Abstract
The phylogenetic diversity of ammonia-oxidizing archaea (AOA) was surveyed in the surface sediments from the northern part of the South China Sea (SCS). The distribution pattern of AOA in the western Pacific was discussed through comparing the SCS with other areas in the western Pacific including Changjiang Estuary and the adjacent East China Sea where high input of anthropogenic nitrogen was evident, the tropical West Pacific Continental Margins close to the Philippines, the deep-sea methane seep sediments in the Okhotsk Sea, the cold deep sea of Northeastern Japan Sea, and the hydrothermal field in the Southern Okinawa Trough. These various environments provide a wide spectrum of physical and chemical conditions for a better understanding of the distribution pattern and diversities of AOA in the western Pacific. Under these different conditions, the distinct community composition between shallow and deep-sea sediments was clearly delineated based on the UniFrac PCoA and Jackknife Environmental Cluster analyses. Phylogenetic analyses showed that a few ammonia-oxidizing archaeal subclades in the marine water column/sediment clade and endemic lineages were indicative phylotypes for some environments. Higher phylogenetic diversity was observed in the Philippines while lower diversity in the hydrothermal vent habitat. Water depth and possibly with other environmental factors could be the main driving forces to shape the phylogenetic diversity of AOA observed, not only in the SCS but also in the whole western Pacific. The multivariate regression tree analysis also supported this observation consistently. Moreover, the functions of current and other climate factors were also discussed in comparison of phylogenetic diversity. The information collectively provides important insights into the ecophysiological requirements of uncultured ammonia-oxidizing archaeal lineages in the western Pacific Ocean.
Electronic supplementary material
The online version of this article (doi:10.1007/s00248-011-9901-0) contains supplementary material, which is available to authorized users.
Introduction
The aerobic microbial ammonia oxidation as the first step of nitrification plays a pivotal role in the global nitrogen cycle because of its wide distribution on the earth not only in the marine ecosystems but also terrestrial environments, carried out by ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) [55, 62]. Since the recognition of AOA which have been proven to be one separated lineage Thaumarchaeota from Crenarchaea lineages [5, 59], this microbial group has been found in a wide variety of habitats including marine water column and sediments [11, 15, 40, 42, 53, 65], the marine sponge [3, 38], soils [21, 30, 45, 54, 69], freshwater and sediments [26, 37, 67], estuaries [6, 14, 43, 51, 52, 65], hydrothermal vents [63] and hot springs [17, 24, 31, 66, 68], activated sludge [58], and groundwater systems [50]. Our knowledge of this type of ammonia oxidizer has expanded rapidly, and more and more studies have been focused on the community structure and abundance of AOA in seawater and sediments all over the world [4, 19]. The wide distribution indicates a likely critical role of them in the N cycle in marine ecosystems. However, our understanding of AOA in the marine sediments is still inadequate.
The ecological features of AOA in the surface sediments of western Pacific Ocean are largely unknown. A few studies have detected this type of microbes in the western Pacific [13, 14, 44, 46]. The quantities and diversity of AOA and AOB amoA genes in the northeastern Japan Sea showed that AOA amoA gene sequences from the water column fell into the Deep Marine Group, while most sequences from pelagic brown sediment were not closely related to any known sequences from the GenBank [44]. The sedimentary AOA diversity, community structure, and spatial distribution in the Changjiang Estuary and the adjacent East China Sea (ECS) shifted along the Changjiang freshwater flow path [14]. It was concluded that the deep-sea sediment of the tropical west Pacific Continental Margin potentially harbors diverse and novel AOA [13]. However, without the information from the largest marginal sea, specifically the South China Sea (SCS), it is difficult to form a general view about AOA. The archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the southern Okinawa Trough were also investigated [46]. These studies in the western Pacific provide an opportunity to combine results from all these studies to form a comprehensive understanding about the ecological distribution pattern of AOA in the western Pacific Ocean.
In the present study, through employing amoA gene as a functional genetic marker, the phylogenetic diversity and the spatial distribution of AOA were surveyed in the northern part of the SCS, which is featured as a deep rhombus-shaped basin in the eastern part and connecting channels, e.g., Luzon Strait, Taiwan Strait, and compared with a selective areas, estuary, continental shelf, slope, deep sea with methane in the western Pacific including Changjiang Estuary and ECS, the deep-sea methane seep sediments in the Okhotsk Sea, the Southern Okinawa Trough, the tropical West Pacific Continental Margins, and the Northeastern Japan Sea to find the dominated lineages and general view about the spatial distributions of AOA in the surface sediments of the western Pacific Ocean. Here, through detections of AOA in the northern SCS and comparison with other ocean ecosystems mentioned above, some general view about distribution of AOA in the western Pacific Ocean sediments might be concluded, and the function of some channels, e.g., Luzon Strait and Taiwan Strait, to shape the AOA community structure could also be discussed.
Materials and Methods
Sampling and Molecular Experiments
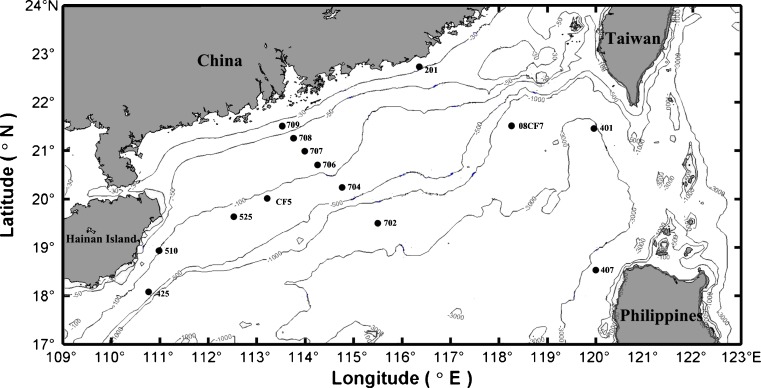
Sampling of sediments was conducted aboard the R/V Shiyan No. 3 during a South China Sea Open Cruise in 2008. A total of seven surface sediment samples were collected from the northern part of the SCS (Fig. 1). Sites 201 and 525 are located in the margin area of the northern part of the SCS and are close to the mainland of China, while site 201 is adjacent to Taiwan Strait. Samples CF5 and 08CF7 were collected from the cold deep sea, site 510 from the upwelling system. Cold deep-sea sites 407 and 425 located in the Luzon strait are close to the Philippines. The environmental physical and chemical parameters of these sites were reported elsewhere [7, 8, 10, 36].
Figure 1.
Sampling map of surface sediments in the northern SCS
One pair of polymerase chain reaction (PCR) primers (Arch-amoAF: 5′-STAATGGTCTGGCTTAGACG-3′ and Arch-amoAR: 5′-GCGGCCATCCATCTGTATGT-3′) [19] was employed to amplify archaeal amoA gene from the total genomic DNA extracted from sediment samples using the SoilMaster DNA Extraction kit (Epicentre Biotechnologies, Madison, WI, USA) as described before [7–10, 36]. Besides the sequences obtained from this current study, other archaeal amoA gene sequences were retrieved from the GenBank based on the published information [8, 13, 14, 44, 46].
