Abstract
Shiga toxins produced by E. coli O157:H7 are responsible for food poisoning and hemolytic uremic syndrome (HUS). The A subunits of Shiga toxins (Stx1A and Stx2A) inhibit translation by depurinating a specific adenine in the large rRNA. To determine if Stx1A and Stx2A require the ribosomal stalk for depurination, their activity and cytotoxicity were examined in the yeast P protein deletion mutants. Stx1A and Stx2A were less toxic and depurinated ribosomes less in a strain lacking P1/P2 on the ribosome and in the cytosol (ΔP2) than in a strain lacking P1/P2 on the ribosome, but containing free P2 in the cytosol (ΔP1). To determine if cytoplasmic P proteins facilitated depurination, Stx1A and Stx2A were expressed in the P0ΔAB mutant, in which the binding sites for P1/P2 were deleted on the ribosome, and P1/P2 accumulated in the cytosol. Stx1A was less toxic and depurinated ribosomes less in P0ΔAB, suggesting that intact binding sites for P1/P2 were critical. In contrast, Stx2A was toxic and depurinated ribosomes in P0ΔAB as in wild type, suggesting that it did not require the P1/P2 binding sites. Depurination of ΔP1, but not P0ΔAB ribosomes increased upon addition of purified P1α/P2β in vitro, and the increase was greater for Stx1 than for Stx2. We conclude that cytoplasmic P proteins stimulate depurination by Stx1 by facilitating the access of the toxin to the ribosome. Although ribosomal stalk is important for Stx1 and Stx2 to depurinate the ribosome, Stx2 is less dependent on the stalk proteins for activity than Stx1 and can depurinate ribosomes with an incomplete stalk better than Stx1.
Keywords: Shiga toxin, E. coli O157:H7, ribosome inactivating protein, ribosomal stalk, P protein, ricin
1. Introduction
Shiga toxin (Stx) producing E. coli (STEC), such as E. coli O157:H7 and other serotypes are the major causes of food poisoning that can lead to either hemorrhagic colitis (HC) or hemolytic uremic syndrome (HUS). Stx-mediated HUS is the common cause of renal failure in children in the US [1]. A recent HUS outbreak in Germany highlighted the public health impact of this emerging pathogen [2]. STEC produce two distinct families of exotoxins, designated Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2) that are major virulence factors, essential to the pathogenesis of E. coli O157:H7 [3, 4]. There are no specific protective measures or therapeutics effective against infection by STEC. Stx1 and Stx2 are AB5 toxins consisting of an enzymatically active A subunit associated with a pentamer of receptor binding B subunits. They are also known as type II ribosome inactivating proteins (RIPs) because their A subunits are N-glycosidases, which remove a specific adenine (A4324 in rat, A3027 in yeast, and A2660 in E. coli) from the universally conserved α-sarcin/ricin loop (SRL) of the large rRNA, inhibiting the elongation step of protein synthesis [5]. The A subunits of Stx1 and Stx2 can be proteolytically cleaved into an enzymatically active A1 chain and an A2 chain, which remains associated with the B pentamer [6]. In the endoplasmic reticulum (ER) lumen, the A1 chain is released from the A2-B5 complex by reduction of the disulfide bond, and undergoes retrotranslocation from the ER into the cytosol [7]. Although Stx1 and Stx2 share a common receptor, globotriaosylceramide (Gb3), and indistinguishable activities, they have only 55% and 57% identical amino acid sequences on the A and B subunits, respectively, and are immunologically distinct. Stx2 is more important than Stx1 in the development of HUS [8, 9]. The German outbreak isolates, which were the deadliest on record, contained only Stx2 and possessed the typical characteristics of enteroaggregative E. coli [2]. The lethal dose of Stx2 is lower than that of Stx1 in animal models [10, 11]. However, it has not been possible to demonstrate the increased cytotoxicity of Stx2 in mammalian cell culture models. For example, Stx1 is more toxic to Vero cells than Stx2, while Stx2 is more toxic to mice and non-human primates [10, 11]. Since Stx1A1 and Stx2A1 are both equally effective in blocking protein synthesis in vitro [10, 12], the basis for the increased potency of Stx2 is not known. The binding affinity of Stx1 is higher than Stx2 to Gb3-mimicking receptors [13, 14] and the B pentamers of Stx1 and Stx2 show differential stability [15, 16].
Accumulating evidence indicates that several RIPs interact with the P proteins of the ribosomal stalk to access the SRL. Trichosanthin (TCS), Stx1 and maize RIP interact with the P proteins in vitro [17–20]. Removal of the last 17 amino acids of P1 or P2 proteins, but not the P0 protein, abolished the interaction between Stx1A1 and human ribosomal stalk proteins, suggesting that the conserved C-terminal domain (CTD) of P1/P2 proteins was critical [19]. TCS binding site on P1/P2 was mapped to the conserved CTD of P proteins by protein crystallography analysis [21]. We have developed a S. cerevisae model to examine ribosome interactions and enzymatic activity of RIPs [22–24], and demonstrated that ricin A chain (RTA) binds to the P proteins of the ribosomal stalk to depurinate the SRL in vivo [25, 26]. Using isolated stalk complexes from yeast, we showed that the stalk is the main landing platform for RTA on the ribosome and multiple copies of the stalk proteins accelerate the recruitment of RTA to the ribosome for depurination [27].
In eukaryotes, the stalk occurs in a pentameric configuration P0-(P1/P2)2 [28, 29], where P0 anchors two P1/P2 dimers [30]. In yeast, P1/P2 proteins have diverged into four different polypeptides, P1α, P1β, P2α and P2β. P1α/P2β and P1β/P2α preferentially form heterodimers prior to binding to P0 [31–33]. Presently, the only ribosomal components that are found free in the cytoplasm are the P1/P2 proteins of the ribosomal stalk [30]. Binding to P2 proteins can prevent P1 proteins from degradation in the cytoplasm. In contrast, P2 proteins are stable in the absence of P1 proteins in vivo [34]. Recent results indicate that the amino terminal end determines the stability of P1 and P2 [35]. The N-terminal domains (NTD) of P1/P2 proteins are responsible for dimerization and binding to P0 via the P1 proteins, while the CTD are mobile in the cytosol and interact with the translational GTPases (tGTPases) [36, 37]. The last 13 amino acids of the C-termini are identical among all five P proteins in yeast [30, 38]. The binding sites for P1α/P2β and P1β/P2α proteins on P0 in yeast have been mapped to amino acids 199–230 and 231–258, respectively [39].
One of the most interesting features of the eukaryotic stalk is its dynamism, where ribosome-bound P1 and P2 are exchanged with the free acidic proteins present in the cytosol and this exchange is increased during protein synthesis [30, 40]. This dynamic property of the stalk results in subpopulations of ribosomes containing different amounts of P1/P2 proteins [28, 41]. The biological significance of this exchange is not well understood. It is thought to play a role in regulating the activity of eukaryotic ribosomes [30, 40]. Since several RIPs access the SRL by binding to the stalk proteins [17–20, 25, 26], understanding how this stalk-dependent regulation affects the activity and toxicity of RIPs at the molecular level would be very important for developing protection strategies.
