Abstract
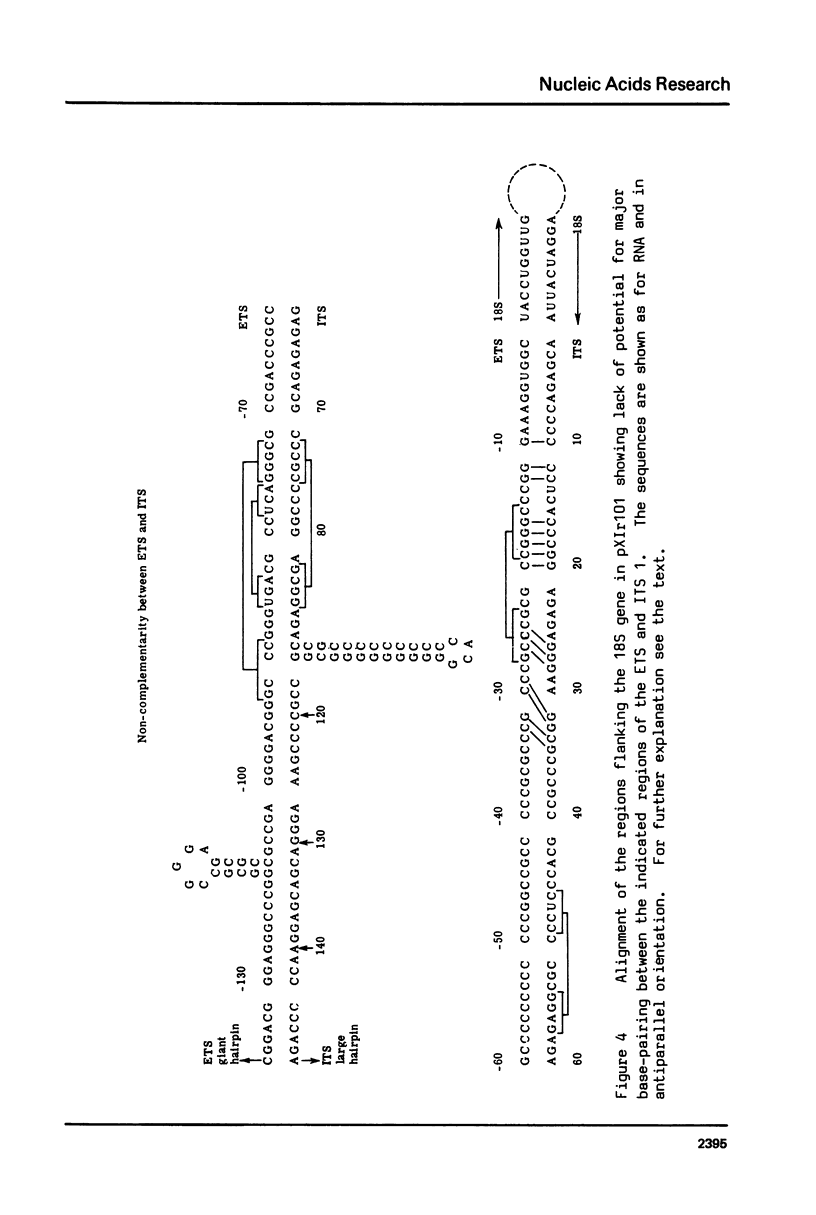
We have sequenced the external transcribed spacer (ETS) of a ribosomal transcription unit from Xenopus laevis, together with sections of the preceding non-transcribed spacer. Our analysis was carried out on the same cloned transcription unit as that from which the internal transcribed spacers (ITS) were previously sequenced. The ETS is approximately 712 nucleotides long and, like the ITS regions, is generally very rich in C plus G. Features of the sequence include an excess of oligo-C tracts over oligo-G tracts and a tract of 37 nucleotides consisting almost entirely of G and A residues. Parts of the sequence can give rise to stable internal secondary structures. However, in contrast to Escherichia coli, there is no potential for major base-pairing between the 18S flanking regions of the ETS and ITS. Further findings are that there are no initiation (ATG) codons in the ETS and that, as in other X.laevis rDNA cloned units, the sequence preceding the ETS is duplicated, with a few changes, in the "Bam island" sequence of the non-transcribed spacer.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botchan P., Reeder R. H., Dawid I. B. Restriction analysis of the nontranscribed spacers of Xenopus laevis ribosomal DNA. Cell. 1977 Jul;11(3):599–607. doi: 10.1016/0092-8674(77)90077-0. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Gerbi S. A. Fine structure of ribosomal RNA. IV. Extraordinary evolutionary conservation in sequences that flank introns in rDNA. Nucleic Acids Res. 1980 Aug 25;8(16):3623–3637. doi: 10.1093/nar/8.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening U. E., Jones K. W., Birnstiel M. L. Properties of the ribosomal RNA precursor in Xenopus laevis; comparison to the precursor in mammals and in plants. J Mol Biol. 1969 Oct 28;45(2):353–366. doi: 10.1016/0022-2836(69)90110-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence encoding the 5' end of Xenopus laevis 18S rRNA. Nucleic Acids Res. 1980 Jul 11;8(13):2871–2884. doi: 10.1093/nar/8.13.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., van Heerikhuizen H., Planta R. J. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon. Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res. 1981 Oct 10;9(19):4847–4862. doi: 10.1093/nar/9.19.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A., Penman S. Processing of 45 s nucleolar RNA. J Mol Biol. 1970 Jan 28;47(2):169–178. doi: 10.1016/0022-2836(70)90337-2. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]



