ G kit (Upstate) as previously (24). For QRT-PCR primers see Supplemental Methods.
DNA based cell cycle and apoptotic flow analysis
Flow analysis was performed as previously (12). Samples were run on FACS Canto machine within 1 hour and analyzed using DIVA flow cytometric analysis software.
Additional experimental detail, methods and figures are included in Supplemental Information
RESULTS
Overexpression and protein interactions among c-REL, ΔNp63α and TAp73 in a subset of head and neck cancer cell lines with mutant TP53
Expression of c-REL, p63, p73, and TP53 was characterized in a panel of cell lines that included 9 HNSCC (UMSCC) and normal primary human epidermal keratinocytes (HEK) (Fig. S1). The UMSCC lines selected were derived from primary and metastatic tumors exhibiting an aggressive clinical course (survival <15 mos.), and representative genetic abnormalities, including inactivation or mutation of TP53 (16–18). c-REL, ΔNp63α, TAp73, and TP53 protein were detected by Western blots, and quantitative RT-PCR (Fig. S1A–F). Total c-REL protein was increased, as were nuclear c-REL and ΔNp63α protein in most UMSCC compared with normal HEK (H) cells (Fig. S1A, Left, Right panels). Interestingly, even greater c-REL expression was detected in 3 of 4 cell lines (UMSCC 22B, 38, 46) with elevated ΔNp63α, TAp73 and mtTP53 (UMSCC 22A, 22B, 38, 46) (Fig. S1A, Left panel; S1B–E). Thus, we find that c-REL, ΔNp63α, TAp73 are overexpressed with mtTP53 in an overlapping subset of HNSCC lines.
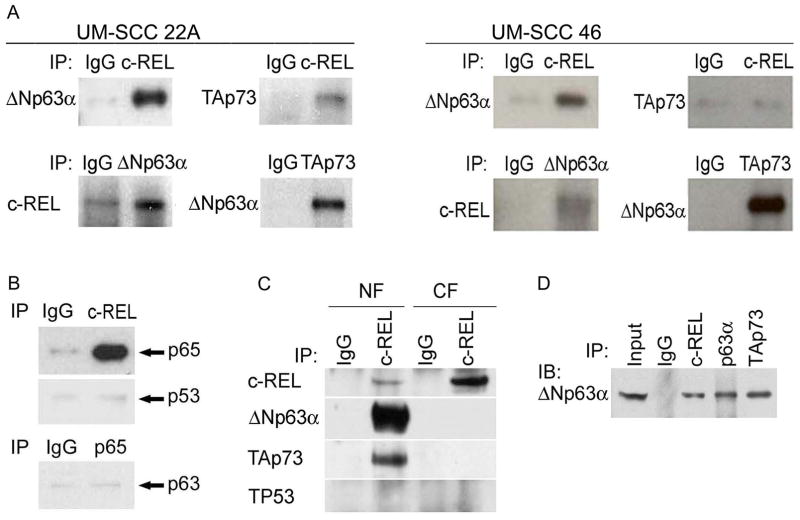
Both c-REL and TAp73 exhibited basal interaction with ΔNp63α in whole cell extracts from two independent cell lines by co-IP analysis (Fig. 1A; UMSCC-22A or −46). However, c-REL and TAp73 exhibited weak or negligible interaction with one another, seen only with supersensitive substrate and prolonged exposure. Further, c-REL, but not RELA(p65), interacted with ΔNp63α (although RELA formed an independent complex with c-REL) (Fig 1B), providing evidence for distinct interactions by these RELs. The complexes between c-REL, ΔNp63α, and TAp73, identified in whole cell extracts above, were found in nuclear, but not cytoplasmic extracts (Fig. 1C). No interaction with mtTP53 was observed (Fig. 1B, C). In the absence of further stimulation, a significant portion of nuclearΔNp63α co-IPed with either c-REL or TAp73, when compared with input (Fig. 1D).
Figure 1.
Interactions among c-REL, ΔNp63α, TAp73, and RELA(p65) in HNSCC with mtTP53. A, left, Whole cell lysates from UM-SCC 22A cells or, right, 46 cells, were immunoprecipitated, then immunoblotted with antibodies as indicated. B, Whole cell lysates of UM-SCC 22A were immunoprecipitated with c-REL or p65 antibodies, then immunoblotted with the antibodies as indicated. C, Cytoplasmic and nuclear fractions were immunoprecipitated with IgG control or c-REL antibody, then probed for c-REL, ΔNp63α, TAp73, and TP53. D, Whole cell lysates of UM-SCC 22A were immunoprecipitated with c-REL, p63 and TAp73 antibodies, and immunoprecipitated proteins and total lysates (Input) were then immunoblotted with p63 antibody.
TNF-αor genetic modulation of c-REL affects nuclear localization and interactions between c-REL, ΔNp63 and TAp73
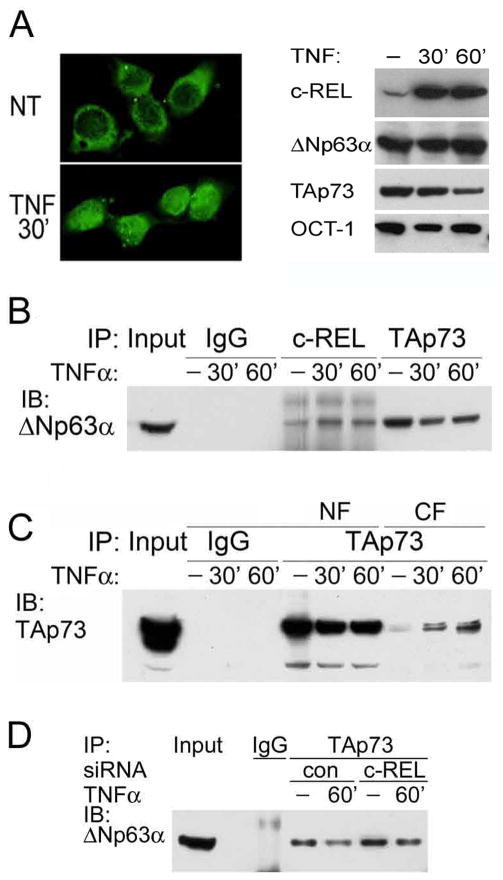
The inflammatory cytokine TNF-α is a canonical inducer of c-REL, and is expressed in the HNSCC microenvironment in vivo, but not by these cell lines in vitro (7, 25). We examined the effect of exogenous TNF-α on the cellular distribution of c-REL, ΔNp63α and TAp73 in the UMSCC 22A cell line, which has lower endogenous c-REL expression, making it amenable to TNF-α or genetic modulation (Fig, S1). TNF-α stimulation increased nuclear localization of c-REL within 30′ (Fig. 2A, left panel). Remarkably, TNF-α not only induced c-REL, but also decreased TAp73, without significantly altering ΔNp63α expression in nuclear extracts (Fig. 2A, right panel). Moreover, TNF-α stimulation increased the interaction between nuclear c-REL and ΔNp63α, and a corresponding dissociation of nuclear ΔNp63 and TAp73 in co-IP (Fig. 2B). This dissociation corresponded with a decrease in TAp73 in the nuclear fraction, and appearance in the cytoplasmic fraction (Fig. 2C). Conversely, c-REL depletion by siRNA enhanced ΔNp63α/TAp73 interaction without or with TNF-α (Fig. 2D).