Molecular details of the interaction of Stx1A1 and Stx2A1 with ribosomes in vivo and contribution of these interactions to the cytotoxicity of each toxin have not been determined. Furthermore, since the B subunit is required for endocytosis and retrograde trafficking in mammalian cells, it has not been possible to study enzymatic activity of the A subunits in the absence of the B subunits in vivo. In a previous study, we identified the amino acids critical for the cytotoxicity of Stx1A and Stx2A and demonstrated that the function of these residues can be differentiated [42]. In this study, using the yeast model, we extend the previous in vitro observations with Stx1 [19] to Stx1 and Stx2 in vivo. We demonstrate that cytoplasmic stalk proteins are critical for Stx1A and Stx2A to access the SRL in vivo and present the first evidence that Stx1 and Stx2 differ in enzymatic activity towards ribosomes, which differ in the composition of the stalk.
2. Materials and Methods
2.1. Yeast strains and plasmids
The S. cerevisiae wild type strain was W303 (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his-3-11,15 can1-100). The stalk mutants ΔP1 and ΔP2 were haploid strains described elsewhere [25], while P0ΔAB mutant in the same genetic background was constructed (Cardenas, Revuelta, Remacha and Ballesta, unpublished results). Yeast strains were grown in YPD medium or minimal SD medium containing 2% glucose. Because the different selection markers are available in the yeast strains, RIPs were cloned into yeast vectors that carry either a URA3 marker (pYES2.1/V5-His TOPO based vector, Invitrogen) or LEU2 marker (Yep351-based NT198). The URA3 plasmids containing Stx1A (NT890) and Stx2A (NT901) with a V5 epitope and a 6×His tag downstream of the GAL1 promoter were previously described [42]. The LEU2 plasmids containing Stx1A (NT821) and Stx2A (NT785) were constructed by amplifying the cDNA from Stx1A (NT819) and Stx2A (NT782), respectively, and cloning them into NT198. The URA3 plasmids were transformed into the wild type, ΔP1 and P0ΔAB strains while the LEU2 plasmids were transformed into the ΔP2 strain.
2.2. Isolation of monomeric ribosome, membrane and cytosol fractions and immunoblot analysis
Yeast cells were grown in YPD media at 30°C overnight and collected at log phase. Monomeric ribosomes and cytosol fraction were isolated as described previously [25]. The cells were re-suspended in lysis buffer (50 mM Tris-HCl, pH 7.9, 5 mM EDTA, 1% (v/v) Triton X-100, 1 mM PMSF and protease inhibitors), glass beads were added to the mixture, followed by vortexing 6 times for 30 sec at maximum speed with 1–2 min on ice between each vortexing cycle. The cell debris was removed by centrifuging at 500 g for 5 min and the supernatant was centrifuged at 100,000 g for 1 hr at 4°C. The pellets containing the membrane fraction and the ribosome fraction were used to detect Stx1 and Stx2 using an immunoblot assay.
For immunoblot analysis the proteins were separated using a 12% SDS-PAGE. Monoclonal antibodies specific for P1β, P2α and P2β were used to detect the P1 and P2 proteins [43]. Polyclonal antibody P0 was obtained by injecting rabbits with recombinant P0 protein lacking the last 21 amino acids to avoid the cross-reaction with the acidic P proteins [44]. Monoclonal antibody against L3 (gift of Dr. J.R. Warner), dolichol-phosphate mannose synthase (Dpm1p; Invitrogen) and 3-phosphoglycerate kinase (Pgk1p; Invitrogen) were used as the loading controls for the ribosome, membrane and cytosol fractions, respectively. Polyclonal antibodies against Stx1A and Stx2A were provided by Dr. C. Thorpe (Tufts University Medical Center, Boston, MA).
2.3. Analysis of ribosome depurination
The depurination of rRNA was determined using either the dual-primer extension assay [45] or the qRT-PCR assay [46]. For in vivo depurination, Stx1A and Stx2A were induced for 10 hr, total rRNA was isolated, followed by the dual-primer extension analysis [45]. For in vitro depurination, yeast ribosomes (20 pmol) were incubated with 0.3 pmol of activated Stx1 or Stx2 in the presence of different amounts of P1α/P2β (gift of Dr. M. Tchorzewski) in a final volume of 100 μl at 30°C for 30 min. Under the conditions of this assay (200 nM ribosome and 3 nM toxin concentration), ribosome depurination was linear with the toxin concentration (Fig. S1). The purified P1α/P2β was used at RIP:P1α/P2β molar ratios of 1:10, 1:100 and 1:1,000, which correspond to ribosome:P1α/P2β ratios of 1:0.15, 1:1.5 and 1:15. The Stx1 and Stx2 holotoxins (Tufts University Medical Center, Boston, MA) were treated with 30 mM DTT at 30°C for 1 hr prior to use to release the A1 chain [25]. For the qRT-PCR assay [46], rRNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) and real-time PCR was carried out using an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Carlsbad, CA) as previously described [46]. Two pairs of primers were designed to amplify a target amplicon (depurinated SRL) and a reference amplicon (25S rRNA) for use in qRT-PCR. The reaction was performed using JumpStart Taq Ready Mix (Sigma) with SYBR Green I dye and ROX reference dye (Invitrogen, Carlsbad, CA). The results were quantified using the comparative ΔCT method (ΔΔCT) comparing RNA from RIP-treated ribosomes to control RNA from non-treated ribosomes [46]. All reactions were done in triplicate on a 96-well plate. The P2β was purified as previously described [46] and purified P1α/P2β heterodimer was a kind gift from Dr. Marek Tchorzewski [33].
2.4. Yeast cell viability assay
The yeast cells were transformed with a plasmid carrying Stx1A or Stx2A genes under the GAL1 promoter. Cells expressing the empty vector were used as controls. The cells were first grown in SD-glucose medium and then transferred to SD-galactose medium. At 10 hr post induction, serial dilutions of 0.1 OD595 of cells were plated on SD-glucose plates and grown at 30°C for 2–3 days.