Phylogenetic and Statistical Analyses
All of the amoA gene sequences retrieved from the present study were transferred into MEGA 5.0 to edit [60] and combined with the most similar amoA gene sequences downloaded from GenBank including the sequences from other sites in the SCS based on the reference information available [8]. And then the alignment file was constructed using Clustal X 1.81 in MEGA 5.0 [60]. Phylogenetic trees were constructed using neighbor-joining criterion in MEGA 5.0 [60] with 1,000 bootstrap tests for every node. Rarefaction analysis, the nonparametric richness estimator (Chao1 and Shannon), and Simpson diversity index were carried out for the gene sequences retrieved from samples in the SCS using DOTUR, and operational taxonomic units (OTUs) were calculated based on a 5% distance cutoff [57].
Principal coordinates analyses (PCoA) and Jackknife Environment Clusters analyses were conducted in an online software UniFrac (http://bmf2.colorado.edu/unifrac/index.psp) [39] which employs the genetic distances to evaluate the community similarity based on the gene sequences data. The archaeal amoA gene sequences from the SCS surface sediments were classified on the basis of the environmental variables. The environment cluster tree was projected in MEGA 5.0 [60]. Canonical correspondence analysis (CCA) was executed in CANOCO 4.5 to determine the correlations between archaeal amoA gene sequence community compositions and physical and chemical parameters [61]. In addition, Pearson moment correlations were also conducted with Microsoft Excel to relate the physical and chemical factors with the diversity and richness indices of the archaeal amoA genes.
Comparative Analyses between the SCS and Other Habitats in the Western Pacific
Comparisons were made between six different types of habitats, including the SCS coast, SCS deep sea, the Philippine deep sea, ESC and Changjiang Estuary, Northeastern Japan Sea, and Hydrothermal fields. All of the archaeal amoA gene sequences from the habitats mentioned above and those from the present study were combined with the most similar sequences downloaded from GenBank and imported into MEGA 5.0 to align using Clustal X1.81 [60]. The phylogenetic tree was constructed using neighbor-joining criteria to explore the phylogenetic diversity of AOA lineages in the western Pacific Ocean. PCoA and Jackknife Environment Clusters analyses were also employed to conduct community similarity analyses based on the genetic distances and phylogenetic relatedness from all archaeal amoA gene sequences in the researched areas through UniFrac [39]. To assess the sources of variations in the UniFrac matrix, permutational MANOVA based on 1,000 permutations [41] with function adonis in vegan package [47] was used.
To compare the phylogenetic diversity between different habitats, all sequences from the different habitats were clustered and homogenized at 95% identity threshold through DOTUR analyses, ended with six clone libraries affiliated to the six different habitats (Supplementary Table S1). Jackknife Environment Clusters analysis was conducted to compare the community similarity on the basis of six clone libraries data via UniFrac [39]. Phylogenetic diversity (PD) indices for each of the six habitats were calculated as the sum of the branch length associated with the amoA gene sequences within each habitat [18], and the mean PD of 1,000 randomized subsamples of each habitat was calculated to correct for unequal number of sequences [1]. Another index, the phylogenetic species variability (PSV), which quantifies how phylogenetic relatedness decreases the variance of a hypothetical neutral trait, was calculated to estimate the phylogenetic structure with the R package picante [1, 25, 33].
The potential environmental factors that could affect the community structures in the variable surface sediments of the western Pacific were deduced through a Multivariate Regression Tree (MRT) analysis with the R package mvpart [1, 16], based on the observed subclades in the above phylogenetic analysis. Because of the heterogeneity of environmental parameters in different studies, a coding for them was used so as to be comparable (Supplementary Table S1). OTUs as the assemblages were related to the environmental matrix.