Figure 2.
TNF-α-induced and overexpressed c-REL interacts with ΔNp63α and modulates TAp73. A, Left, Distribution of c-REL detected by IF staining in nuclear and cytoplasmic cellular counterparts, non-treated (NT), or with 20 ng/ml TNF-α for 30 min (200X). Right, nuclear c-REL, ΔNp63α and TAp73 proteins were detected before and after 20ng/ml TNF-α treatment for 30 and 60 minutes. OCT-1:loading control. B, Increased nuclear interaction between c-REL and ΔNp63α, and decreased nuclear interaction between TAp73 and ΔNp63α were observed 30 and 60 min after TNF-α treatment. C, Decreased nuclear and increased cytoplasmic TAp73 were observed 30 and 60 min after TNF-α treatment. D, Knockdown of c-REL increased the interaction between TAp73 and ΔNp63α without and with TNF-α stimulation.
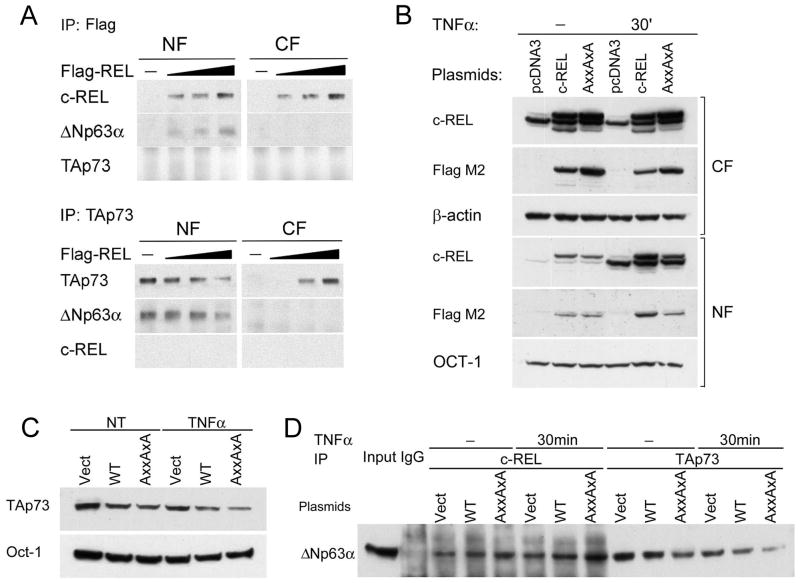
Similar to TNF-α-induced c-REL, overexpressing c-REL-Flag in UMSCC-22A resulted in increased interaction between c-REL and ΔNp63α in the nuclear fraction (Figure 3A, upper panels). Concurrently, TAp73 dissociated from ΔNp63α in the nuclear fraction, and appeared in the cytoplasmic fraction (Fig. 3A, lower panels). Further supporting that c-REL and TAp73 form alternative complexes with ΔNp63α, no significant interaction between c-REL and TAp73 was detected in co-IP. Overall, the above findings indicate that TNF-α-induced or overexpressed c-REL is involved in the dissociation of TAp73 from ΔNp63α in the nucleus to the cytoplasm.
Figure 3.
Overexpressed c-REL and c-REL κB binding mutant interacts with ΔNp63α and modulates TAp73. A, After transfection with increasing amount of the expression plasmid for Flag-c-REL, interaction between exogenous c-REL and endogenous ΔNp63α was increased (Upper), and interaction between emdogenous ΔNp63α and TAp73 was decreased, with redistribution of TAp73 from the nucleus to the cytoplasm (Lower). B, Detection of endogenous c-REL 48h after transfection with empty pCDNA3 vector, recombinant Flag-c-REL wild-type, and Flag c-REL with AxxAxA mutations of the RxxRxR N-terminal κB DNA-binding residues, in cytoplasmic and nuclear fractions (CF, NF). β-actin and OCT-1 were used as loading controls. C, Nuclear TAp73 expression detected 48h after pCDNA3 and c-REL plasmid transfection −/+ TNF-α treatment (20ng/ml, 60min). D, Overexpression of recombinant c-REL wild type and c-REL AxxAxA mutant enhanced interaction between c-REL and ΔNp63α, and decreased interaction between ΔNp63α and TAp73. TNF-α treatment enhanced the effect of wild type and AxxAxAmutant c-REL in decreasing ΔNp63α-TAp73 interactions.
Previous studies have indicated that c-REL can bind κB motifs bound by other NF-κB/REL members, as well as distinct promoter sites and synthetic oligonucleotides (26). We compared the effect of overexpressing a wild-type (wt)c-REL and a N-terminal RxxRxR to AxxAxA mutant reported to abrogate c-REL binding to classical REL/κB DNA motifs (27). Overexpression and TNF-α enhanced nuclear translocation of exogenous wt and AxxAxA mutant c-REL (Fig. 3B). Interestingly, both wt and AxxAxA mutant c-REL reduced total nuclear TAp73 and ΔNp63-TAp73 interaction, and this reduction was preferentially enhanced with TNF-α-stimulated AxxAxA mutant c-REL (Fig. 3C, D). These results suggested that TNF-α-induced or overexpressed c-REL interacts withΔNp63 and modulates bound TAp73 in a manner distinct from those previously shown to mediate c-REL binding to κB DNA recognition sequences.