3. Results
3.1. Stx1A and Stx2A depurinate ribosomes more in ΔP1 mutant than in ΔP2 mutant in vivo
To determine if P proteins of the ribosomal stalk are critical for ribosome depurination by Stx1A and Stx2A in vivo, they were introduced into the yeast strains lacking the acidic P1 (ΔP1) or P2 (ΔP2) proteins and to the isogenic wild type strain. Fig. 1A shows a model of the P proteins on ribosomes and in the cytosol in the wild type and in the P protein mutants based on immunoblot analysis shown in Fig. 1B. The P proteins were present on the ribosome, as well as in the cytosolic fraction in the wild type (Fig. 1B). In contrast, only P0 protein was detected on the ribosomes isolated from the P protein mutants (Ribosome fraction, Fig. 1B). Free P2 proteins were detected in the cytosol, but not on the ribosomes in the ΔP1 mutant, while free P1 proteins were not detected in the cytosol or on the ribosomes in the ΔP2 mutant (Fig. 1B). These results are consistent with previously reported studies, which indicated that in the absence of P2 proteins P1 proteins are rapidly degraded in yeast, suggesting that P2 protects P1 from degradation, possibly through the formation of a P1/P2 heterodimer [34]. In contrast, P2 proteins are stable in the absence of P1 proteins in vivo [34]. Notably, relatively higher amount of P2 proteins were detected in the cytosolic fraction of the ΔP1 mutant than in the wild type, since all P2 remained in the cytosol in this mutant. Stx1A and Stx2A containing the V5 and His epitope tags on a URA3 plasmid were transformed into the ΔP1 mutant and wild type, while Stx1A and Stx2A without the epitope tags on a LEU2 plasmid were transformed into the ΔP2 mutant. Previous results indicated that V5 and His epitope tags do not affect the activity of Stx1A or Stx2A [42]. Polyclonal antibodies against Stx1A and Stx2A were used to detect expression in the transformed strains (Fig. 2A and B). Antibody against the integral ER membrane protein, Dpm1, was used as the loading control. Stx1A and Stx2A were expressed at a slightly higher level in the ΔP2 strain than in the ΔP1 strain or in the isogenic wild type strain (Fig. 2A and B) and migrated faster in ΔP2, since they did not contain the V5 and the His tags (Fig. 2A and B).
Fig. 1.
Stalk composition of yeast mutants. (A) Schematic representation of the ribosomal stalk in the yeast mutants used in this study. P proteins on the ribosome and in the cytosol are shown in each yeast strain. (B) Ribosomes (10 pmol) and the cytosolic fraction (40 μg) isolated from the ΔP1, ΔP2 and P0ΔAB (ΔAB) mutants and the isogenic wild type were analyzed by immunoblot analysis using polyclonal antibody against P0 and monoclonal antibodies against P1β, P2α and P2β. Anti-L3 and anti-Pgk1 were used as loading controls for the ribosome and cytosol fractions, respectively. The immunoblot analysis was repeated three times using different ribosome preparations.
Fig. 2.
Depurination of ribosomes from the P protein mutants by Stx1A and Stx2A in vivo. Yeast cells were transformed with Stx1A or Stx2A under the GAL1 promoter. URA3 plasmids were transformed into the wild type (WT), ΔP1 and P0ΔAB (ΔAB) mutants while LEU2 plasmids were transformed into the ΔP2 mutant as described in the Experimental Procedures. Cells transformed with the empty vector (VC) were used as the control. Total protein was isolated at 10 h post induction and used in immunoblot analysis with polyclonal antibodies against Stx1A (A) or Stx2A (B). Anti-Dpm1 was used as the loading control. Total RNA (1 μg) isolated from the same cells expressing Stx1A (C) or Stx2A (D) was analyzed at 10 h post induction using dual-primer extension assay to detect depurination. The depurination levels were quantified by dividing the intensity of the depurination bands by the intensity of the 25S bands and multiplied by 100. Primer extension products using the 25S primer only (25S) and the depurination primer only (Dep) are shown. The analysis was repeated three times with different transformants. Results shown are from a representative experiment.
Total RNA was isolated from the transformed yeast cells and the level of depurination was examined by dual-primer extension analysis at 10 h post induction as previously described [45]. Ribosome depurination by Stx1A and Stx2A was reduced more in the ΔP2 mutant, which did not contain any P1/P2 proteins on the ribosome or in the cytosol, than in the ΔP1 mutant, which contained free P2 proteins in the cytosol (Fig. 2C and D). The level of depurination by Stx1A and Stx2A in the ΔP1 mutant was similar to or higher than the level of depurination in the wild type strain.
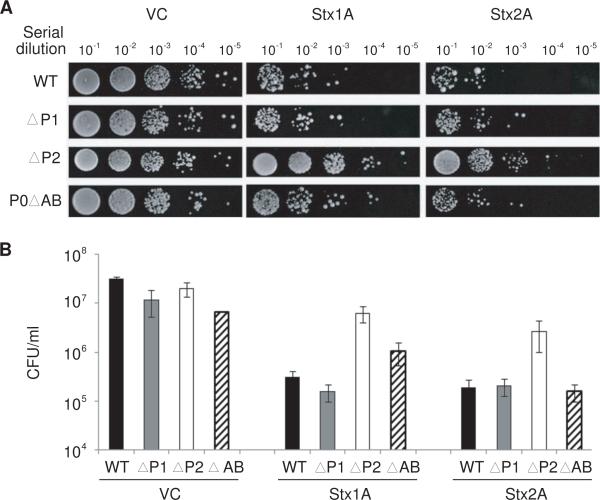
To determine if differences in the level of depurination affected the cytotoxicity of Stx1A and Stx2A, the effect of each toxin on viability of the different stalk mutants was determined. An equal number of cells were spotted on selective media after 10 h of galactose induction (Fig. 3A) and quantified by determining the CFU/ml (Fig. 3B). Consistent with the depurination results, ΔP2 expressing Stx1A or Stx2A was more viable than ΔP1 or the isogenic wild type strain (Fig. 3A and B). In contrast, ΔP1 was as sensitive to Stx1A and Stx2A as the wild type (Fig. 3A and B). These results indicated that both RIPs required P1/P2 proteins for ribosome depurination and suggested that cytoplasmic P proteins facilitated depurination in vivo and contributed to the higher toxicity of Stx1A and Stx2A in the ΔP1 mutant than in the ΔP2 mutant.
Fig. 3.
Viability of P protein mutants expressing Stx1A or Stx2A. (A) Yeast cells were transformed with a plasmid carrying Stx1A or Stx2A under the GAL1 promoter. Cells carrying the empty vector (VC) were used as controls. The cells were first grown in SD medium supplemented with 2% glucose and then transferred to SD medium supplemented with 2% galactose. At 10 hr post induction, a series of 10-fold dilutions were plated on media containing 2% glucose and grown at 30°C for 2–3 days. (B) Colony forming units per ml (CFU/ml) were calculated using at least 4 different colonies from three independent transformations.
