Abstract
The recently developed Coherent Anti-stokes Raman Scattering (CARS) microscopy and Stimulated Raman Scattering (SRS) microscopy have provided new methods to visualize the localization and regulation of biological molecules without the use of invasive and potentially perturbative labels. They allow rapid imaging of specific molecules with high resolution and sensitivity. These tools have been effectively applied to the study of lipid metabolism using Caenorhabditis elegans as a genetic model, unraveling new lipid storage phenotypes and their regulatory mechanisms. Here we review the underlying principle of CARS and SRS microscopy, as well as their recent applications in lipid biology research in C. elegans.
Introduction
The ability to track specific biological molecules following their spatial distribution and temporal dynamics in vivo is essential for understanding their physiological impacts and regulatory mechanisms. To this end, fluorescence microscopy is currently the most popular imaging contrast used in biological studies. Various fluorescence-based techniques have flourished such as confocal laser scanning, two-photon excited fluorescence and single-molecule microscopy and super-resolution imaging (1–6). However, due to the physical size and chemical invasiveness of fluorescent tags, fluorescence microscopy is not well suited for visualizing many essential biological molecules in vivo, such as lipids.
The worldwide epidemic of obesity is a major cause for concern with excess adiposity being associated with type II diabetes, cardiovascular disease, and some types of cancer. The prevalence of obesity has spurred increased interest in the molecular pathways and physiological mechanisms regulating lipid metabolism (7). However, it has been challenging to detect lipid distribution and dynamics in vivo at both cellular and organismal levels. Lipids are intrinsically non-fluorescent and difficult to tag with fluorophores, which limits the use of fluorescence microscopy. To better understand the biology of obesity, new imaging methods are desirable to directly and specifically visualize lipids and analyze their regulation.
Recently emerged Coherent Anti-Stokes Raman Scattering (CARS) microscopy and Stimulated Raman Scattering (SRS) microscopy are fundamentally different from fluorescence microscopy (8). These new chemical imaging techniques capture biological molecules based on their intrinsic chemical-group vibrational contrast. They have been utilized to visualize protein, DNA, lipids and small metabolites in vivo without any labeling (9) (10–16). Due to the strongest signal arising from lipid molecules, one area in which CARS and SRS have quickly been applied is the study of lipid metabolism. CARS and SRS are capable of detecting lipid molecules directly in vivo in a label-free, highly selective and quantitative manner (14, 16). Moreover, their recent applications in the model organism Caenorhabditis elegans have also set the stage to uncover the most important players in lipid metabolism and its regulation (17–20). In this review, the first section introduces the underlying principle of CARS and SRS microscopy, and the second part overviews the recent applications of these new technologies in lipid biology research in C. elegans.