Nucleotide Sequences Accession Numbers
The archaeal amoA gene sequences retrieved in this study from the seven surface sediment samples were deposited in GenBank with the accession numbers from JF924520 to JF924865.
Results
Environmental Parameters and the Diversity and Richness of amoA Gene Sequences
The environmental features of the surface sediments in these investigated sites are summarized in Table 1. The seawater depth above sediment ranged from 30 m at site 201 located in the coastal area to 1,900 m at site 407, which is the deep-sea margin. The temperature could be divided into two distinct groups: cold and warm. Within the seven sites, site CF5 exhibited extremely high concentration of inorganic nitrogen, especially NH+4, site 201 showing the second highest. Among these samples from the present study, the higher diversities of amoA gene were shown at sites CF5 and 407, while the lowest one was at site 525 based on a group of indices (Table 1). Within each site clone library, the number of OUT showed high variability from 10 to 27 on the basis of the rarefaction analysis (Fig. S1).
Table 1.
Physicochemical parameters and amoA gene diversity and richness indices from the surface sediments in the northern SCS
| Sample name | Sampling position | Seawater depth (m) | Temperature (°C) | NH+4 (μmol/L) | NO−2 (μmol/L) | NO−2 + NO−3 (μmol/L) | Clone numbers | OTUs (95%) | Diversity and richness | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Simpson | Shannon | Chao1 | |||||||||
| 407 | 18°29N/120°00E | 1,900 | 2–4 | 16.50 | 0.37 | 9.2 | 68 | 26 | 0.13 | 2.72 | 34.8 |
| 525 | 19°23N/114°36E | 1,100 | 2–4 | 25.67 | 1.06 | 13.3 | 46 | 10 | 0.18 | 1.86 | 12.0 |
| CF5 | 19°55N/115°13E | 1,153 | 2–4 | 275.18 | 0.68 | 259.7 | 59 | 27 | 0.05 | 2.97 | 40.0 |
| 08CF7 | 22°70N/119°17E | 1,301 | 2–4 | 42.24 | 2.70 | 17.1 | 57 | 22 | 0.07 | 2.76 | 38.5 |
| 425 | 18°31N/110°59E | 1,470 | 2–4 | 38.79 | 0.40 | 16.5 | 42 | 16 | 0.08 | 2.48 | 25.3 |
| 510 | 19°30N/110°46E | 76 | 19.8 | 43.11 | 0.38 | 10.3 | 42 | 18 | 0.07 | 2.64 | 22.7 |
| 201 | 21°41N/116°18E | 30 | 21.6 | 137.03 | 0.59 | 39.9 | 32 | 17 | 0.06 | 2.64 | 23.5 |
Phylogenetic Analysis of Archaeal amoA Gene in the SCS Surface Sediments
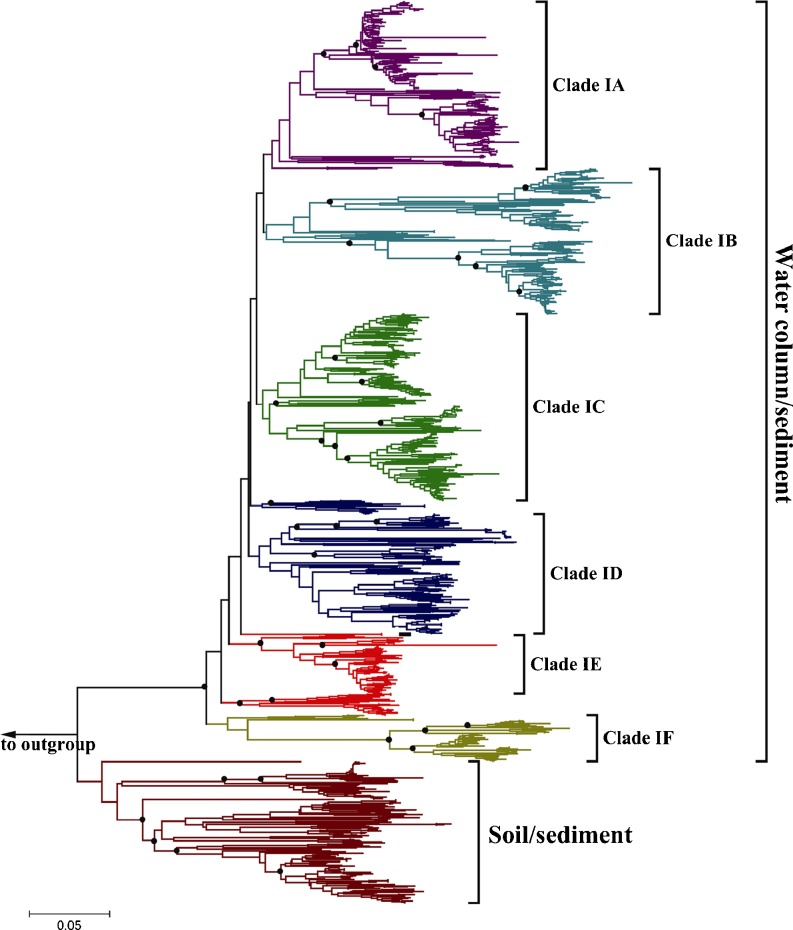
Based on the phylogenetic analysis on all the sequences from the surface sediments in the western Pacific and the sequences from one former study in the northern SCS (Figs. 2 and S3), a comparatively slim soil/sediment clade was evident, while most sequences clustered into the marine water column/sediment clade. The sequences in the soil/sediment clade were from sites 407 (ca. 7), 525 (ca. 17), 201 (ca. 3), and 08CF7 (ca. 1), and they shared higher similarity with those mostly from the tropical West Pacific Continental Margin sediment [13] and some retrieved from ESC [14] or the Bahia del Tobari Mexico Gulf [19] (Fig. S3).
Figure 2.
Phylogenetic tree based on archaeal amoA gene sequences from the variable surface sediments in the western Pacific and most similar sequences using the neighbor-joining criterion. Filled circles at the nodes represent the significant bootstrap values (>50) through 500 resampling test. Branch lengths correspond to sequence differences as indicated by the scale bar
In the marine water column/sediment clade, most of the sequences could be grouped with the closest lineages. Six subclades were identified in the present phylogenetic tree based on the phylogenetic analyses of all the sequences in the western Pacific and most similar sequences as references (Figs. 2 and S3). Clade IA was mostly constituted with sequences from the northern SCS surface sediments and also included some sequences from other environments. In Clade IB, sequences were mostly retrieved from the shallow surface sediments of the northern SCS and ESC [14]. A few sequences from the deep-sea site CF5 and some from the San Francisco Bay and Bahia del Tobari Mexico Gulf [19] were also included in this clade. Clades IC and Clade ID were mixture clades including sequences shared by the deep-sea and shallow marine surface sediments. Clade IE also consisted of sequences from the deep-sea sediments, the tropical West Pacific Continental Margin sediment [13], Okhotsk cold deep-sea sediments [15], a few sequences from the hydrothermal vents sediments in Southern Okinawa trough [46], and Juan de Fuca field hydrothermal vents [63]. Most sequences in Clade IF were retrieved from the hydrothermal vents and deep-sea sediments and rooted in the marine water column/sediment clade.