3.2. Cytoplasmic P proteins cannot facilitate ribosome depurination by Stx1A if ribosomes do not contain intact binding sites for P1/P2
To determine how cytoplasmic P proteins facilitated depurination by Stx1A and Stx2A, we analyzed the level of depurination in the P0ΔAB mutant (labeled as ΔAB in the figures), in which the two binding sites for the P1/P2 heterodimers on P0 were deleted. As shown in Fig. 1B, P0 migrated faster in the ribosomal fraction from P0ΔAB and these ribosomes did not contain any P1 or P2 proteins. P1 and P2 proteins were detected in the cytosolic fraction of P0ΔAB mutant at a higher level than in the isogenic wild type strain, since all P1 and P2 remain in the cytosol in this mutant (Fig. 1B). Stx1A and Stx2A were expressed at similar levels in P0ΔAB as in ΔP1 and the isogenic wild type (Fig. 2A and B). The rRNA was purified and used in the dual primer extension assay to determine the level of depurination. Stx1A showed only 10% depurination in P0ΔAB compared to 54% depurination in the isogenic wild type strain at 10 h post induction (Fig. 2C). In contrast, Stx2A showed 60% depurination in P0ΔAB compared to 65% depurination in the wild type (Fig. 2D). P0ΔAB expressing Stx1A was more viable than the ΔP1 or wild type expressing Stx1A, while viability of P0ΔAB expressing Stx2A was reduced to the same level as the wild type (Fig. 3A and B). These results indicated that cytoplasmic P1/P2 proteins facilitated ribosome depurination by Stx1A only if their binding sites on the ribosomes were intact. In contrast, Stx2A was able to depurinate ribosomes better than Stx1A in the P0ΔAB strain, suggesting that it did not require the P1/P2 binding sites.
3.3. Purified P1α/P2β stimulates ribosome depurination by Stx1A1 more than by Stx2A1 if P0 contains an empty binding site for P1α/P2β
The differences in the sensitivity of P0ΔAB mutant to Stx1A and Stx2A suggested that the cytoplasmic P proteins played a differential role in the activity of each toxin. To confirm the in vivo results and to determine if free P1/P2 proteins can stimulate the depurination activity of the RIPs, ribosomes isolated from ΔP1, P0ΔAB and the isogenic wild type (WT) were treated with activated Stx1 and Stx2 in vitro in the presence or absence of the purified P1α/P2β heterodimer. The level of depurination by Stx1A1 and Stx2A1 was quantified by a qRT-PCR assay recently developed for yeast [46]. The fold increase in depurination in toxin-treated ribosomes relative to untreated ribosomes is shown in Fig. 4. Since the depurination level of each ribosome (WT, ΔP1 or P0ΔAB) after toxin treatment was calibrated against itself in the absence of the toxin, the depurination levels can be compared for each ribosome, but not across different ribosomes, since different calibrators were used. As shown in Fig. 4A, wild type ribosomes treated with Stx2A1 showed a higher level of depurination than wild type ribosomes treated with Stx1A1. Similar results were observed using ribosomes from the ΔP1 and P0ΔAB mutants (Fig. 4B and C), indicating that WT ribosomes and ribosomes from the stalk mutants were more sensitive to Stx2A1 than Stx1A1.
Fig. 4.
Effect of the purified P1α/P2β heterodimer on the depurination activity of Stx1A1 and Stx2A1 on yeast ribosomes. Yeast ribosomes (20 pmol) from the wild type (A), ΔP1 (B) and P0ΔAB (ΔAB) mutants (C) were incubated with 0.3 pmol of activated Stx1 (black bars) or Stx2 (open bars) in the presence of different amounts of P1α/P2β dimer in a final volume of 100 μl at 30°C for 30 min. The rRNA was isolated and 500 ng was used to quantify the relative level of depurination using qRT-PCR by the comparative ΔCT method (ΔΔCT). The y-axis shows the fold change of toxin-treated samples over the control samples without toxin treatment (No toxin control, NC). RIP:P1α/P2β ratios were 1:10, 1:100 and 1:1,000, which corresponded to ribosome:P1α/P2β ratios of approximately 1:0.15, 1:1.5 and 1:15 as indicated on the x-axis. The analysis was repeated three times using different preparations of ribosomes.
When wild type ribosomes, which contain the P0 and P1α/P2β and P1β/P2α dimers, were treated with RIPs in the presence of increasing amounts of purified P1α/P2β, a very minor effect was observed in the level of depurination by Stx1A1 or Stx2A1. A slight reduction in depurination was observed at a ribosome:P1α/P2β ratio of 1:15, possibly because the excess P1α/P2β competes with the ribosomal stalk for binding to the RIPs. When ribosomes from ΔP1, which contain empty binding sites for P1/P2 dimers, were treated at a ribosome:P1α/P2β ratio of 1:0.15, the level of depurination by Stx1A1 and Stx2A1 increased by 46% and 8%, respectively, over the level of depurination by each toxin alone (100%) (Fig. 4B). At a ribosome:P1α/P2β ratio of 1:1.5, the level of depurination of ΔP1ribosomes by Stx1A1 and Stx2A1 increased by 288% and 44%, respectively, and at a ratio of 1:15, depurination increased by 156% and 22% for Stx1A1 and Stx2A1, respectively, over the level of depurination by each toxin alone (100%) (Fig. 4B). These results showed that the free P1α/P2β dimer increased ribosome depurination by each toxin and had more impact on the activity of Stx1A1 than on the activity of Stx2A1.
When ribosomes from the P0ΔAB mutant, which do not contain binding sites for P1/P2 dimers on P0, were treated with each RIP, we observed a gradual reduction in depurination with increasing amounts of P1α/P2β dimers (Fig. 4C). At a ribosome:P1α/P2β ratio of 1:15, the depurination level of the P0ΔAB ribosomes by Stx1A1 and Stx2A1 was reduced to 36% and 45%, respectively, compared to the depurination levels observed in the absence of P1α/P2β (100%) (Fig. 4C). These results suggested that if the binding sites for P1/P2 were deleted on the ribosome, exogenous P1α/P2β was unable to increase the level of depurination by Stx1A1 or Stx2A1. Instead, depurination of P0ΔAB ribosomes was reduced, suggesting that P1α/P2β binds to the toxins and prevents them from depurinating the ribosomes.