TNF-α or genetically modulated c-REL reciprocally affects binding of TAp73 with ΔNp63α on p63 sites of p21WAF1, NOXA and PUMA gene promoters
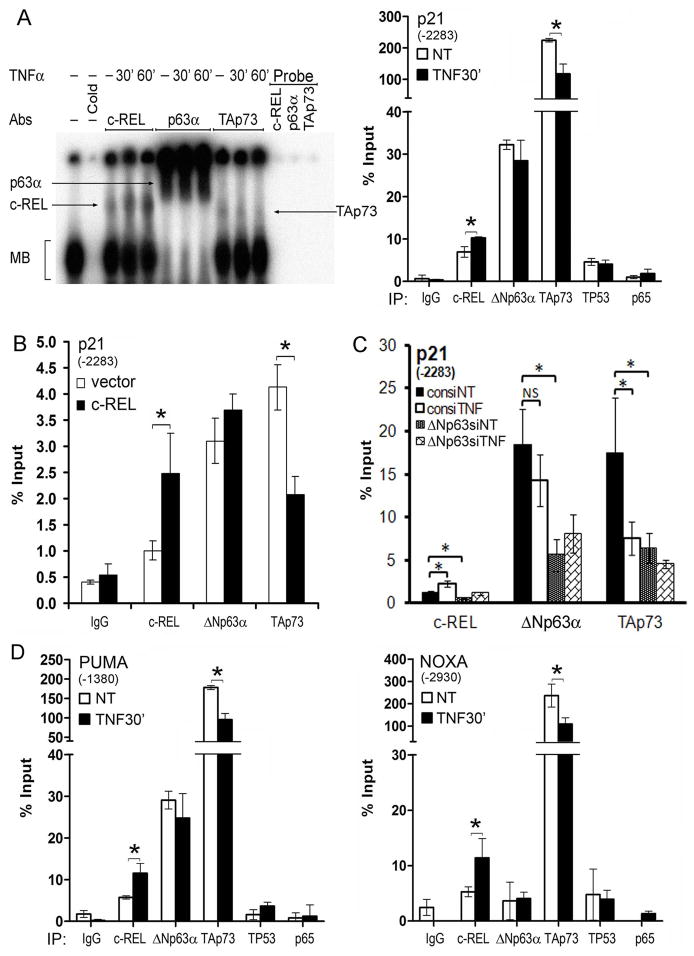
To explore whether TNF-α modulates interactions between c-REL, ΔNp63α and TAp73 on p63/p73 regulatory sites, we examined their binding to an oligonucleotide encoding a well characterized p63/p73 binding site (−2283 to −2273; Fig. S2A), from the promoter of the p63/p73 regulated growth arrest gene p21WAF1 (15, 23). Using this p63 site-specific radiolabeled oligonucleotide probe, a major band was revealed by electrophoretic mobility shift assay (EMSA) in human UMSCC 22A cells (Fig. 4A, left panel, MB). Binding specificity was confirmed by unlabeled probe competition, and the complex contained c-REL, ΔNp63α or TAp73, demonstrated by supershift with specific antibodies. Similar to our findings in nuclear extracts, TNF-α stimulated c-REL binding, while decreasing binding of TAp73, without significantly affecting ΔNp63α.
Figure 4.
c-REL, ΔNp63α and TAp73 complex binding to the p21WAF1, NOXA and PUMA gene promoters A, Left, EMSA with nuclear extracts from UM-SCC 22A cells showing protein bound to oligonucleotide containing the established p63 binding site sequence (−2283bp) for the p21WAF1 promoter. MB: Main band. Cold: unlabeled oligonucleotide competition (200X); Arrows, protein supershifts using antibodies against ΔNp63α, TAp73 or c-REL. c-REL binding gradually increased while TAp73 binding decreased after TNF-α treatment (20ng/ml for 30′ and 60′). Probe only (no extract), indicates the antibodies lack inherent probe shifting activity. A, Right, ChIP assay indicates c-REL, ΔNp63α, TP53, and TAp73 bound to (−2283bp) p63 site of p21WAF gene promoter, while p65 showed no significant binding activity. TNF-α (20ng/ml for 30′) increased the binding of c-REL, but decreased the binding of TAp73. B, Overexpression of c-REL increased c-REL but decreased TAp73 binding to the p21WAF1 promoter. C, ΔNp63 siRNA decreased the binding activities of c-REL, ΔNp63α, and TAp73. D, Left, PUMA and Right, NOXA p63 site ChIP binding for antibodies indicated, −/+ TNF-α 20ng/ml for 30′ (* p<0.05).
Next, we examined the endogenous binding of c-REL, ΔNp63 and TAp73 to p21WAF1 promoter DNA using chromatin immunoprecipitation (ChIP) assays. All three factors exhibited basal binding to the p63 binding site region of the p21WAF1 promoter in UM-SCC 22A, −46 or −1 cells (Fig. 4A, right panel; S2B, C, D). TNF-α induced c-REL binding and decreased TAp73 binding, without significantly affecting ΔNp63α binding to the p63 site in two different cell lines (Fig. 4B, S2C). Similarly, c-REL overexpression also induced the same effects in either UMSCC 22A or −1 lines (Fig. 4B, S2D). Conversely, c-REL depletion by siRNA significantly decreased c-REL and ΔNp63 binding, and reciprocally enhanced TAp73 binding activity in UM-SCC 22A cells (Fig. S3E). Importantly, ΔNp63α siRNA knockdown attenuated basal and TNF-α inducible binding of c-REL, ΔNp63α and TAp73, implicating ΔNp63α as a key anchor for c-REL and TAp73 binding activity (Fig. 4C). Together, these findings in 3 different UMSCC lines indicate that TNF-α or genetic modulation of c-REL results in reciprocal modulation of TAp73 binding with ΔNp63α on the established p63 regulatory site of the p21WAF1 promoter.
To examine the broader role of TNF-α-induced c-REL, ΔNp63 and TAp73 interactions, we performed ChIP assays for p63 binding sites identified by bioinformatic analysis (28; suppl methods) on the promoters of two key pro-apoptotic genes, NOXA and PUMA (Fig. S2A, arrows). As with p21WAF1, stimulation with TNF-α increased c-REL and decreased TAp73 binding without affecting ΔNp63 bound to the predicted p63 sites in the PUMA and NOXA promoters (Fig. 4D). No significant binding by RELA(p65) or TP53 was detected on the p63 promoter sites of any of these 3 genes (Fig. 4A, right panel; D), consistent with co-IP results above. Together, our results indicate that TNF-α or genetically modulated c-REL reciprocally affects binding of ΔNp63α with TAp73 on p63 sites of multiple gene promoters.