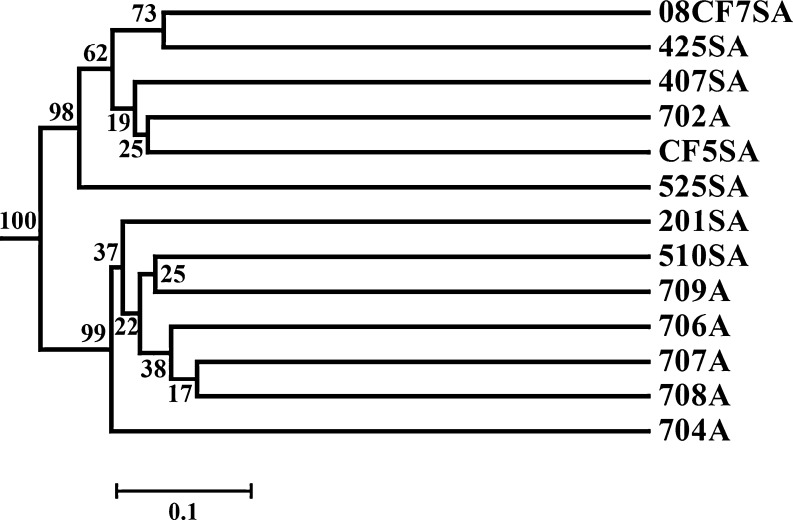
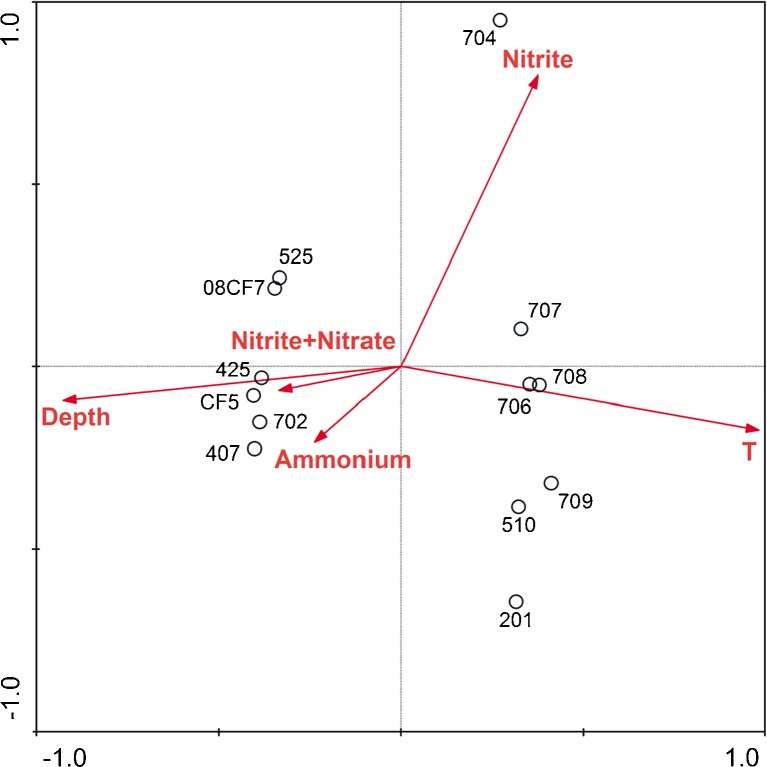
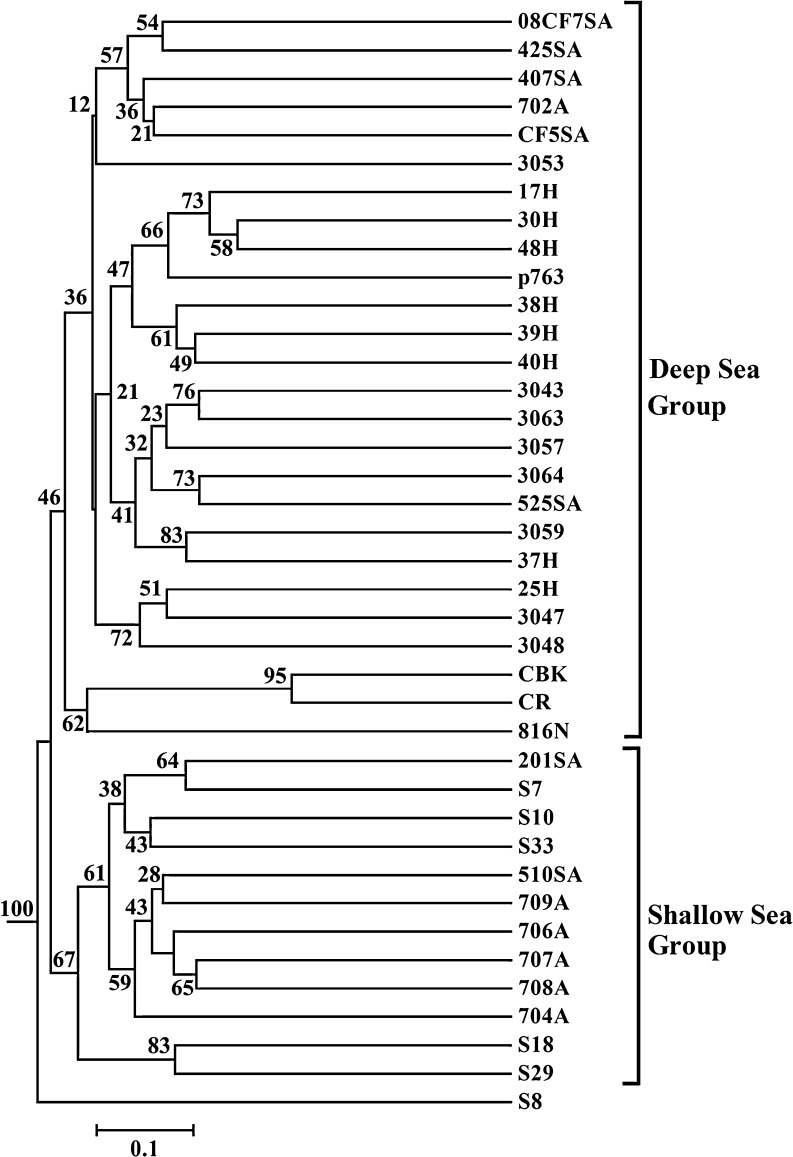
Two groups were identified in the UniFrac analyses (Figs. 3 and S4). All the shallow samples were clustered together and separated with another group containing all the samples from the deep-sea sediments. In each group, the lower statistical support indicated that the similarity between the samples within the same group was higher. These two groups supported that depth and the resulting temperature might be the important factors to shape the distinct AOA lineages. Although no single factor was distinguished to determine the diversity of archaeal amoA gene in the Pearson moment analyses (Table S2), CCA could compensate for the lack of information in the Pearson moment analyses. Depth and temperature were proposed as the main contributors for the diversity of archaeal amoA gene (Fig. 4). The concentration of inorganic nitrogen contributed to the distribution of the deep-sea sediments, especially for 407, 702, 425, and CF5, while the separation of 704 was a result of the concentration of nitrite (Fig. 4).
Figure 3.
Hierarchical clustering analysis (UPGMA algorithm with Jackknife supporting values) for the archaeal amoA gene sequences from the whole northern SCS based on the online UniFrac software
Figure 4.
CCA ordination plots for the physicochemical parameters and the ammonia oxidizing archaeal lineages represented by amoA gene sequences
Community of AOA Based on Phylogenetic Diversity in the Western Pacific
Samples from 43 archaeal amoA gene clone libraries were clustered on the basis of the phylogenetic community similarity (Figs. 5 and S5). Shallow marine sediments were clearly separated from the deep-sea and hydrothermal vent sediments (Figs. 5 and S5), and depth was the strongest and the only significant environmental factor. Although the statistical support was not strong enough, two groups were present in the UniFrac analyses as in the former analyses constrained to the SCS (Figs. 5 and S5). All the shallow samples including SCS and ESC grouped together, while all the samples from the SCS clustered together, and ESC samples divided into two sub-groups (Figs. 5 and S5). Sample 201 shared higher similarity with those from ESC than with others from SCS. All the deep-sea sediment samples were separated from the shallow samples and formed another group. The deep-sea samples from the SCS shared higher similarity with those from the Philippine deep sea than Northern Japan Sea and Okhotsk Sea (Figs. 5 and S5). These two groups as shown in the UniFrac analyses of the SCS were also proposed to be affected by depth and the resulting temperature. A MRT analysis was carried out in order to link the abundance of the lineages to environmental data. The analysis showed a two-leaf tree ordination primarily based on their origin environments, deep sea vs. shallow water (Fig. S6).
Figure 5.
Hierarchical clustering analysis (UPGMA algorithm with 100 replicates Jackknife supporting test) for the all archaeal amoA gene sequences from the western Pacific represented every clone library according to the online UniFrac software
Rarefaction curves (Fig. S2) and diversity indices were determined for the six types of habitats (Fig. 6). PD values were higher in the Philippine deep-sea sediments and the ESC, whereas hydrothermal vents hold the lowest PD value (Fig. 6). Particularly, the Philippine deep sea and the ESC showed the highest PSV values (that is, more over-dispersed), whereas SCS coast, hydrothermal vent, and Okhotsk Sea with the lowest (that is, more phylogenetically clustered).
Figure 6.