The principle of CARS and SRS microscopy
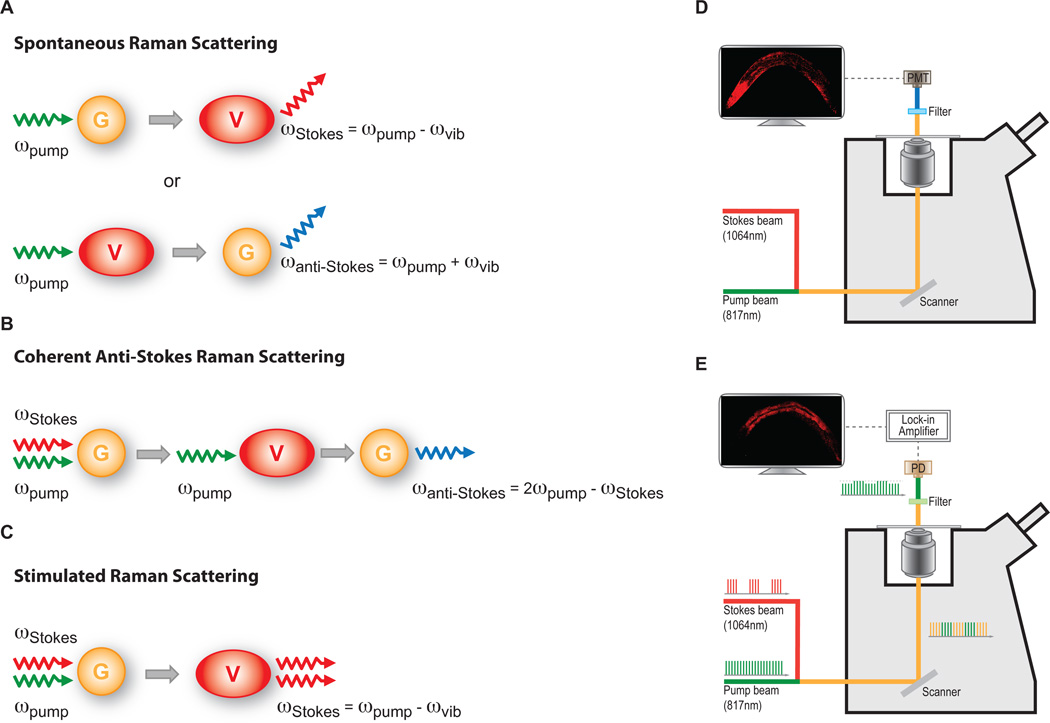
When light interacts with matter, the energy is both elastically scattered and inelastically scattered (Figure 1A). When a laser beam illuminates a sample and interacts with individual molecules, most photons will elastically scatter (Rayleigh scatter), maintaining the energy of the incident light. However a much smaller fraction of photons undergo inelastic (i.e., energy exchange) scattering by interacting with the vibrational state of molecules. Different vibrational states are dictated by the composition of the molecules and the chemical bonds that are present. The scattering photons (normally called Stokes Raman photons) will have less energy than the incoming photons if the chemical bonds get excited to a higher energy vibrational level. In contrast, by interacting with the chemical bonds in an excited vibrational state, the scattering photons (anti-Stokes Raman photons) will have higher energy than the incident ones. The frequency difference (Raman shifts) between the incoming and scattering light is determined by the vibrational energy levels of the chemical bonds. A complex molecule will generate a complicated Raman spectrum consisting of the combined Raman shift peaks from all the consisting chemical bonds. The Raman spectra can be acquired by Raman microscopy, and provides a characteristic fingerprint of specific molecules in the sample, allowing their identification without labeling.
Figure 1. Principle and diagram of CARS and SRS microscopy systems.
A. Spontaneous Raman scattering. Incident photons interacting with molecules normally scatter elastically. However, a very small fraction of photons are inelastically scattered. When an incident photon (pump photons) interacts with a molecule carrying specific chemical bonds in the vibrational ground state (G), the inelastic scattering photon (Stokes photon) will have lower energy than the incident ones. The energy difference between the pump and Stokes photons is equal to the vibrational energy of the chemical bond, ϖpump - ϖStokes = ϖvib. In contrast, a scattering photon with a higher energy (Anti-Stokes photons) will generate, by interacting with the chemical bonds in an excited vibrational state (V), ϖanti-Stokes - ϖpump= ϖvib.
B. Coherent anti-Stokes Raman scattering. This multiphoton process is achieved by using higher energy “pump” photons and lower energy “Stokes” photons, with energy difference being resonant with the vibrational frequency of specific chemical bonds in a molecule. The non-linear interaction between pump and Stokes photons will excite the vibration coherence of the chemical bonds in the molecule. This excited vibration coherence will further interact with a second pump photon, resulting in the coherent emission of an anti-Stokes photon that is more energetic (ϖanti-Stokes = 2ϖpump - ϖStokes = ϖpump + ϖvib). Unlike spontaneous Raman scattering, where inelastic photons scatters in all directions, the anti-Stokes light in CARS emits in a certain specific direction.