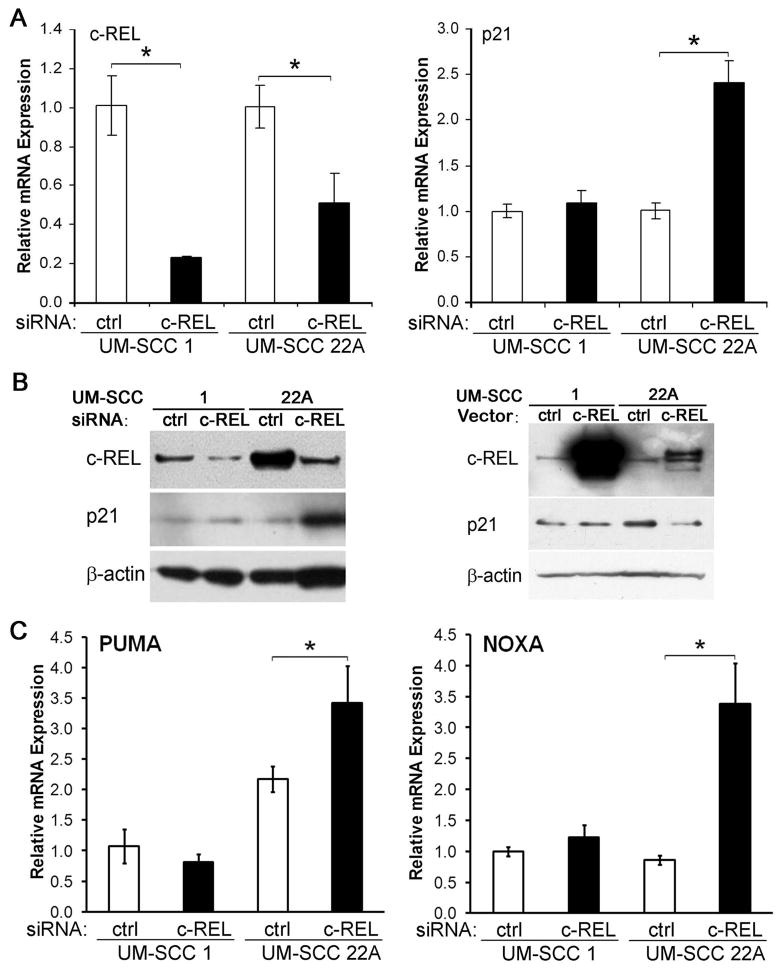
c-REL differentially modulates p21WAF1, NOXA and PUMA gene expression in UMSCC cell lines differing in expression of TP53 family members
We next examined how c-REL modulated ΔNp63α-TAp73 interactions affect expression of growth arrest mediator p21WAF1. The UM-SCC cell subsets express lower or higher baseline levels p21WAF1 mRNA and protein (Fig. S3A,B), similar to relative differences in expression of wt or mt TP53, ΔNp63α, and TAp73 shown above (Fig. S1A,B,D). Depletion of endogenous c-REL by siRNA significantly reduced levels of c-REL mRNA in two cell lines, UMSCC 1 and UMSCC 22A (Fig. 5A, left panel), from these subsets that differ in baseline expression of p21WAF1 and TP53 family proteins. However, c-REL depletion in UMSCC 22A cells with abundant mtTP53, ΔNp63 and TAp73 resulted in a strong increase in p21WAF1 mRNA and protein levels, when compared to UMSCC 1 cells lacking wtTP53 and TAp73 (Fig. 5A, right panel; 5B, left panel). Conversely, overexpression of c-REL by transient transfection repressed p21WAF1 expression in UMSCC 22A cells, but not in UMSCC 1 cells (Fig. 5B, right panel). Similarly, TNF-α induced nuclear c-REL, while inhibiting expression of TAp73 target protein p21WAF1 (Fig. S3C). Increased p21WAF1 expression after c-REL depletion in UMSCC 22A was attenuated by co-transfection with TAp73 siRNA (Fig. S3D), indicating the contribution of TAp73. Transfection of TAp73-deficient UMSCC 1 cells with TAp73 increased, while co-transfection with c-REL suppressed an increase in p21WAF1 expression (Fig. S3E). Depletion of c-REL by siRNA also enhanced PUMA and NOXA expression in the mtTP53 cell line UMSCC 22A, but not in UMSCC 1 cells deficient in TP53 and TAp73 (Fig. 5C). Together, these results indicate that c-REL represses three important growth arrest and pro-apoptotic genes in HNSCC expressing ΔNp63 and TAp73 with mtTP53.
Figure 5.
c-REL modulates p21WAF1, PUMA, NOXA expression in HNSCC lines. A, Left, c-REL siRNA significantly decreased c-REL mRNA expression in UM-SCC 1 (deficient wtTP53) and 22A (mtTP53) cells. A, Right, significant induction of p21WAF1 mRNA was detected only in UM-SCC 22A cells. B, Western blots, Left, c-REL protein expression decreased after c-REL siRNA knockdown in UM-SCC 1 and 22A cells, but p21WAF1 protein increased only in UM-SCC 22A. B, Right, Transfection with plasmid to overexpress c-REL decreased p21WAF1 protein expression in UM-SCC 22A cells. C, Left, PUMA and Right, NOXA, mRNA expression after c-REL siRNA depletion in UM-SCC 1 and 22A cells, with significant induction of PUMA and NOXA in UM-SCC 22A cells. The mean ± SD is shown. * P < 0.05.