Hierarchical clustering analysis (UPGMA algorithm with 100 replicates Jackknife supporting test) for the all amoA gene sequences from the western Pacific represented six different habitats calculated by the online UniFrac software. The number of sequence (n), number of libraries (Nlib), phylogenetic diversity with SD (PD ± SD), and phylogenetic species variability (PSV) in each habitat are given. SD for PSV index was 0.001 for all habitats
Discussion
Molecular Ecological Pattern of AOA in the Northern SCS
The surface sediments of coastal, deep sea, upwelling, and estuarine were used to delineate the ecological pattern of AOA lineages so as to gain a comprehensive understanding about this newly known ammonia-oxidizing prokaryote in the SCS. A few features could be derived from the present analyses. Firstly, the soil/sediment clade lineages were absent in most samples from the SCS surface sediments except for a few clones retrieved from sites 201, 407, 08CF7, and 525, but site 525 contributed to the most clones (ca. 17), indicating this site was mostly affected by terrestrial influence. Similar to reported studies, only lineages related to the marine water column/sediment clade, but not the soil/sediment clade related lineages, was detected at deep-sea hydrothermal fields of the Southern Mariana Trough [32], or a small number of the marine water column/sediment clade-related lineages was present in the present study. This could be resulted from the fact that these samples without soil/sediment clade lineages were not affected by the terrestrial influence. In contrast, samples 201, 407, and 525 could be affected by the terrestrial influence as the soil/sediment clade of AOA was detected. Another possibility is the bias of the PCR primers used which might not be efficient enough to amplify all AOA lineages inhabiting in this environment because of the lower similarity between the potential soil/sediment lineages and the known sequences deposited in the GenBank, which were used to design the primers in this study. Improvement in sensitivity of detection methods could resolve this problem including the use of pyrosequencing [35] or new genetic markers, e.g., nirK (nitrite reductases) [2] and hcd (putative 4-hydroxybutyrate-CoA dehydratase) genes [48, 69].
In the study areas, based on the UniFrac and CCA analyses, two psychrophile and mesophile groups could be distinguished. Water depth and the associated temperature might be potential important factors to drive the distribution patterns of these two large groups of AOA. In the marine water column/sediment clade, most of the sequences could be grouped with their closest lineages in the phylogenetic tree. Six subclades were identified in the present phylogenetic tree based on the analyses of all sequences in the western Pacific and closest sequences from other environments. Except for the Clade 1F, the AOA lineages were broadly distributed into all other clades in the marine water column/sediment clade indicating the marine features. Both psychrophile and mesophile detected in this study could be grouped with the relatives distributed in other sediments of the Pacific, indicating some geographical factors could be responsible for this similarity [13, 14]. For example, in Clade IB, the sequences were mostly retrieved from the shallow surface sediments in the northern SCS and ESC [14]. Some endemic lineages were also present in the SCS, e.g., some lineages in Clade IA indicating the geographical distance resulting in the observed differences from other environments. However, no sequence from the SCS was affiliated with the base subclade (Clade 1F), which included sequences retrieved from the hydrothermal vents in the marine water column/sediment clade. It seems that the different geographical distribution of the AOA lineages in the SCS may be related to the history of the environments.
General Features of AOA in the Western Pacific
Though studies have been carried out to explore the diversity or the abundance of AOA in the western Pacific [7, 8, 13–15, 32, 44], the scattered information could not provide a concrete comprehensive view about this newly revealed group of microorganism. For the first time, results from different studies in the western Pacific were gathered and analyzed to discuss the molecular ecological patterns of the AOA, and this could provide a model to investigate the role of AOA in the marine ecosystem.
As in the analyses of sequences from the SCS, the soil/sediment clade was slimly present in the whole phylogenetic tree. The highest phylogenetic diversities of AOA were found in the Philippine deep-sea sediments and the ESC, which were affected by the terrestrial discharges, e.g., the Changjiang Estuary and the Philippines Island. Previous studies indicated that the AOA communities in terrestrial environments are distinct from those in marine environments [49], whereas other studies indicated that estuaries and coasts might harbor mixed populations of AOA from soil and sediment sources [4, 14]. The deposition of archaea from terrestrial source could potentially explain the existence of the putative terrestrial or estuarine AOA-related sequences in some of the deep-sea sites [13]. The lowest PD values of hydrothermal vents sediments suggest that the phylotypes were constrained and were mostly endemic or thermophilic (Fig. 6). The hydrothermal vent environments were grouped into the deep-sea sediments group, indicating that the ammonia-oxidizing archaeal lineages exhibited higher deep-sea features than the thermophilic feature.
A number of factors could be responsible for shaping the diversity and distribution patterns of AOA. Firstly, sedimentological conditions including pH values, temperature, nutrient, organic matter contents, O2 concentrations, and pore-water redox may play an important role affecting the ecological distribution patterns [13–15, 29, 40]. However, as the environmental parameters were highly variable in different studies, it was impossible to carry out the same multivariate statistic analysis on all of them. One MRT analyses was employed, and the water depth was singled out to be a potential important factor. On the other hand, currents, tides, upwelling, hydrothermal vents, water mixture, and the intensity and dynamics of these activities may also influence the sediment AOA assemblages [15]. In this area, current in the Taiwan Strait and the Philippines should not be ignored [13]. However, the relationships between AOA communities in the marine surface sediments and the ecological conditions still need further confirmation. About the thermophilic AOA [17, 24, 32, 68] and the comprehensive review about AOA [49], the basic position of AOA in the whole phylogeny and the marine water column/sediment clade indicates a thermophilic evolutionary history of AOA, and this could shed light on the evolutionary study of ammonia oxidation. Otherwise, some geothermal events, for example, volcanoes, hydrothermal vents, or hot geofluids from deep subsurface biosphere or oil reservoirs, might be responsible for some dispersal of thermophilic microorganisms to the deep-sea surface sediments [27, 28].
Shallow Marine AOA in the Western Pacific
In the AOA community cluster analysis, the marine surface sediment cluster of shallow water was grouped together and in the shallow marine groups and the SCS coastal marine samples shared high similarity with some from the ESC than with other deep-sea sediment samples. These two habitats with shallow water shared some lineages as shown in the phylogenetic analysis, e.g., Clade 1A (Figs. 2 and S3). In a former study, the current influence from the Taiwan Strait was not strong [14], but the UniFrac analysis indicated that site 201 close to the Taiwan Strait was grouped together with samples from the ESC, indicating the higher phylogenetic similarity between them (Figs. 5 and S5). One possible explanation was that the Taiwan Strait current might be responsible for the similarity between site 201 and the ESC samples through carrying them into the mixing zone between them or the close geographical distance between them resulting in such phylogenetic similarity.