C. Stimulated Raman scattering. When the energy difference between pump and Stokes photons resonant with the vibrational frequency of a type of chemical bonds in a molecule. The non-linear interaction between two photons stimulates the chemical bonds into excited vibrational state. The energy transfer from the pump beam to the Stokes beam accompanies this, vibrational excitation, resulting in the disappearance of one pump photon and the creation of a Stokes photon.
D. Diagram of CARS microscope. The Stokes laser beam has a fixed wavelength at 1064nm, while the pump laser beam is tunable. Pump beam at 817nm for lipid imaging is used an example here. The combined pump and Stokes laser beams is scanned over the sample by a XY scanner. CARS signal is collected using a red-sensitive photomultipier tube (PMT). In front of the PMT, filters are used to block the pump and Stokes beams and any induced two-photon fluorescence.
E. Diagram of SRS microscope. The laser set up is very similar to CARS, except that the pump laser beam is modulated at a high frequency (~10MHz). The filter is used to block the Stoke beam completely. The transmitted pump beam containing SRL signals is detected by a photodiode (PD). An electronic device, Lock-in Amplifier, is used to demodulate the SRL signals carried on the pump beam.
In the conventional Raman microscopy, a single frequency of excitation laser beam is utilized to generate spontaneous Raman scattering. Unfortunately, owing to signal weakness of the spontaneous vibrational transition (10 orders of magnitudes weaker than fluorescence), conventional Raman microscopy images require high laser power and long data acquisition time, which limit its application in biomedical studies (21). Raman signals can be amplified via using two coherent excitation laser beams and non-linear interaction between two laser beams and the molecules. This provides the principle for coherent non-linear optical imaging techniques including CARS and SRS.
I. Coherent anti-Stokes scattering (CARS) microscopy
In CARS microscopy, two laser beams, pump and Stokes, focus on a common focal spot (Figure 1 B and D). One laser beam has a fixed frequency, while the other is tunable. When their energy difference matches with the vibrational frequency of a type of chemical bond, a dramatically enhanced coherent anti-Stokes Raman scattering (CARS) signal is generated at the anti-Stokes frequency via a third-order non-linear process (Figure 1B) (21). The newly emitted light has a blue-shifted color that is different from those of the incidence laser beams and background fluorescence. This spectral separation makes it easy to detect the CARS signals. Utilization of near-infrared pump and Stokes laser beams allows deep penetration in tissues, as well as significantly reducing photodamage in samples. Moreover, the non-linear dependence on excitation intensities provides CARS the inherent capability to generate 3-dimensional images. Thus CARS microscopy offers a direct and noninvasive way to image biological samples with chemical selectivity. In 1999, three-dimensional imaging of living cells by CARS was achieved in live bacterial and Hela cells (22). This work triggered CARS applications in detection of distinct biological molecules in various living cells and organisms, including DNA, protein and lipids using phosphate stretching vibration, amide I vibration and CH stretching vibration respectively (9–12). Among these, the strongest signal arises from the lipid CH stretching mode.
Although the CARS signal is much stronger than the spontaneous Raman scattering, it has a significant drawback. CARS imaging has a non-resonant background at the anti-Stokes frequency. This non- resonant background is independent of the laser frequency tuning and is not derived from the target molecular vibration. Instead, it has a complicated dependence on the geometrical structure and local concentration of the object, and can cause image artifacts especially for complex biological samples (8, 20).
II. Stimulated Raman scattering microscopy
Recently, SRS microcopy has emerged as a new label-free imaging technique, overcoming limitations of CARS microscopy (14). Similar to CARS, SRS microscopy relies on two laser beams coincided on the sample. When their energy difference matches the molecular vibrational frequency, the molecular vibrational excitation is stimulated after non-linear interaction (Figure 1C). This is accompanied by the energy transfer from the pump beam to the Stokes beam, and results in the intensity loss of the pump beam called stimulated Raman loss (SRL), and the intensity gain of the Stokes beam called stimulated Raman gain (SRG). When the energy difference between the two laser beams does not match with the target molecule vibrational frequency, there is no energy transfer between the pump and Stokes beams, and therefore no SRG or SRL signals. Thus SRS microscopy is free from the non-resonant background.