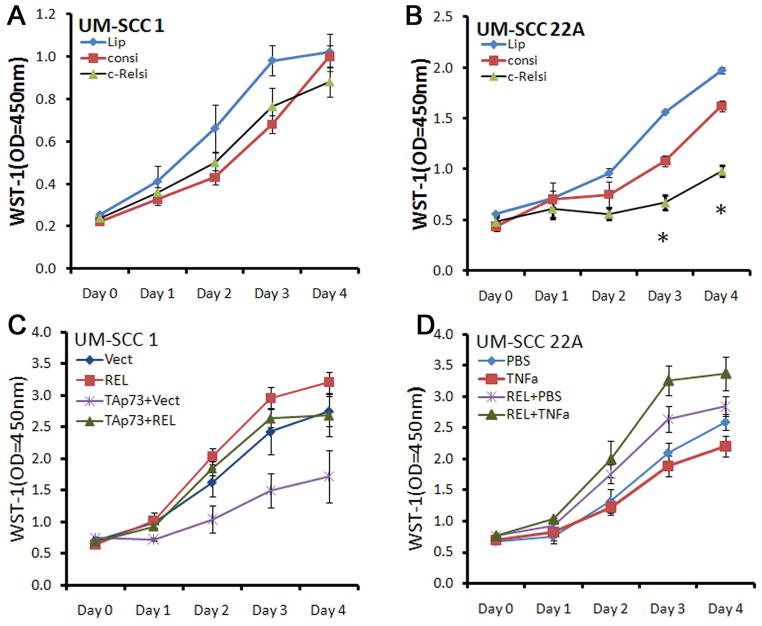
c-REL promotes cellular proliferation and survival in UMSCC cells expressing ΔNp63, TAp73 with mtTP53
We further examined the function of c-REL in proliferation and survival of HNSCC cells in vitro. c-REL depletion with siRNA had no significant effect on proliferation of UMSCC 1 cells that express lower levels of wtTP53 and other family members, but inhibited proliferation and density of UMSCC 22A cells that exhibit high protein levels of ΔNp63α, TAp73 and mtTP53 and inducible p21WAF1 (Fig. 6A,B; Fig. S4A). c-REL siRNA depletion also increased sub-G0 DNA-positive dead cells in UMSCC 22A, but not in UMSCC 1 cells (Fig. S4B), whereas c-REL overexpression reduced the sub-G0 fraction in UMSCC 22A cells (Fig. S4C). The results showing that c-REL promotes cell proliferation and survival are consistent with the effect of c-REL in repressing p21WAF1, NOXA and PUMA gene expression (Fig. 5), which mediate growth arrest and apoptosis in HNSCC (29).
Figure 6.
c-REL differentially affects cellular density of UM-SCC lines exhibiting differences in TP53 family protein expression. A, UM-SCC 1 (deficient for wtTP53 and TAp73), or B UM-SCC 22A (increased mtTP53, TAp73) following knockdown of c-REL by siRNA. C, UM-SCC 1, over-expression of TAp73 decreased proliferation, while co-transfection of c-REL restored cell proliferation. D, UM-SCC 22A proliferation after overexpression of c-REL was increased by TNF-α. Lip: Lipofectamine alone; consi, control siRNA; c-RELsi, c-REL siRNA; REL, c-REL vector; TAp73, TAp73 vector; TNFα, 20ng/ml. Proliferation data were normalized against Day 0 WST-1 optical density (OD450nm) value.
To further examine the effects of c-REL on the suppressor function of TAp73, UMSCC 1 lacking TAp73 was transfected with control vector, c-REL, TAp73, or a combination of c-REL and TAp73. c-REL enhanced, and TAp73 inhibited proliferation, while co-transfection of c-REL with TAp73 abrogated the inhibitory function of TAp73 (Fig. 6C). As HNSCC are relatively resistant to TNF-α-mediated inhibition of growth and survival (8), we examined the functional effect of TNF-α treatment or c-REL transfection alone, or in combination, on UM-SCC 22A cells (Fig. 6D). TNF-α alone weakly inhibited and c-REL enhanced proliferation, but the combination of TNF-α and c-REL overexpression further enhanced proliferation. Together, these data support the role of endogenous or exogenously overexpressed c-REL in repressing the anti-proliferative function of TNF-α and TAp73.
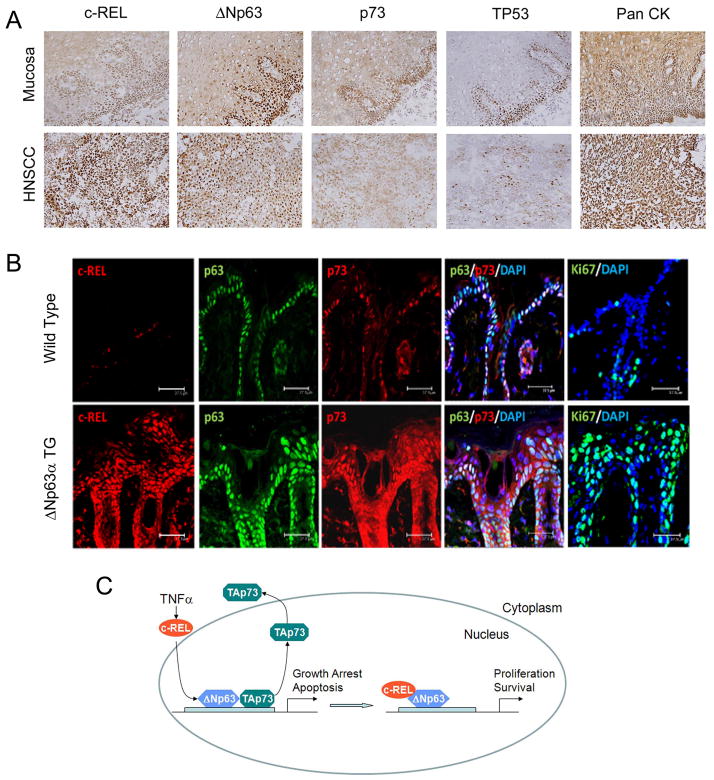
Aberrant expression and localization of c-REL, ΔNp63α, and TAp73 in HNSCC and ΔNp63α transgenic mice exhibiting squamous hyperplasia
To further confirm the biological significance in human HNSCC, we compared the expression and cellular distribution of these proteins in human HNSCC and squamous epithelia. Human HNSCC tumors displayed increased (2+~3+) nuclear co-staining for c-REL (24/24; 100%) and ΔNp63α (20/24; 83%), and decreased TAp73 staining distributed in the cytoplasm and nucleus throughout the malignant epithelia, when compared with a basilar pattern of nuclear staining for these proteins in nonmalignant squamous mucosas (5/5; 100%) (Figs. 7A, S5A,B). Interestingly, greater nuclear c-REL and cytoplasmic TAp73 were detected in HNSCC tumors than in unstimulated UMSCC cultured in vitro (Fig. S1A), consistent with overexpression of inflammatory factors such as TNFα previously demonstrated in the HNSCC tumor environment (7).
Figure 7.