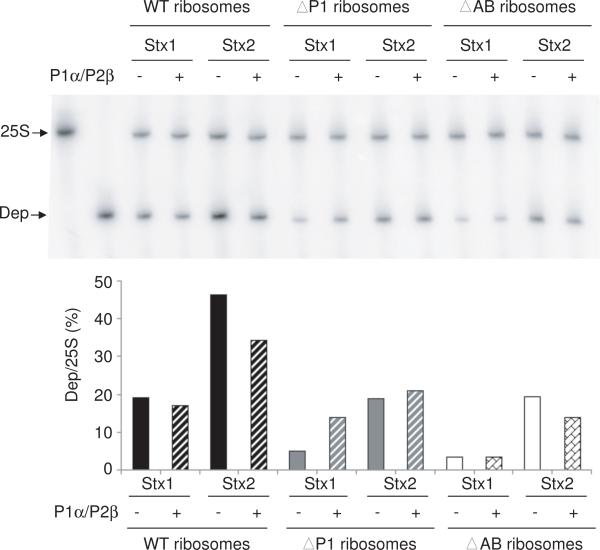
To compare the level of depurination of ribosomes from the different stalk mutants by the RIPs, the dual primer extension assay was used [45]. Each RIP depurinated WT ribosomes at a higher level than ribosomes from either the ΔP1 or the P0ΔAB mutant (Fig. 5). Stx2A1 depurinated WT ribosomes and ribosomes from the P protein mutants at a higher level than Stx1A1. At a ribosome:P1α/P2β ratio of 1:1.5, Stx1A1 and Stx2A1 depurinated ribosomes from the ΔP1 mutant at a higher level in the presence of P1α/P2β than in the absence of P1α/P2β, consistent with the qRT-PCR results (Fig. 4B). Overall, a greater increase in depurination was observed for Stx1A1 than Stx2A1. Stx2A1 showed less increase in depurination activity, since it was able to depurinate ΔP1 ribosomes at a higher level than Stx1A1 in the absence of P1α/P2β. At a ribosome:P1α/P2β ratio of 1:1.5, the level of depurination of P0ΔAB ribosomes by Stx1A1 did not increase and the level of depurination by Stx2A1 decreased. The dual primer extension results confirmed the qRT-PCR results and indicated that P1α/P2β heterodimer does not stimulate the enzymatic activity of the toxins directly, and increases the level of depurination only when P0 on the ribosome contains an empty binding site for P1α/P2β.
Fig. 5.
Effect of P1α/P2β heterodimers on the depurination activity of Stx1A1 and Stx2A1 on yeast ribosomes by the dual-primer extension assay. The same rRNAs (1 μg) from the reactions in Fig. 4 were used in the dual primer-extension assay. Yeast ribosomes (20 pmol) from the wild type, ΔP1 and P0ΔAB mutant (ΔAB) were incubated with 0.3 pmol of activated Stx1 or Stx2 in the presence of 30 pmol of P1α/P2β at 30°C for 30 min (Ribosome:P1α/P2β ratio of 1:1.5). The level of depurination was quantified by dual primer extension analysis. The first two lanes correspond to the primer extension products with the 25S rRNA primer only (25S) and the depurination primer only (Dep).
4. Discussion
4.1. Cytoplasmic P proteins are critical for ribosome depurination by Stx1A and Stx2A in vivo
Previous results showed that RTA interacts with the ribosomal stalk to access the SRL and this interaction with the stalk is required for the depurination activity and cytotoxicity of RTA in yeast [25]. The interaction between the yeast ribosomes and RTA was characterized by a two-step binding model, where the slower electrostatic interactions with the ribosome facilitated the faster, more specific interactions with the ribosomal stalk [26]. We show here that when Stx1A and Stx2A were expressed in the ΔP2 mutant, which lacks P1/P2 proteins on the ribosome and in the cytosol, ribosomes were depurinated at reduced levels, and ΔP2 mutant expressing Stx1A or Stx2A was more viable than the wild type. These results indicated that the ribosomal stalk is critical for ribosome depurination by Stx1A and Stx2A in vivo. However, in contrast to previous results with RTA [25], Stx1A and Stx2A depurinated ribosomes at a similar level in the ΔP1 mutant as in the wild type in vivo. Since ΔP1 and ΔP2 mutants differ only by the presence of cytoplasmic P2 proteins in the ΔP1mutant, but not in the ΔP2 mutant, these results suggested that cytoplasmic P2 proteins facilitated ribosome depurination by Stx1A and Stx2A. Stx1A and Stx2A were more toxic in the ΔP1 mutant than in the ΔP2 mutant, indicating that cytoplasmic P proteins also contributed to the higher toxicity of Stx1A and Stx2A.
4.2. P1/P2 heterodimers facilitate access of Stx1A and Stx2A to the ribosomal stalk
To determine if the cytoplasmic P proteins stimulate the depurination activity, Stx1A and Stx2A were expressed in the P0ΔAB mutant, in which the binding sites for P1/P2 heterodimers were deleted on the ribosome, and P1/P2 heterodimers accumulated only in the cytosol, but not on the ribosome. P0ΔAB expressing Stx1A was more viable than the wild type. Stx1A depurinated ribosomes at a very low level in the P0ΔAB mutant, suggesting that cytoplasmic P1/P2 proteins facilitated depurination by Stx1A only if their binding sites on ribosomes were intact. In contrast, viability of P0ΔAB expressing Stx2A was reduced as the wild type, and Stx2A was able to depurinate ribosomes close to the wild type levels in the P0ΔAB mutant.
The higher level of depurination of the P0ΔAB mutant by Stx2A suggested that either P0ΔAB ribosomes were more sensitive to Stx2A or cytoplasmic P1/P2 proteins stimulated the depurination activity of Stx2A even if they were not able to bind to the ribosome. To distinguish between these possibilities, we examined the activity of Stx1 and Stx2 on ribosomes isolated from the ΔP1 and P0ΔAB mutants and the wild type strain in vitro. Ribosomes from the wild type strain, ΔP1 and P0ΔAB mutants showed higher levels of depurination by Stx2A1 compared to Stx1A1 in vitro (Fig. 4A–C). These results provided further evidence that Stx2A1 was less dependent on P1/P2 than Stx1A1. It is possible that Stx2A may bind to another ribosomal protein besides the ribosomal stalk or to another region in addition to the CTD of P0, allowing ribosomes to be depurinated by Stx2A1 at a higher level than by Stx1A1.
To confirm the effect of free P1/P2 on toxin activity in vitro, we examined depurination of ribosomes from the P protein mutants in the presence of increasing amounts of purified P1α/P2β. Recent results indicate that P1α and P2β form a heterodimer independently of the other stalk components [29]. The P1α/P2β heterodimer is the most stable form involved in stalk formation [31, 32]. As shown in Fig. S2, the purified P1α/P2β heterodimer migrated as two bands by SDS-PAGE and as a single band by native PAGE, suggesting that this heterodimer appears uniform in solution. In contrast, purified P1α or P2β migrated as a single band by SDS-PAGE, but appeared nonuniform by native PAGE (Fig. S2), consistent with earlier reports indicating that when P1α and P2β are separated from one another, they lack a stable tertiary structure [47, 48].
Addition of P1α/P2β caused an increase in the level of depurination by each RIP on ribosomes from the ΔP1 mutant, which contains wild type P0 with empty binding sites for the P1/P2 heterodimers. The increase was greater for Stx1A1 than for Stx2A1. There are two possibilities for the increase in the level of depurination of ΔP1 ribosomes. The first is that P1α/P2β might stimulate the enzymatic activity of the RIPs. However, addition of P1α/P2β to P0ΔAB ribosomes failed to increase the level of depurination. Instead, the level of depurination of P0ΔAB ribosomes by each RIP was gradually attenuated with increasing level of P1α/P2β. Furthermore, addition of P1α/P2β did not increase the depurination level of the wild type ribosomes, which contain wild type P0 with P1α/P2β and P1β/P2α dimers. These observations showed that the increase in depurination was not due to the stimulation of the enzymatic activity of the RIPs by P1α/P2β.