The continental margin is the most important interface between the terrestrial and marine environments, especially in the cycling of nutrients [19]. SCS is the largest marginal sea in the western Pacific with a deep section named China Sea Basin, which is surrounded by broad continental shelves less than 100 m in depth and the margin areas from an important interface of the terrestrial and marine environments. The AOA community at this area might be a mixed population of both soil/sediment clade, and marine water column/sediment clade. However, only site 201 showed the soil/sediment-related AOA resulting from the long distance transport from the shore or estuaries of other shallow sites in the SCS. On the other hand, the ECS close to the Changjiang estuary mixing zone harbored a distinct AOA community including the soil/sediment-related AOA lineages [14]. The distribution of AOA communities correlated significantly with the gradients of surface water salinity and sediment sorting coefficient. The spatial distribution of putative soil-related AOA in certain sampling stations indicated a strong impact of Changjiang freshwater discharge on the marine benthic microbial ecosystem [14]. Besides the freshwater, nutrients, organic matter, and suspended particles, the Changjiang water might also contribute to the transport of terrestrial source archaea into the marine water column and sediments in the mixing zone along its flow path [14]. This could explain the existence of the putative soil-related AOA lineages in the coastal sedimentary environments.
Deep-cold Sea Lineages in the Western Pacific
AOA may contribute to the deep-sea nitrate reservoirs [34], but their ecological features were largely undetected for most of the deep-sea sediments of the oceans [15]. A few deep-sea sediments were compared in this study to understand the ecological pattern of psychrophilic AOA including the SCS deep sea, the Philippine deep sea, and the Northern Japan deep sea (Okhotsk sea and another one in Northern Japan Sea). The hydrothermal vents samples were buried into the deep-sea group (Fig. 6). Recent studies also showed that diverse and unique AOA lineages could inhabit in the continental margin deep-sea sediments [13, 44]. By comparison, AOA phylogenetic diversity was higher in the Philippines cold deep-sea sediments than that of the hypernutrified subtropical Bahía del Tóbari Estuary [4], and similar to the temperate Changjiang estuary and its adjacent East China Sea (Fig. 6), indicating in the tropical West Pacific Continental Margin, the stations in the Philippines inland seas (including station 3043) contained AOA of various terrestrial sources, whereas the other stations connected directly to the open Philippine Sea showed AOA dominated by marine origin [13, 14]. Ocean current may have a significant impact on the regional marine ecosystem, as the Philippine Sea is a marginal sea of the West Pacific and the Kuroshio Current, the largest western boundary current of the Pacific Ocean [13]. The importance of climatic events in the transport of terrestrial microorganisms to the deep-sea sedimentary environments is almost totally neglected previously. However, the present study show that most AOA lineages have their closest relatives in the northern SCS, indicating that these lineages could be the endemic ones found in the western Pacific, not only in the Philippines alone.
Okhotsk Sea represents of another type of deep-sea sediments, which is a marginal sea located on the continental slope offshore Sakhalin Island in the northwestern Pacific Ocean and is one of the largest reservoir of methane hydrates in the world [20]. Although the cold seep sediments may have a limited O2 supply, archaeal amoA genes were detected with lower diversity and abundance [15]. Through the present comparative analysis, the diversity in this area was similar to the SCS deep-sea sediments and higher than the hydrothermal vent sediments (Fig. 6). In a former study, AOA lineages from this area were affiliated to the GenBank sequences originally obtained from deep-sea hydrothermal vents and were proposed that the related AOA lineages either have a wide range of temperature adaption or they have a thermophilic evolutionary history in the modern cold deep-sea sediments of the Okhotsk Sea [15]. This is consistent with the current study in which the samples from the hydrothermal vent sediments were clustered with the other deep-sea habitats (Fig. 6). As in the former study, most of the amoA sequences shared highest similarity with those retrieved from these western Pacific areas in the present study [15], especially the sequences from the Philippine deep-sea sediments sequences (68.3%) [13], consistent with the present study (Fig. 6). Though a few studies have explored the ecology of AOA in the deep-sea sediments, the physiology of AOA is less known in the O2-limited environments, considering they are better known as the aerobic AOA [12, 13, 15, 32, 34, 44, 63]. Another point is that the hydroxylamine oxidoreductase encoding hao gene responsible for the ammonia oxidation step is absent in the genome of AOA [22, 23, 64], but the nitrite reductase gene nirK has been identified widely in AOA [2, 55]. Based on such information, recent available genomic data of Nitrosopumilus maritimus indicated that nitroxyl could also serve as electron acceptor in a similar way as O2 to compensate for the less availability which differs from the HAO employed in AOB [56, 64]. Additionally, a few endemic psychrophilic or O2-repressed AOA lineages have been found, especially from the Northern Japan deep sea showing larger genetic distance with others from the water column/sediment clade, indicating the psychrophiles could adapt to these habitats and could provide evolutionary significance through adaptive evolutionary analysis in future.
Conclusions
The marine water/sediment dominant distribution and a few habitat-specific AOA subclades were identified in the AOA clade of the phylogenetic tree. The distinctness of community composition between shallow and deep-sea sediment was observed, indicating the water depth and associated temperature could be the main factors resulting in the present phylogenetic diversity of AOA. Higher phylogenetic diversity was observed in the Philippines while lower diversity in the hydrothermal vent habitat. Additionally, current and other climate factors could also contribute to the AOA distribution patterns based on comparison of phylogenetic diversity. The current research provides a comprehensive understanding the AOA in the western Pacific and set an example for the study of the AOA in the whole marine environments.