Immunohistochemical staining and protein expression of TP53, p63, p73 and c-REL in HNSCC and ΔNp63α transgenic mice. Proposed model for interaction of c-REL, ΔNp63α, and TAp73. A, Immunostaining for c-REL, ΔNp63, p73, and TP53 in representative matched squamous mucosa and HNSCC tumor specimens. Pancytokeratin staining was used as a positive control to highlight epithelia. Original, 200X. B, Dorsal skin sections from adult transgenic (TG) K5-ΔNp63α mice were stained with specific antibodies for c-REL, ΔNp63α, and TAp73 by immunofluorescence as indicated. Ki67, marker of proliferation and Keratin 14, marker of epidermal differentiation. Bars, 37.5 μM. C, Model of TNF-α modulated c-REL, ΔNp63α, and TAp73 interactions in HNSCC. In response to TNF-α c-REL undergoes nuclear translocation, binding with ΔNp63α, while TAp73 dissociates from the promoters of growth arrest and apoptotic genes, and the nucleus, enhancing cell proliferation and survival. c-REL mediated effects are pronounced in the subset the HNSCC with increased expression of c-REL, ΔNp63α, and TAp73 with mtTP53.
We also examined a recently developed transgenic mouse model in which overexpression of ΔNp63α targeted to skin under a tet-K5 promoter results in inflammation, overexpression of TNF-α, other proinflammatory cytokines, and squamous hyperplasia (28, 30). By one month, these mice developed cutaneous hyperplasia, with increased suprabasilar nuclear c-Rel and ΔNp63α, and both cytoplasmic and nuclear staining for TAp73 in squamous epithelia (Fig. 7B). Thus, overexpression of ΔNp63α results in suprabasilar enhancement of nuclear c-REL and ΔNp63α, and cytoplasmic/nuclear distribution of TAp73, similar to that of orthologous proteins throughout malignant human squamous epithelia.
Discussion
Here we unveil evidence for a new paradigm (Fig. 7C), whereby an inflammatory factor, TNF-α, can promote nuclear interaction of c-REL with ΔNp63α, and TAp73 translocation to the cytoplasm, inhibiting the compensatory ability of TAp73 to activate key genes that mediate growth arrest and apoptosis in HNSCC with mutant TP53. Our findings establish a novel link between TNF-α expression and resistance (6–8, 10), nuclear activation of proto-oncogene c-REL (13–15), and dysregulation of p63/p73 family transcription factors (4, 5, 23, 28, 29), which have been independently implicated in cancer progression. TNF-α-induced c-REL/ΔNp63α interactions and TAp73 dissociation were demonstrated at unique p63 DNA regulatory sites within the promoters of important growth arrest and pro-apoptotic genes. The role of TNF-α induced c-REL in these specific interactions with ΔNp63α/TAp73, did not appear to involve RELA, nor c-REL transactivation domain residues involved in binding to κB enhancers shared with other NF-κB/REL family members. These findings suggest that TNF-α-induced c-REL has a distinct function in inactivating TAp73 in p63/p73 promoter-regulated proapoptotic gene expression, complementing that previously demonstrated for RELA in promoting κB-regulated prosurvival genes important in the malignant phenotype (10, 31). Furthermore, our findings suggest that targeting TNF-α signaling, c-REL, or other regulators of these c-REL-ΔNp63α-TAp73 interactions, could enhance TAp73 function, potentially helping in prevention or treatment of cancers with altered TP53 status.
A novel finding of this study is the demonstration of the role of TNF-α in coordinating interactions between nuclear c-REL/ΔNp63α and reciprocal dissociation of ΔNp63α/TAp73 complexes, linking these previously reported components and interactions into a common dynamic mechanism. Previously, we discovered nuclear interactions between murine c-Rel and overexpressed ΔNp63α in keratinocytes, and human c-REL and ΔNp63α in HNSCC (15). In murine keratinocytes, c-Rel and ΔNp63α were shown to prevent growth arrest, suggesting that corresponding c-REL/ΔNp63α complexes in HNSCC may also contribute to loss of growth control and promote the malignant phenotype. Independently, overexpressed ΔNp63α was shown to interact with TAp73, and promote survival in HNSCC (29). However, the relationship between these components, factor(s) modulating them, and basis for TAp73 inactivation were unknown. Here, we found that either c-REL or TAp73 interact with ΔNp63α, but we detected minimal interaction between c-REL and TAp73, suggesting that c-REL or TAp73 alternately complex withΔNp63α. Supporting this model, we further established that TNF-α dynamically promotes nuclear c-REL/ΔNp63α interaction and reciprocal dissociation of ΔNp63α/TAp73. This is accompanied by nuclear-cytoplasmic translocation of TAp73, thus providing a basis for the inactivation of TAp73. TNF-α-induced c-REL was an important component of this mechanism, as c-REL siRNA attenuated dissociation ofΔNp63α/TAp73, while c-REL overexpression had a similar promoting effect on the dissociation and cytoplasmic translocation of TAp73. These effects of TNF-α or c-REL in promoting increased nuclear c-REL/ΔNp63α and cytoplasmic TAp73 in a subset of HNSCC lines may help explain the similar distribution we observed in HNSCC tumors, where increased TNF-α expression, as well as amplification and nuclear localization of c-REL have been detected (7, 13, 14).
These dynamic nuclear c-REL-ΔNp6α-TAp73 interactions modulated by TNFα or c-REL, were mirrored on established and novel predicted p63 regulatory sites in the promoters of several growth arrest and apoptotic genes. EMSA results indicated TNF-α induces reciprocal modulation between c-REL or TAp73 in binding to an established p63 site sequence of the p21WAF1 promoter (15, 23). ΔNp63α binding remained relatively unchanged by TNF-α, but ΔNp63α knockdown inhibited ChIP binding of both c-REL and TAp73 to the p21WAF1 promoter, indicating that ΔNp63α bound to this p63 site is a key anchor for c-REL and TAp73 transcription factors. While NOXA and PUMA were also previously identified as p63/p73 target genes (29), specific binding sites for p63/p73 were not previously resolved. Using a bioinformatics approach, we predicted p63 binding sites in these genes. We observed similar specificity in binding and modulation by TNF-α of c-REL and TAp73 co-bound with ΔNp63α on p21WAF1, NOXA, and PUMA promoters by ChIP assay. We have recently demonstrated the capability of TNF-α to modulate c-REL/ΔNp63α interaction on p63 sites of additional genes by ChIP and EMSA (28), and obtained further evidence for the specificity of c-REL in interacting withΔNp63α at p63 regulatory sites. Preliminary ChIP sequencing results indicate that c-REL, p63 and/or p73 factors bind at many additional loci (H. Lu, unpublished data). Together, these findings suggest that the dynamic modulation of nuclear interactions involving these transcription factors observed in co-IP analysis, are likely related to their specific interactions on p63/p73 sites of multiple gene promoters.