The second possibility is that P1α/P2β may interact with the RIPs in the cytosol and deliver them to the stalk. We observed that P1α/P2β increased the level of depurination by the RIPs only on ΔP1 ribosomes, indicating that the empty binding sites for P1/P2 proteins were critical for the increase in depurination. Previous studies showed that the CTDs of P1/P2 proteins interact with several RIPs [19, 21], while the NTDs of P1/P2 are responsible for dimerization and interaction with P0 proteins [36]. These results suggest that the cytosolic P1/P2 proteins may interact with the RIPs through their CTD and facilitate access of the toxins to the stalk by binding to P0 through their NTD. The toxins may bind to the stalk assembled P proteins in the cytosol to more easily get to their target, the SRL. Since the rate of toxin binding to the ribosome is much higher than expected from simple diffusion [26], the cytosolic P1/P2 proteins might deliver the toxins to the ribosome to create a toxin pool near the SRL. At a high concentration of P1α/P2β (ribosome:P1α/P2β of 1:15), the increase was diminished on ΔP1 ribosomes, possibly because the heterodimer competed with the stalk for binding to the toxins.
The higher level of depurination by Stx1A and Stx2A observed in the ΔP1 mutant than in the ΔP2 mutant suggested that free P2 proteins might also stimulate ribosome depurination by Stx1A and Stx2A in vivo. Since the results with P0ΔAB mutant indicate that stimulation requires the ability to bind to the ribosomes, these results suggest that the P2 proteins alone may be able to bind to the ribosome. However, this is contrary to the available experimental data that clearly show the requirement of P1 proteins for the binding of P2 [29, 35]. A possible explanation for these results could be that a P2 homodimer might be able to weakly associate with the ribosome in ΔP1 in vivo, but may be washed out during the ribosome preparation. Supporting this possibility, the formation of small proportion of homodimers in purified P2 preparations have been reported [47]. The addition of purified P2β caused an increase the level of depurination of ΔP1 ribosomes by Stx1A and Stx2A (Fig. S3). However, the increase was much less than that observed with P1α/P2β, possibly because P2β exists in a partially folded conformation when it is not bound to P1α [47] and might not be able to interact with the RIPs as well as P1α/P2β in vitro.
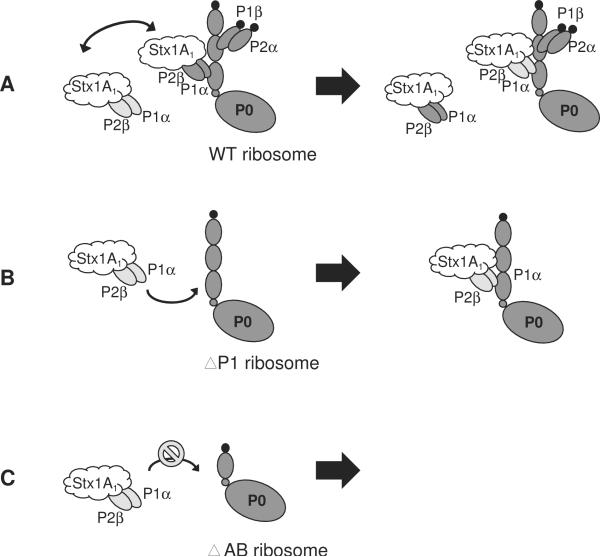
A model that illustrates the interaction of Stx1A1 with the purified P1α/P2β is shown in Fig. 6A. Stx1A1 could either bind to exogenous P1α/P2β or to ribosome-bound P1α/P2β on wild type ribosomes to access the SRL. Addition of P1α/P2β had a minor effect on the depurination activity of Stx1A1 on wild type ribosomes, because binding sites for P1/P2 were occupied (Fig. 6A). Since the binding sites for P1/P2 proteins were empty on ΔP1 ribosomes, the exogenous P1α/P2β was able to recruit Stx1A1 to P0 (Fig. 6B). Therefore, this resulted in increased depurination, even at a low level of P1α/P2β. In the P0ΔAB mutant, since the binding sites for the heterodimer were deleted, P1α/P2β was unable to bind to ribosomes. Therefore, P1α/P2β failed to deliver Stx1A1 to the ribosomal stalk on the P0ΔAB ribosomes (Fig. 6C). Instead, binding of toxin to P1α/P2β reduced the amount of toxin that targeted ribosomes. Consequently, we observed an inhibitory effect on depurination, especially at the higher levels of P1α/P2β (Fig. 4C).
Fig. 6.
Model representing the interaction of Stx1A1 with ribosomes from the stalk mutants in the presence of purified P1α/P2β in vitro. (A) Stx1A1 is able to interact with both the free P1α/P2β and the ribosome-bound P1α/P2β on the wild type ribosomes. The cytoplasmic P1α/P2β can exchange with the ribosome-bound P1α/P2β. The addition of P1α/P2β had a minor effect on the depurination activity of Stx1A1 on the wild type ribosomes because binding sites for P1/P2 were occupied. (B) The free P1α/P2β can interact with Stx1A1 and deliver this toxin to the stalk on ΔP1 ribosomes, which contain empty binding sites for P1α/P2β, causing a dramatic increase in the level depurination of ΔP1 ribosomes. (C) Since the binding sites for P1/P2 dimers are deleted on the ribosomes from P0ΔAB, P1α/P2β was not able to bind to these ribosomes, leading to protection of P0ΔAB ribosomes from depurination by Stx1A1.
4.3. Stx2A1 is less dependent on P1/P2 than Stx1A1
Although Stx1 and Stx2 act on the same substrate, the SRL, and share a common receptor, Gb3, they are two immunologically distinct toxins. Stx2 is more associated with severe disease and the development of HUS [8, 9]. It is not clear if the A or the B subunit or both are responsible for mediating the increased cytotoxicity of Stx2, since Stx1 and Stx2 inhibit protein synthesis at similar levels in vitro [10, 12]. We show here that it is possible to distinguish between the depurination activity of the A subunits of Stx1 and Stx2 in the P0ΔAB mutant in vivo. Stx2A depurinated ribosomes at a higher level in the P0ΔAB mutant than Stx1A. The addition of purified P1/P2 did not increase the depurination level of P0ΔAB ribosomes by Stx2A1 in vitro, suggesting that the higher level of depurination by Stx2A in the P0ΔAB strain was not due to the cytoplasmic P1/P2. Since P0ΔAB ribosomes were more sensitive to Stx2A in vivo than in vitro, these results suggest that there may be a factor weakly associated with P0ΔAB ribosomes, which increases their sensitivity to Stx2A in vivo.