In an SRS microscope (as shown in Figure 1E) (14), the intensity of the incoming Stokes laser beam is temporarily modulated at a high frequency, whereas the incident pump beam (before interacting with the sample) remains unmodulated. After interacting with the samples, the pump beam intensity change (SRL) is detected and amplified via a lock-in amplifier. The extracted signals will be utilized for image construction. Owing to the non-linear dependence, SRS has inherent 3-D sectioning capability. The SRL signal intensity is linearly associated with the concentration of the targeted molecules in the sample (14), which is crucial for quantification. SRS microscopy was first demonstrated in biological imaging in 2008 (14), which was followed immediately by works from two other groups in 2009 (23, 24). This new label-free microscopy technique allows imaging biological molecules in living cells with high resolution, sensitivity, and speed (8). It has been broadly applied to lipid measurement, drug delivery monitoring and tumor cell detection (14, 20, 25).
In the following section, we will focus on the application of CARS and SRS microscopy in studying lipid metabolism using C. elegans as a model system.
Genetic understanding of lipid metabolism in C. elegans using CARS and SRS
Lipids contain large amounts of fatty acid side chains that have abundant C-H bonds and specific CH2 stretching frequency at a Raman shift of 2845cm−1. CARS and SRS microscopy can specifically detect the CH2 stretching vibration, and are well suited to analyze lipid molecules directly in vivo. C. elegans is a well-established multicellular organism for high-throughput genetic studies due to its small size, rapid development, available genetic tools, and optical transparency. It also emerged as a promising model to understand genetic and environmental regulation of fat storage (26, 27). However, canonical lipid analysis methods have limited specificity, sensitivity and spatial information. Recent advances using the power of CARS and SRS microscopy have overcome these hurdles, and provided noninvasive, sensitive and quantitative ways to study lipid metabolism in C. elegans (17–20).
I. Overview of canonical lipid analysis methods in C. elegans
Analysis of lipid storage in C. elegans can be performed using traditional biochemical methods such as thin layer chromatography (TLC) or solid phase exchange (SPE) chromatography with gas chromatography /mass spectrometry(GC/MS) (28–30). Using a 13C isotope feeding assay, these techniques were extended to analysis of fat synthesis versus dietary absorption (30). However, biochemical methods are limited by the large number of worms required and the lack of spatial information achieved by analysis on whole worms.
Alternatively, simple feeding protocols have led to wide use of Nile red and C1-C12-BODIPY vital dyes in screening for and characterizing lipid storage phenotypes in C. elegans. However, several groups have demonstrated, by comparison with biochemical quantification, fixative-based staining, and label-free imaging techniques, that these vital dyes are poor assays for both localization and quantification of lipid storage (19, 20, 31–33). The vital dyes strongly stain lysosome-related organelles rather than the main fat stores in neutral lipid droplets in wild-type C. elegans (31, 33). In addition, with feeding-based protocols, the vital dyes are dependent on absorption and trafficking to lipid compartments through intestinal cells, preventing staining of germline and peripheral hypodermal lipid stores (17). Since the vital dyes have been shown to be such poor markers of lipid storage, fixative-based protocols have been increasingly relied upon.
Oil red O, Sudan black, and LipidTOX red and green are fixative-based dyes. Unfortunately, organic solvents used for fixation often interfere with the lipid storage structures. Detection of Oil red O and Sudan black staining uses bright field microscopy under white light illumination, which greatly reduces spatial resolution. LipidTOX green and red suffer from low efficiency in staining and rapid photobleaching (33). Fixative-based protocols using Nile red were also recently developed to localize major lipid stores (32, 34). However, this lipophilic dye may stain lipofuscin, a hydrophobic aging-related accumulation of protein and lipid (19, 35). Moreover, although the intensity of these dyes is correlated with the amount of lipid molecules, there is no direct linear relationship between them. Therefore, these methods only reveal the trend of alterations in lipid levels, but often their results are not consistent with biochemical quantification (19).