Our results further revealed the important and reversible function of nuclear c-REL in attenuating the compensatory ability of TAp73 to promote expression of these key growth arrest and apoptotic genes. Knockdown of c-REL potentiated the expression of p21WAF1, NOXA and PUMA, further supporting the biologic and potential therapeutic relevance of c-REL-ΔNp63α-TAp73 interactions observed on their promoters. Moreover, the modulation of these growth arrest and apoptotic genes was specifically observed in UMSCC 22A from a subset overexpressing TAp73 andΔNp63α with mtTP53. This effect was not seen in UMSCC 1, from a subset with attenuation of expression and function of both TAp73 and wtTP53, which we have shown can result from other therapeutically reversible mechanisms (17, 32). Consistent with these findings, c-REL modulation affected proliferation and apoptosis in UMSCC22A but not UMSCC 1. Conversely, overexpressed c-REL inhibited the expression of p21WAF1 and anti-proliferative effects when TAp73 was re-expressed in TAp73-deficient UMSCC 1, supporting an important role for c-REL in inhibiting the compensatory ability of TAp73. Together, these results highlight the functional importance and potentially reversible nature of c-REL-mediated inhibition of TAp73 function in HNSCC overexpressing TAp73, ΔNp63α and mtTP53.
Our findings in UMSCC lines are likely to be of broader relevance in HNSCC and other cancers. Increased nuclear c-REL/ΔNp63α and nuclear and cytoplasmic TAp73 was observed in a majority of HNSCC tumor specimens. Similarly, increased ΔNp63α and TAp73 was previously seen in an independent panel of HNSCC tumors and lines, and linked with inactivation of TAp73 (5, 29). Breast cancer specimens also show increased ΔNp63 and TAp73 immunostaining, and this most often seen in specimens with mtTP53 status (33). Subsets exhibiting increased expression of ΔNp63 in HNSCC and breast cancer have also been reported to be more sensitive to the chemotherapy drug cisplatin (33, 34), underscoring the potential clinical relevance of identifying and selecting agents active in these tumor subsets.
Inflammatory mediator TNF-α is identified as a key modulator of dynamic interactions of c-REL with ΔNp63α, and inactivation of TAp73. This mechanism could therefore contribute to the acquired resistance and promoting effects of TNF-α produced by inflammatory cells during tumorigenesis and metastatic tumor progression of HNSCC and other cancers (6–8, 10). Supporting this hypothesis, we show here that combining TNF-α with overexpression of c-REL not only negated the inhibitory effect of TNF-α, but enhanced proliferation over that observed with either alone. Further, human HNSCC tumors and epithelia of K5-ΔNp63α transgenic mice that exhibit increased TNF-α expression, inflammation, and epithelial proliferation (7, 28), showed greater nuclear c-REL/ΔNp63α, and distribution of TAp73 between the nucleus and the cytoplasm. Previous findings support targeting TNF-α, or the canonical signal pathway which activates c-REL, for prevention or therapy of SCC. Knockout of TNF-α or TNF receptor-1, or TNF-α inhibitors, have been shown to reduce chemical carcinogenesis and malignant progression of SCC of the skin and other epithelial cancers (6). We previously showed that blocking canonical pathway signaling increased TNF-α cytotoxicity in HNSCC in vitro, and induced apoptosis and regression of established murine and human SCC in vivo (8). However, early phase clinical trials with TNF-α or proteasome inhibitors, which inhibit canonical pathway activation, have demonstrated limited potential to slow disease progression in patients with advanced cancers (6, 13).
Consequently, other molecular requirements for coordinated modulation of c-REL, ΔNp63α and TAp73 interactions on the promoters of target genes merit investigation as targets for therapy. Unique structural characteristics of c-REL have previously been implicated in TNF-α-induced, TBK and IKKepsilon phosphorylation, dimerization, DNA binding, or transactivation (35, 36). One of our laboratories found that the ΔNp63 α-domain, critical for oligomerization (5), is necessary for interaction with c-Rel (15). Cisplatin induced IKKα and β activation have been reported to promote degradation ofΔNp63α and stabilization of p73, suggesting DNA damaging agents and these kinases may be important modulators of these important components of the mechanism described herein (37, 38).
Supplementary Material
Acknowledgments
Grant support: NIDCD Intramural Research Projects Z01-DC-000016 and DC-000073. C.A. supported by the NIH-Pfizer Clinical Research Training Program; J.C. supported by the HHMI-NIH Scholars Program.