Stx2A1 could depurinate wild type yeast ribosomes and ribosomes from the stalk mutants that did not contain any P1 and P2 proteins at a higher level than Stx1A1 in vitro, suggesting that Stx2A1 can depurinate ribosomes much more efficiently than Stx1A1, even when ribosomes do not contain an intact stalk. Addition of purified P1α/P2β had a minor effect on depurination of ΔP1 ribosomes by Stx2A1, suggesting that P1/P2 binding sites were less critical for Stx2A1. These results demonstrated that Stx2A1 is less dependent on the P1/P2 proteins for depurination activity than Stx1A1.
Taken together, these results provide the first evidence that Stx1 and Stx2 differ in enzymatic activity due to differences in their requirement for the stalk proteins. The free P1/P2 proteins facilitate access of Stx1A1 to the ribosomal stalk, while Stx2A1 is less dependent on the stalk proteins for activity. The differences in the role of the stalk proteins in toxin activity contribute to their relative toxicity, suggesting that the A subunits may mediate the variation in the potency of Stx1 and Stx2.
Supplementary Material
Acknowledgements
We would like to acknowledge Dr. Marek Tchórzewski and Przemyslaw Grela for providing the purified P1α/P2β heterodimer, Dr. Rong Di for constructing NT821 and NT781, Drs. Cheleste Thorpe and Anne Kane for providing polyclonal antibodies against Stx1A and Stx2A, and the Stx1 and Stx2 holotoxins, Dr. Jonathan Warner for monoclonal antibody against L3 and Drs. Jennifer Nielsen Kahn and Michael Pierce for helpful comments. This work was supported by National Institutes of Health Grant AI072425 (to N.E.T.) and grant BFU-2007-64280 (to M.R.) and BFU-2009-09738 to (J.P.G.B.) from the Spanish Ministry of Science and Innovation (MICINN) and an Institutional grant to the Center of Molecular Biology Severo Ochoa (CBMSO) from Fundacion Areces.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Siegler R, Oakes R. Hemolytic uremic syndrome; pathogenesis, treatment, and outcome. Curr. Opin. Pediatr. 2005;17:200–204. doi: 10.1097/01.mop.0000152997.66070.e9. [DOI] [PubMed] [Google Scholar]
- [2].Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A, Bauwens A, Peters G, Karch H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 2011 doi: 10.1016/S1473-3099(11)70165-7. in press. [DOI] [PubMed] [Google Scholar]
- [3].Paton JC, Paton AW. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tesh VL. Induction of apoptosis by Shiga toxins. Future Microbiol. 2010;5:431–453. doi: 10.2217/fmb.10.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 1988;171:45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x. [DOI] [PubMed] [Google Scholar]
- [6].Garred O, van Deurs B, Sandvig K. Furin-induced cleavage and activation of Shiga toxin. J. Biol. Chem. 1995;270:10817–10821. doi: 10.1074/jbc.270.18.10817. [DOI] [PubMed] [Google Scholar]
- [7].Sandvig K, van Deurs B. Delivery into cells: lessons learned from plant and bacterial toxins. Gene Ther. 2005;12:865–872. doi: 10.1038/sj.gt.3302525. [DOI] [PubMed] [Google Scholar]
- [8].Orth D, Grif K, Khan AB, Naim A, Dierich MP, Wurzner R. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn. Microbiol. Infect. Dis. 2007;59:235–242. doi: 10.1016/j.diagmicrobio.2007.04.013. [DOI] [PubMed] [Google Scholar]
- [9].Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tesh VL, Burris JA, Owens JW, Gordon VM, Wadolkowski EA, O'Brien AD, Samuel JE. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 1993;61:3392–3402. doi: 10.1128/iai.61.8.3392-3402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Siegler RL, Obrig TG, Pysher TJ, Tesh VL, Denkers ND, Taylor FB. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic uremic syndrome. Pediatr. Nephrol. 2003;18:92–96. doi: 10.1007/s00467-002-1035-7. [DOI] [PubMed] [Google Scholar]
- [12].Head SC, Karmali MA, Lingwood CA. Preparation of VT1 and VT2 hybrid toxins from their purified dissociated subunits. Evidence for B subunit modulation of a subunit function. J. Biol. Chem. 1991;266:3617–3621. [PubMed] [Google Scholar]
- [13].Takeda T, Yoshino K, Adachi E, Sato Y, Yamagata K. In vitro assessment of a chemically synthesized Shiga toxin receptor analog attached to chromosorb P (Synsorb Pk) as a specific absorbing agent of Shiga toxin 1 and 2. Microbiol. Immunol. 1999;43:331–337. doi: 10.1111/j.1348-0421.1999.tb02413.x. [DOI] [PubMed] [Google Scholar]
- [14].Nakajima H, Kiyokawa N, Katagiri YU, Taguchi T, Suzuki T, Sekino T, Mimori K, Ebata T, Saito M, Nakao H, Takeda T, Fujimoto J. Kinetic analysis of binding between Shiga toxin and receptor glycolipid Gb3Cer by surface plasmon resonance. J. Biol. Chem. 2001;276:42915–42922. doi: 10.1074/jbc.M106015200. [DOI] [PubMed] [Google Scholar]
- [15].Kitova EN, Mulvey GL, Dingle T, Sinelnikov I, Wee S, Griener TP, Armstrong GD, Klassen JS. Assembly and stability of the shiga toxins investigated by electrospray ionization mass spectrometry. Biochemistry. 2009;48:5365–5374. doi: 10.1021/bi9003155. [DOI] [PubMed] [Google Scholar]
- [16].Conrady DG, Flagler MJ, Friedmann DR, Vander Wielen BD, Kovall RA, Weiss AA, Herr AB. Molecular basis of differential B-pentamer stability of shiga toxins 1 and 2. PLoS One. 2010;5:e15153. doi: 10.1371/journal.pone.0015153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chan DS, Chu LO, Lee KM, Too PH, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chan SH, Hung FS, Chan DS, Shaw PC. Trichosanthin interacts with acidic ribosomal proteins P0 and P1 and mitotic checkpoint protein MAD2B. Eur. J. Biochem. 2001;268:2107–2112. doi: 10.1046/j.1432-1327.2001.02091.x. [DOI] [PubMed] [Google Scholar]
- [19].McCluskey AJ, Poon GM, Bolewska-Pedyczak E, Srikumar T, Jeram SM, Raught B, Gariepy J. The Catalytic Subunit of Shiga-like Toxin 1 Interacts with Ribosomal Stalk Proteins and is Inhibited by Their Conserved C-Terminal Domain. J. Mol. Biol. 2008;378:375–386. doi: 10.1016/j.jmb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- [20].Yang Y, Mak AN, Shaw PC, Sze KH. Solution structure of an active mutant of maize ribosome-inactivating protein (MOD) and its interaction with the ribosomal stalk protein P2. J. Mol. Biol. 2010;395:897–907. doi: 10.1016/j.jmb.2009.10.051. [DOI] [PubMed] [Google Scholar]