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
Statistical analysis of diversity and abundance (DOC 39 kb)
Statistical analysis of physicochemical parameters and the diversity and abundance of nitrifying prokaryotes (DOC 229 kb)
Rarefaction curves of archaeal amoA gene sequences retrieved from the northern SCS on the basis of 5% distance cut-off calculated from DOTUR software (DOC 499 kb)
Rarefaction curves of archaeal amoA gene sequences retrieved from the western Pacific on the basis of 5% distance cut-off calculated from DOTUR software (DOC 167 kb)
Principal coordinate analysis (PCoA) (a) for the archaeal amoA gene sequences from the whole northern SCS based on the online UniFrac software (DOC 145 kb)
Principal coordinate analysis (PCoA) for the all archaeal amoA gene sequences from the western Pacific represent of every clone library according to the online UniFrac software (DOC 336 kb)
Multivariate regression tree (MRT) analysis of the interaction between archaeal amoA lineage abundance (in term of sequence number) and environmental parameters. Columns under each leaf represent the mean of normalized AOA lineage abundance for each lineage significantly correlated with environmental parameters (DOC 144 kb)
Acknowledgements
This research was supported in part by a Ph.D. studentship (H-LC) from The University of Hong Kong and in part by grants from Agriculture, Fisheries and Conservation Department of the Hong Kong Government (J-DG), and National Natural Science Foundation of China (41076095), Knowledge Innovation Key Project of the Chinese Academy of Sciences (KZCX2-YW-QN207) (Y-GH). We would like to thank Ms. Jessie Lai and Dr. Zhen-ye Zhao for the laboratory assistance at The University of Hong Kong and Dr. Jean-Christophe Auguet from Centre d'Estudis Avançats de Blanes, CEAB-CSIC, Accés Cala Sant Francesc, Girona, Spain to help calculate the mean PD values.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Auguet JC, Barberan A, Casamayor EO. Global ecological patterns in uncultured archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- 2.Bartossek R, Nicol GW, Lanzen A, Klenk HP, Schleper C. Homologues of nitrite reductases in ammonia-oxidizing archaea: diversity and genomic context. Environ Microbiol. 2010;12:1075–1088. doi: 10.1111/j.1462-2920.2010.02153.x. [DOI] [PubMed] [Google Scholar]
- 3.Bayer K, Schmitt S, Hentschel U. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ Microbiol. 2008;10:2942–2955. doi: 10.1111/j.1462-2920.2008.01582.x. [DOI] [PubMed] [Google Scholar]
- 4.Beman JM, Francis CA. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol. 2006;72:7767–7777. doi: 10.1128/AEM.00946-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 6.Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 2007;1:660–662. doi: 10.1038/ismej.2007.79. [DOI] [PubMed] [Google Scholar]
- 7.Cao H, Hong Y, Li M, Gu J-D (2011) Lower abundance of ammonia-oxidizing archaea than ammonia-oxidizing bacteria detected in the subsurface sediments of the Northern South China Sea. Geomicrobiol J (in press)
- 8.Cao H, Hong Y, Li M, Gu J-D (2011) Diversity and abundance of ammonia-oxidizing prokaryotes in sediments from the coastal margin to the South China Sea. Antonie van Leeuwenhoek. doi:10.1007/s10482-011-9610-1 [DOI] [PMC free article] [PubMed]
- 9.Cao H, Li M, Dang H, Gu JD. Responses of aerobic and anaerobic ammonia/ammonium oxidizing microorganisms to anthropogenic pollution in coastal marine environments. In: Klotz MG, Stein LY, editors. Methods in enzymology, part B: research on nitrification and related processes. San Diego, California: Academic Press; 2011. pp. 35–62. [DOI] [PubMed] [Google Scholar]
- 10.Cao H, Li M, Hong Y, Gu J-D (2011) Diversity and abundance of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in polluted mangrove sediment. Syst Appl Microbiol. doi:10.1016/j.syapm.2010.11.023 [DOI] [PubMed]
- 11.Church MJ, Wai B, Karl DM, DeLong EF. Abundance of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coolen MJ, Abbas B, van Bleijswijk J, Hopmans EC, Kuypers MM, Wakeham SG, Sinninghe Damste JS. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol. 2007;9:1001–1016. doi: 10.1111/j.1462-2920.2006.01227.x. [DOI] [PubMed] [Google Scholar]
- 13.Dang H, Li J, Zhang X, Li T, Tian F, Jin W. Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J Appl Microbiol. 2009;106:1482–1493. doi: 10.1111/j.1365-2672.2008.04109.x. [DOI] [PubMed] [Google Scholar]
- 14.Dang H, Zhang X, Sun J, Li T, Zhang Z, Yang G. Diversity and spatial distribution of sediment ammonia-oxidizing Crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiology. 2008;154:2084–2095. doi: 10.1099/mic.0.2007/013581-0. [DOI] [PubMed] [Google Scholar]
- 15.Dang H, Luan XW, Chen R, Zhang X, Guo L, Klotz MG. Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol Ecol. 2010;72:370–385. doi: 10.1111/j.1574-6941.2010.00870.x. [DOI] [PubMed] [Google Scholar]
- 16.De'Ath G. Multivariate regression trees: a new technique for modeling species-environment relationships. Ecology. 2002;83:1105–1117. [Google Scholar]
- 17.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 18.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 19.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginsburg GD, Soloviev VA, Cranston RE, Lorenson TD, Kvenvolden KA. Gas hydrates from the continental slope, offshore Sakhalin Island, Okhotsk Sea. Geo-Mar Lett. 1993;13:41–48. doi: 10.1007/BF01204391. [DOI] [Google Scholar]
- 21.Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 22.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, Preston C, de la Torre J, Richardson PM, DeLong EF. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–21349. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmus MR, Bland TJ, Williams CK, Ives AR. Phylogenetic measures of biodiversity. Am Nat. 2007;169:68–83. doi: 10.1086/511334. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann M, Saunders AM, Schramm A. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl Environ Microbiol. 2009;75:3127–3136. doi: 10.1128/AEM.02806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubert C, Loy A, Nickel M, Arnosti C, Baranyi C, Brüchert V, Ferdelman T, Finster K, Christensen FM, Rezende JR, Vandieken V, Jørgensen BB. A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science. 2009;325:1541–1544. doi: 10.1126/science.1174012. [DOI] [PubMed] [Google Scholar]
- 28.Inagaki F, Takai K, Komatsu T, Kanamatsu T, Fujioka K, Horikoshi K. Archaeology of Archaea: geomicrobiological record of Pleistocene thermal events concealed in a deep-sea subseafloor environment. Extremophiles. 2001;5:385–392. doi: 10.1007/s007920100211. [DOI] [PubMed] [Google Scholar]
- 29.Jackson CR, Weeks AQ. Influence of particle size on bacterial community structure in aquatic sediments as revealed by 16S rRNA gene sequence analysis. Appl Environ Microb. 2008;74:5237–5240. doi: 10.1128/AEM.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia Z, Conrad R. Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 31.Jiang H, Huang Q, Dong H, Wang P, Wang F, Li W, Zhang C. RNA-based investigation of ammonia-oxidizing archaea in hot springs of Yunnan Province, China. Appl Environ Microbiol. 2010;76:4538–4541. doi: 10.1128/AEM.00143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato S, Kobayashi C, Kakegawa T, Yamagishi A. Microbial communities in iron-silica-rich microbial mats at deep-sea hydrothermal fields of the Southern Mariana Trough. Environ Microbiol. 2009;11:2094–2111. doi: 10.1111/j.1462-2920.2009.01930.x. [DOI] [PubMed] [Google Scholar]
- 33.Kembel S, Ackerly D, Blomberg S, Cowan P, Helmus M, Webb C (2008) Picante: tools for integrating phylogenies and ecology. Version 0.4.0. Available at http://picante.r-forge.r-project.org/ [DOI] [PubMed]