II. Label-free imaging of lipid dynamics using CARS and SRS
To overcome the limitations of canonical lipid analysis methods, several groups have used high-speed CARS microscopy coupled with fingerprint confocal Raman analysis to enable label-free imaging of lipids in C. elegans (17, 18). These studies have revealed several interesting phenotypes that were previously unknown.
Dauer is an alternative C. elegans developmental stage that takes place in stressful conditions, e.g. starvation and overcrowding. Dauer formation is under the control of several signaling pathways, including insulin/IGF-1 and TGF-β signaling (36). In the dauer mutants, lipid storage levels are dramatically increased (37). With the high resolution of CARS microscopy, Hellerer et al. observed a more than 4 fold increase in small-sized lipid droplets in the hypodermis of the dauer mutants. Interestingly, this hypodermal lipid accumulation did not exhibit a significant reduction even after 3 weeks of dauer arrest, however the total lipid levels were decreased substantially (17). This suggests that energy mobilization in the intestinal and the hypodermal cells may be differentially regulated.
More fat in general is bad, but not all fat is the same. Protective effects of unsaturated fat, especially polyunsaturated fat are well documented. However specific analysis of unsaturated fat in vivo was previously impossible using the canonical methods. Le et al. coupled CARS microscopy with spontaneous Raman microspectroscopy to enable quantification of the unsaturation ratio in single lipid droplets in live worms (18). They confirmed that lipid unsaturation levels were reduced by half in the mutants of Δ9 desaturases that catalyze the first step in fatty acid unsaturation (38).
Although CARS represents an advance in label-free imaging of lipid in vivo, it is still hindered by significant non-resonant background and autofluorescence (8, 19, 20, 25). In addition, CARS signals have a complicated nonlinear relationship to the target molecule concentration (21). As a result, CARS signals display a partial overlap with non-lipid-related autofluorescence signals (18, 19), and are not suited for straightforward quantification (20).
The use of SRS microscopy has overcome these limitations (8, 14, 23, 24). First, SRS is free from both non-resonant and autofluorescence backgrounds. Together with strict linear concentration dependence, SRS avoids most of the image artifacts and allows direct and reliable quantification (20). Quantification of lipid levels by SRS in vivo is no different from that using the in vitro chromatography method. However, the SRS method only requires five worms instead of 5 × 103 in the biochemical analysis, and the experimental processing time is shortened to a few minutes (20). Second, SRS is demonstrated to exhibit much higher detection sensitivity than CARS. The detection limit is only 50 µM for retinol, corresponding to 3000 molecules in focus (14). Third, like CARS, SRS microscopy is capable of three-dimensional optical sectioning in a thick biological sample, and its spatial resolution is similar to two-photon fluorescence microscopy, 200 ~ 300 nm in the x and y-plane and 500 ~ 800 nm in the z-axis (14). Finally, the most recent technical advances have raised the imaging speed of SRS to twenty-five frames (512 × 512 pixels) per second, and achieved its light collection in the epi direction (25). These enable fast detection of molecule dynamics even in non-transparent samples like mouse and human tissues.
The video-rate imaging speed and label-free nature also make SRS well-suited for high-throughput genetic screening. We have applied SRS microscopy in C. elegans RNA interference (RNAi) screening (20). At first, SRS microscopy allowed us to detect neutral lipid distribution in different tissues, identify lipid droplets at subcelluar levels and accurately quantify lipid levels in vivo. Next, we chose about 300 genes that encode cell-surface and nuclear hormone receptors, and have measured neutral lipid storage at the organism level upon RNAi inactivation of those genes. This allowed us to identify several new genetic regulators of lipid storage that are well conserved in human. These studies have set the stage for genome-wide analysis to uncover important genes regulating lipid metabolism and their underlying mechanisms.