- [21].Too PH, Ma MK, Mak AN, Wong YT, Tung CK, Zhu G, Au SW, Wong KB, Shaw PC. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009;37:602–610. doi: 10.1093/nar/gkn922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hudak KA, Parikh BA, Di R, Baricevic M, Santana M, Seskar M, Tumer NE. Generation of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: evidence that ribosome depurination is not sufficient for cytotoxicity. Nucleic Acids Res. 2004;32:4244–4256. doi: 10.1093/nar/gkh757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li XP, Baricevic M, Saidasan H, Tumer NE. Ribosome depurination is not sufficient for ricin-mediated cell death in Saccharomyces cerevisiae. Infect. Immun. 2007;75:417–428. doi: 10.1128/IAI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hur Y, Hwang DJ, Zoubenko O, Coetzer C, Uckun FM, Tumer NE. Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: identification of residues important for toxicity. Proc. Natl. Acad. Sci. U S A. 1995;92:8448–8452. doi: 10.1073/pnas.92.18.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol. Microbiol. 2008;70:1441–1452. doi: 10.1111/j.1365-2958.2008.06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li XP, Chiou JC, Remacha M, Ballesta JP, Tumer NE. A two-step binding model proposed for the electrostatic interactions of ricin a chain with ribosomes. Biochemistry. 2009;48:3853–3863. doi: 10.1021/bi802371h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li XP, Grela P, Krokowski D, Tchorzewski M, Tumer NE. Pentameric organization of the ribosomal stalk accelerates recruitment of ricin a chain to the ribosome for depurination. J. Biol. Chem. 2010;285:41463–41471. doi: 10.1074/jbc.M110.171793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guarinos E, Santos C, Sanchez A, Qiu DY, Remacha M, Ballesta JP. Tag-mediated fractionation of yeast ribosome populations proves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol. Microbiol. 2003;50:703–712. doi: 10.1046/j.1365-2958.2003.03733.x. [DOI] [PubMed] [Google Scholar]
- [29].Grela P, Krokowski D, Gordiyenko Y, Krowarsch D, Robinson CV, Otlewski J, Grankowski N, Tchorzewski M. Biophysical properties of the eukaryotic ribosomal stalk. Biochemistry. 2010;49:924–933. doi: 10.1021/bi901811s. [DOI] [PubMed] [Google Scholar]
- [30].Ballesta JP, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog. Nucleic Acid. Res. Mol. Biol. 1996;55:157–193. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- [31].Lalioti VS, Perez-Fernandez J, Remacha M, Ballesta JP. Characterization of interaction sites in the Saccharomyces cerevisiae ribosomal stalk components. Mol. Microbiol. 2002;46:719–729. doi: 10.1046/j.1365-2958.2002.03179.x. [DOI] [PubMed] [Google Scholar]
- [32].Krokowski D, Tchorzewski M, Boguszewska A, Grankowski N. Acquisition of a stable structure by yeast ribosomal P0 protein requires binding of P1A-P2B complex: in vitro formation of the stalk structure. Biochim. Biophys. Acta. 2005;1724:59–70. doi: 10.1016/j.bbagen.2005.03.009. [DOI] [PubMed] [Google Scholar]
- [33].Tchorzewski M, Boguszewska A, Dukowski P, Grankowski N. Oligomerization properties of the acidic ribosomal P-proteins from Saccharomyces cerevisiae: effect of P1A protein phosphorylation on the formation of the P1A-P2B hetero-complex. Biochim. Biophys. Acta. 2000;1499:63–73. doi: 10.1016/s0167-4889(00)00108-7. [DOI] [PubMed] [Google Scholar]
- [34].Nusspaumer G, Remacha M, Ballesta JP. Phosphorylation and N-terminal region of yeast ribosomal protein P1 mediate its degradation, which is prevented by protein P2. EMBO J. 2000;19:6075–6084. doi: 10.1093/emboj/19.22.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Camargo H, Nusspaumer G, Abia D, Briceno V, Remacha M, Ballesta JP. The amino terminal end determines the stability and assembling capacity of eukaryotic ribosomal stalk proteins P1 and P2. Nucleic Acids Res. 2011;39:3735–3743. doi: 10.1093/nar/gkq1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jose MP, Santana-Roman H, Remacha M, Ballesta JP, Zinker S. Eukaryotic acidic phosphoproteins interact with the ribosome through their amino-terminal domain. Biochemistry. 1995;34:7941–7948. doi: 10.1021/bi00024a019. [DOI] [PubMed] [Google Scholar]
- [37].Bargis-Surgey P, Lavergne JP, Gonzalo P, Vard C, Filhol-Cochet O, Reboud JP. Interaction of elongation factor eEF-2 with ribosomal P proteins. Eur. J. Biochem. 1999;262:606–611. doi: 10.1046/j.1432-1327.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- [38].Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- [39].Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- [40].Remacha M, Jimenez-Diaz A, Santos C, Briones E, Zambrano R, Rodriguez Gabriel MA, Guarinos E, Ballesta JP. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem. Cell. Biol. 1995;73:959–968. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]
- [41].Saenz-Robles MT, Remacha M, Vilella MD, Zinker S, Ballesta JP. The acidic ribosomal proteins as regulators of the eukaryotic ribosomal activity. Biochim. Biophys. Acta. 1990;1050:51–55. doi: 10.1016/0167-4781(90)90140-w. [DOI] [PubMed] [Google Scholar]
- [42].Di R, Kyu E, Shete V, Saidasan H, Kahn PC, Tumer NE. Identification of amino acids critical for the cytotoxicity of Shiga toxin 1 and 2 in Saccharomyces cerevisiae. Toxicon. 2011;57:525–539. doi: 10.1016/j.toxicon.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vilella MD, Remacha M, Ortiz BL, Mendez E, Ballesta JP. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44' have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- [44].Santos C, Ballesta JP. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 1994;269:15689–15696. [PubMed] [Google Scholar]
- [45].Parikh BA, Coetzer C, Tumer NE. Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the alpha-sarcin/ricin loop of rRNA. J. Biol. Chem. 2002;277:41428–41437. doi: 10.1074/jbc.M205463200. [DOI] [PubMed] [Google Scholar]
- [46].Pierce M, Kahn JN, Chiou J, Tumer NE. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA. 2011;17:201–210. doi: 10.1261/rna.2375411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zurdo J, Sanz JM, Gonzalez C, Rico M, Ballesta JP. The exchangeable yeast ribosomal acidic protein YP2beta shows characteristics of a partly folded state under physiological conditions. Biochemistry. 1997;36:9625–9635. doi: 10.1021/bi9702400. [DOI] [PubMed] [Google Scholar]
- [48].Zurdo J, Parada P, van den Berg A, Nusspaumer G, Jimenez-Diaz A, Remacha M, Ballesta JP. Assembly of Saccharomyces cerevisiae ribosomal stalk: binding of P1 proteins is required for the interaction of P2 proteins. Biochemistry. 2000;39:8929–8934. doi: 10.1021/bi000362j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.