- 34.Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MM. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee OO, Wang Y, Yang J, Lafi FF, Al-Suwailem A, Qian PY. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J. 2011;5:650–664. doi: 10.1038/ismej.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Cao H, Hong Y, Gu J-D. Spatial distribution and abundance of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol. 2011;89:1243–1254. doi: 10.1007/s00253-010-2929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Huang S, Sun G, Xu Z, Xu M (2011) Diversity and abundance of ammonia-oxidizing archaea in the Dongjiang River, China. Microbiol Res. doi:10.1016/j.micres.2010.08.002 [DOI] [PubMed]
- 38.López-Legentil S, Erwin PM, Pawlik JR, Song B. Effects of sponge bleaching on ammonia-oxidizing archaea: distribution and relative expression of ammonia monooxygenase genes associated with the barrel sponge Xestospongia muta. Microb Ecol. 2010;60:561–571. doi: 10.1007/s00248-010-9662-1. [DOI] [PubMed] [Google Scholar]
- 39.Lozupone C, Hamady M, Knight R. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinforma. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 41.McArdle B. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. doi: 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2. [DOI] [Google Scholar]
- 42.Molina V, Belmar L, Ulloa O. High diversity of ammonia-oxidizing archaea in permanent and seasonal oxygen-deficient waters of the eastern South Pacific. Environ Microbiol. 2010;12:2450–2465. doi: 10.1111/j.1462-2920.2010.02218.x. [DOI] [PubMed] [Google Scholar]
- 43.Mosier AC, Francis CA. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T, Mori K, Kato C, Takahashi R, Tokuyama T. Distribution of cold-adapted ammonia-oxidizing microorganisms in the deep-ocean of the northeastern Japan Sea. Microbes Environ. 2007;22:365–372. doi: 10.1264/jsme2.22.365. [DOI] [Google Scholar]
- 45.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 46.Nunoura T, Oida H, Nakaseama M, Kosaka A, Ohkubo SB, Kikuchi T, Kazama H, Hosoi-Tanabe S, Nakamura K, Kinoshita M, Hirayama H, Inagaki F, Tsunogai U, Ishibashi J, Takai K. Archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the Southern Okinawa trough. Appl Environ Microbiol. 2010;76:1198–11211. doi: 10.1128/AEM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oksanen J, Kindt R, Legendre P, O'hara B, Simpson GL, Stevens MHH (2008) Vegan: community ecology package. Version 1.11.14. Available at http://vegan.r-forge.r-project.org
- 48.Offre P, Nicol GW, Prosser JI (2011) Community profiling and quantification of putative autotrophic thaumarchaeal communities in environmental samples. Environ Microbiol Reports. doi:10.1111/j.1758-2229.2010.00217.x [DOI] [PubMed]
- 49.Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- 50.Rogers DR, Casciotti KL. Abundance and diversity of archaeal ammonia oxidizers in a coastal groundwater system. Appl Environ Microbiol. 2010;76:7938–7948. doi: 10.1128/AEM.02056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahan E, Muyzer G. Diversity and spatio-temporal distribution of ammonia-oxidizing archaea and bacteria in sediments of the Westerschelde estuary. FEMS Microbiol Ecol. 2008;64:175–186. doi: 10.1111/j.1574-6941.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 52.Santoro AE, Francis CA, de Sieyes NR, Boehm AB. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol. 2008;10:1068–1079. doi: 10.1111/j.1462-2920.2007.01547.x. [DOI] [PubMed] [Google Scholar]
- 53.Santoro AE, Casciotti KL, Francis CA. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- 54.Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, Sharma S, Wilke BM, Matthies M, Smalla K, Munch JC, Amelung W, Kaupenjohann M, Schloter M, Schleper C. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol. 2009;11:446–456. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- 55.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 56.Schleper C, Nicol GW. Ammonia-oxidising archaea-physiology, ecology and evolution. Adv Microb Physiol. 2010;57:1–41. doi: 10.1016/B978-0-12-381045-8.00001-1. [DOI] [PubMed] [Google Scholar]
- 57.Schloss PD, Handelsman J. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl Environ Microbiol. 2006;72:6773–6779. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonthiphand P, Limpiyakorn T. Change in ammonia-oxidizing microorganisms in enriched nitrifying activated sludge. Appl Microbiol Biotechnol. 2011;89:843–853. doi: 10.1007/s00253-010-2902-y. [DOI] [PubMed] [Google Scholar]
- 59.Spang A, Hatzenpichler R, Brochler-Armanet C, Rattei T, Tischler P, Spieck E, Streit W, Stahl DA, Wagner M, Schleper C. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using likelihood, distance, and parsimony methods. Mol Biol Evol. doi:10.1093/molbev/msr113 [DOI] [PMC free article] [PubMed]
- 61.ter Braak CJF, Smilauer P. CANOCO reference manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (Version 4.5) Ithaca: Microcomputer Power; 2002. [Google Scholar]
- 62.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Xiao X, Jiang L, Peng X, Zhou H, Meng J, Wang F. Diversity and abundance of ammonia-oxidizing archaea in hydrothermal vent chimneys, Juan de Fuca Ridge. Appl Environ Microbiol. 2009;75:4216–4220. doi: 10.1128/AEM.01761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PS, Chan PP, Gollabgir A, Hemp J, Hügler M, Karr EA, Könneke M, Shin M, Lawton TJ, Lowe T, Martens-Habbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine Crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wankel SD, Mosier AC, Hansel CM, Paytan A, Francis CA. Spatial variability in nitrification rates and ammonia-oxidizing microbial communities in the agriculturally-impacted Elkhorn Slough Estuary. Appl Environ Microbiol. 2011;77:269–280. doi: 10.1128/AEM.01318-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weidler GW, Gerbl FW, Stan-Lotter H. Crenarchaeota and their role in the nitrogen cycle in a subsurface radioactive thermal spring in the Austrian Central Alps. Appl Environ Microbiol. 2008;74:5934–5942. doi: 10.1128/AEM.02602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Y, Xiang Y, Wang J, Zhong J, He J, Wu Q. Heterogeneity of archaeal and bacterial ammonia-oxidizing communities in Lake Taihu, China. Environ Microbiol Reports. 2010;2:569–576. doi: 10.1111/j.1758-2229.2010.00146.x. [DOI] [PubMed] [Google Scholar]
- 68.Zhang CL, Ye Q, Huang Z, Li W, Chen J, Song Z, Zhao W, Bagwell C, Inskeep WP, Ross C, Gao L, Wiegel J, Romanek CS, Shock EL, Hedlund BP. Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol. 2008;74:6417–6426. doi: 10.1128/AEM.00843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci USA. 2010;107:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analysis of diversity and abundance (DOC 39 kb)
Statistical analysis of physicochemical parameters and the diversity and abundance of nitrifying prokaryotes (DOC 229 kb)
Rarefaction curves of archaeal amoA gene sequences retrieved from the northern SCS on the basis of 5% distance cut-off calculated from DOTUR software (DOC 499 kb)
Rarefaction curves of archaeal amoA gene sequences retrieved from the western Pacific on the basis of 5% distance cut-off calculated from DOTUR software (DOC 167 kb)
Principal coordinate analysis (PCoA) (a) for the archaeal amoA gene sequences from the whole northern SCS based on the online UniFrac software (DOC 145 kb)
Principal coordinate analysis (PCoA) for the all archaeal amoA gene sequences from the western Pacific represent of every clone library according to the online UniFrac software (DOC 336 kb)
Multivariate regression tree (MRT) analysis of the interaction between archaeal amoA lineage abundance (in term of sequence number) and environmental parameters. Columns under each leaf represent the mean of normalized AOA lineage abundance for each lineage significantly correlated with environmental parameters (DOC 144 kb)