Based on the presence of C=C double bonds in unsaturated lipid, SRS microscopy enables to differentiate the distributions of unsaturated from saturated lipid in living cells (14). In the unpublished work, we also examined unsaturated lipid distribution in live C. elegans. Early this year, Freudinger et al. developed spectrally tailored excitation-SRS (STE-SRS) to further improve the selectivity of SRS for specific chemical molecules (13). This method showed the potential to selectively image different types of fatty acids in live C. elegans.
Advancing beyond fluorescence microscopy, CARS and SRS optical imaging provide new mechanisms to probe biological molecules in a selective yet non-invasive manner. SRS microscopy overcomes most of the limitations inherent to CARS. CARS and SRS are flexible methodologies that can be utilized to study other molecules, including DNA, protein, and a number of small molecules (9, 13–16). CARS and SRS not only allow scientists to see molecules that were previously invisible, but also enable mechanistic studies to identify what was previously unknown about their regulation. They have broad applications, ranging from tumor detection to monitoring drug delivery. Recent applications of CARS and SRS in C. elegans have moved forward the research on lipid metabolism, and unraveled very important phenotypes and mechanisms. Further technological advancement is expected to increase the detection sensitivity and specificity, which will allow in vivo imaging of low-abundant small molecules and distinct molecules with closely related chemical structure. Currently, the expensive instruments and complexity to set up them are the two major limitations that prevent CARS and SRS from being used by more groups. We are expecting new technology development and commercialization will lower the cost and ease the use, which will accelerate the widespread use of CARS and SRS microscopy systems in the near future.
Table 1.
Methods of lipid detection and measurement in C. elegans.
| Biochemical Methods | Fixative-Based Staining | CARS | SRS | |
|---|---|---|---|---|
| Quantitative | Yes | No | Semi | Yes |
| Spatial Resolution | - | Low | High | High |
| Lipid Differentiation | Yes | No | Yes | Yes |
| Detection Sensitivity | Low | Low | Medium | High |
| Measurement Consistency | High | Low | Medium | High |
| Non-Invasive Analysis | No | No | Yes | Yes |
| Major Limitations | •No spatial information | •Inconsistency between samples | •Non-resonant background | |
| •Large number of samples | •Indirect quantification | •Autofluorescence interference |
||
| •Long processing time | •No detection specificity | •Indirect quantification | ||
Acknowledgements
We would like to thank Dr. J. Wang for critical reading of the manuscript. A.F. received support from National Institute of Health grants through the Program in Development Biology and Medical Scientist Training Program (T32 HD055200, T32 GM007330). M.C.W. is supported by a National Institute of Health grant (AG034988).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 2.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betzig E, Patterson GH, Sougrat R, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 5.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 7.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- 8. Min W, Freudiger CW, Lu S, Xie XS. Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annu Rev Phys Chem. 2011;62:507–530. doi: 10.1146/annurev.physchem.012809.103512. ‥ The authors review the underlying principle and excitation and detection schemes of coherent nonlinear optical imaging techniques including CARS and SRS, and their biomedical applications. This is an outstanding resource to learn the physical properties of these new imaging mechanisms, as well as other biomedical applications beyond studying lipid biology.
- 9.Cheng JX, Jia YK, Zheng G, Xie XS. Laser-scanning coherent anti-Stokes Raman scattering microscopy and applications to cell biology. Biophys J. 2002;83:502–509. doi: 10.1016/S0006-3495(02)75186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conovaloff A, Wang HW, Cheng JX, Panitch A. Imaging growth of neurites in conditioned hydrogel by coherent anti-stokes raman scattering microscopy. Organogenesis. 2009;5:149–155. doi: 10.4161/org.5.4.10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nan X, Cheng JX, Xie XS. Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy. J Lipid Res. 2003;44:2202–2208. doi: 10.1194/jlr.D300022-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Wang HW, Fu Y, Huff TB, et al. Chasing lipids in health and diseases by coherent anti-Stokes Raman scattering microscopy. Vib Spectrosc. 2009;50:160–167. doi: 10.1016/j.vibspec.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freudiger CW, Min W, Saar BG, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. ‥ This is the first study to demonstrate SRS microscopy as a new lable-free biochemical imaging technology.