The authors thank Drs. James F. Battey (NIDCD/NIH), Victor Zhurkin (NCI/NIH), and Cheng-Ming Chiang (University of Texas, Southwestern Medical Center) for serving as readers and for helpful comments.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2:a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–61. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King KE, Weinberg WC. P63: Defining roles in the morphogenesis, homeostasis and neoplasia of the epidermis. Cancer Res. 2008;68:5122–31. doi: 10.1002/mc.20337. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbluth JM, Pietenpol JA. The jury is in: p73 is a tumor suppressor afterall. Genes Dev. 2008;22:2591–5. doi: 10.1101/gad.1727408. [DOI] [PubMed] [Google Scholar]
- 5.DeYoung, Ellisen LW. P63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–83. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 6.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–71. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 7.Younes F, Quartey EL, Kiguwa S, Partridge M. Expression of TNF and the 55-kDa TNF receptor in epidermis, oral mucosa, lichen planus and squamous cell carcinoma. Oral Dis. 1996;2:25–31. doi: 10.1111/j.1601-0825.1996.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 8.Duffey DC, Crowl-Bancroft CV, Chen Z, Ondrey FG, Nejad-Sattari M, Dong G, et al. Inhibition of transcription factor nuclear factor-kappaB by a mutant inhibitor-kappaBalpha attenuates resistance of human head and neck squamous cell carcinoma to TNF-alpha caspase-mediated cell death. Br J Cancer. 2000;83:1367–74. doi: 10.1054/bjoc.2000.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beg AA, Baldwin AS., Jr Activation of multiple NF-kappaB/Rel DNA binding complexes by tumor necrosis factor. Oncogene. 1994;9:1487–92. [PubMed] [Google Scholar]
- 10.Wang CY, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 11.Duan J, Friedman J, Nottingham L, Chen Z, Ara G, Van Waes C. Nuclear factor-kappaB p65 small interfering RNA or proteasome inhibitor bortezomib sensitizes head and neck squamous cell carcinomas to classic histone deacetylase inhibitors and novel histone deacetylase inhibitor PXD101. Mol Cancer Ther. 2007;6:37–50. doi: 10.1158/1535-7163.MCT-05-0285. [DOI] [PubMed] [Google Scholar]
- 12.Lee TL, Yeh J, Friedman J, Yan B, Yang X, Yeh NT, et al. A signal network involving coactivated NF-kappaB and STAT3 and altered p53 modulates BAX/BCL-XL expression and promotes cell survival of head and neck squamous cell carcinomas. Int J Cancer. 2008;122:1987–98. doi: 10.1002/ijc.23324. [DOI] [PubMed] [Google Scholar]
- 13.Allen C, Saigal K, Nottingham L, Arun P, Chen Z, Van Waes C. Bortezomib-induced apoptosis with limited clinical response is accompanied by inhibition of canonical but not alternative nuclear factor-{kappa}B subunits in head and neck cancer. Clin Cancer Res. 2008;14:4175–85. doi: 10.1158/1078-0432.CCR-07-4470. [DOI] [PubMed] [Google Scholar]
- 14.Liu CJ, Lin SC, Chen YJ, Chang KM, Chang KW. Array-comparative genomic hybridization to detect genomewide changes in microdissected primary and metastatic oral squamous cell carcinomas. Mol Carcinog. 2006;45:721–31. doi: 10.1002/mc.20213. [DOI] [PubMed] [Google Scholar]
- 15.King KE, Ponnamperuma RM, Allen C, Lu H, Duggal P, Chen Z, et al. The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res. 2008;68:5122–31. doi: 10.1158/0008-5472.CAN-07-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan B, Yang X, Lee TL, Friedman J, Tang J, Van Waes C, et al. Genome-wide identification of novel expression signatures reveal distinct patterns and prevalence of binding motifs for p53, nuclear factor-kappaB and other signal transcription factors in head and neck squamous cell carcinoma. Genome Biol. 2007;8:R78. doi: 10.1186/gb-2007-8-5-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman J, Nottingham L, Duggal P, Pernas FG, Yan B, Yang XP, et al. Deficient TP53 expression, function, and cisplatin sensitivity are restored by quinacrine in head and neck cancer. Clin Cancer Res. 2007;13:6568–78. doi: 10.1158/1078-0432.CCR-07-1591. [DOI] [PubMed] [Google Scholar]
- 18.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32:417–26. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TL, Yang XP, Yan B, Friedman J, Duggal P, Bagain L, et al. A Novel Nuclear Factor-{kappa}B Gene Signature Is Differentially Expressed in Head and Neck Squamous Cell Carcinomas in Association with TP53 Status. Clin Cancer Res. 2007;13:5680–91. doi: 10.1158/1078-0432.CCR-07-0670. [DOI] [PubMed] [Google Scholar]
- 20.Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, et al. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–75. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thurfjell N, Coates PJ, Vojtesek B, Benham-Motlagh P, Eisold M, Nylander K. Endogenous p63 acts as a survival factor for tumour cells of SCCHN origin. Int J Mol Med. 2005;16:1065–70. [PubMed] [Google Scholar]
- 22.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 23.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–76. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine- methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–15. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, Malhotra PS, Thomas GR, Ondrey FG, Duffey DC, Smith CW, et al. Expression of proinflammatory and proangiogenic cytokines in patients with head and neck cancer. Clin Cancer Res. 1999;5:1369–79. [PubMed] [Google Scholar]
- 26.Kunsch C, Rubin SM, Rosen CA. Selection of optimal kappa B/Rel DNA-binding motifs: interaction of both subunits of NF-kappa B with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–21. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Gelinas CA. mutant Rel-homology domain promotes transcription by p50/NFkappaB1. Oncogene. 1997;14:1521–30. doi: 10.1038/sj.onc.1200985. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J, et al. ΔNp63 versatilely regulates a broad NF-κB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71:3688–700. doi: 10.1158/0008-5472.CAN-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Romano RA, Smalley K, Liu S, Sinha S. Abnormal hair follicle development and altered cell fate of follicular keratinocytes in transgenic mice expressing DeltaNp63alpha. Development. 2010;137:1431–39. doi: 10.1242/dev.045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–3. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 32.Ehsanian R, Brown M, Lu H, Yang XP, Pattatheyil A, Yan B, et al. YAP dysregulation by phosphorylation or ΔNp63-mediated gene repression promotes proliferation, survival and migration in head and neck cancer subsets. Oncogene. 2010;29:6160–71. doi: 10.1038/onc.2010.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007;117:1370–80. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zangen R, Ratovitski E, Sidransky D. DeltaNp63alpha levels correlate with clinical tumor response to cisplatin. Cell Cycle. 2005;4:1313–15. doi: 10.4161/cc.4.10.2066. [DOI] [PubMed] [Google Scholar]
- 35.Gilmore TD, Kalaitzidis D, Liang MC, Starczynowski DT. The c-REL transcription factor and B-cellproliferation: a deal with the devil. Oncogene. 2004;23:2275–86. doi: 10.1038/sj.onc.1207410. [DOI] [PubMed] [Google Scholar]
- 36.Gilmore TD, Garbati MR. Inhibition of NF-kB signaling as a strategy in disease therapy. In: Karin M, editor. NF-kB in health and disease. Berlin: Springer-Verlag; 2010. pp. 245–63. [Google Scholar]
- 37.Chatterjee A, Chang X, Sen T, Ravi R, Bedi A, Sidransky D. Regulation of p53 family member isoform DeltaNp63 alpha by the nuclear factor kappaB targeting kinase IkappaB kinase beta. Cancer Res. 2010;70:1419–1429. doi: 10.1158/0008-5472.CAN-09-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuya K, Ozaki T, Hanamoto T, Hosoda M, Hayashi S, Barker PA, et al. Stabilization of p73 by nuclear IkappaB kinase-alpha mediates cisplatin-induced apoptosis. J Biol Chem. 2007;282:18365–78. doi: 10.1074/jbc.M610522200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.