- 14. Freudiger CW, Min W, Holtom GR, et al. Highly specific label-free molecular imaging with spectrally tailored excitation-stimulated Raman scattering (STE-SRS) microscopy. Nature Photonics. 2011;5:103–109. doi: 10.1038/nphoton.2010.294. ‥The authors report spectrally tailored excitation-stimulated Raman scattering (STE-SRS) microscopy. This advance in SRS will enhance its spectral specificity and open the door for broader application of SRS in detecting closely related molecules.
- 15.Nan X, Potma EO, Xie XS. Nonperturbative chemical imaging of organelle transport in living cells with coherent anti-stokes Raman scattering microscopy. Biophys J. 2006;91:728–735. doi: 10.1529/biophysj.105.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nan X, Yang WY, Xie XS. CARS microscopy: lights up lipids in living cells. Biophotonics International. 2004;11:44. [Google Scholar]
- 17.Hellerer T, Axang C, Brackmann C, et al. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc Natl Acad Sci U S A. 2007;104:14658–14663. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Le TT, Duren HM, Slipchenko MN, et al. Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans. J Lipid Res. 2010;51:672–677. doi: 10.1194/jlr.D000638. ‥The authors report the application of CARS micrscopy in studying lipid metabolism in the model organism, Caenorhabditis elegans.
- 19.Yen K, Le TT, Bansal A, et al. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang MC, Min W, Freudiger CW, et al. RNAi screening for fat regulatory genes with SRS microscopy. Nat Methods. 2011;8:135–138. doi: 10.1038/nmeth.1556. …Using SRS microscopy, the authors demonstrate a new RNA interference screening platform to unravel important players in lipid metabolism and its regulation. This is the first study to apply SRS microscopy in high-throughput genetic screens.
- 21.Evans CL, Xie XS. Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:883–909. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 22.Zumbusch A, Holtom GR, Xie XS. Three-Dimensional Vibrational Imaging by Coherent Anti-Stokes Raman Scattering. Physical Review Letters. 1999;82:4142–4145. [Google Scholar]
- 23.Ozeki Y, Dake F, Kajiyama S, et al. Analysis and experimental assessment of the sensitivity of stimulated Raman scattering microscopy. Opt Express. 2009;17:3651–3658. doi: 10.1364/oe.17.003651. [DOI] [PubMed] [Google Scholar]
- 24.Nandakumar P, Kovalev A, Volkmer A. Vibrational imaging based on stimulated Raman scattering microscopy. New Journal of Physics. 2009;11:1–9. [Google Scholar]
- 25. Saar BG, Freudiger CW, Reichman J, et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. ‥In this study, the authors have reported video-rate molecular imaging and epi dectetion of scattering light using SRS microscopy. These are crucial for biomedical applications of SRS in mice and human.
- 26.Ashrafi K. Obesity and the regulation of fat metabolism. WormBook. 2007:1–20. doi: 10.1895/wormbook.1.130.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watts JL. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol Metab. 2009;20:58–65. doi: 10.1016/j.tem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watts JL, Browse J. Dietary manipulation implicates lipid signaling in the regulation of germ cell maintenance in C. elegans. Dev Biol. 2006;292:381–392. doi: 10.1016/j.ydbio.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder LK, Kremer S, Kramer MJ, et al. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PLoS One. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang SO, Trimble R, Guo F, Mak HY. Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol. 2010;11:96. doi: 10.1186/1471-2121-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung T, Hohn A, Grune T. Lipofuscin: detection and quantification by microscopic techniques. Methods Mol Biol. 2010;594:173–193. doi: 10.1007/978-1-60761-411-1_13. [DOI] [PubMed] [Google Scholar]
- 36.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogg S, Paradis S, Gottlieb S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 38.Watts JL, Browse J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;272:263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]