Abstract
The NAD-dependent histone deacetylase SIRT1 is overexpressed and catalytically activated in a number of human cancers, but recent studies argue have actually suggested that it may function as a tumor suppressor and metastasis inhibitor in vivo. In breast cancer, SIRT1 stabilization has been suggested to contribute to the oncogenic potential of the estrogen receptor α (ERα), but SIRT1 activity has also been associated with ERα deacetylation and inactivation. In this study, we show that SIRT1 is critical for estrogen to promote breast cancer. ERα physically interacted and functionally cooperated with SIRT1 in breast cancer cells. ERα also bound to the promoter for SIRT1 and increased its transcription. SIRT1 expression induced by ERα was sufficient to activate anti-oxidant and pro-survival genes in breast cancer cells, such as catalase and glutathione peroxidase, and to inactivate tumor suppressor genes such as cyclin G2 (CCNG2) and p53. Moreover, SIRT1 inactivation eliminated estrogen/ERα-induced cell growth and tumor development, triggering apoptosis. Taken together, these results indicated that SIRT1 is required for estrogen-induced breast cancer growth. Our findings imply that the combination of SIRT1 inhibitors and anti-estrogen compounds may offer more effective treatment strategies for breast cancer.
Keywords: Breast cancer, Estrogen, Estrogen receptor, SIRT1, p53
Introduction
SIRT1 (Silent information regulator 1) is a NAD+-dependent histone deacetylase, which is implicated in multiple biologic processes in several organisms (1, 2). All seven members of the sirtuin family (SIRT1–7) catalyze either protein deacetylation or ADP-ribosylation. SIRT1 has protein deacetylase activity, but no ADP-ribosyl transferase activity. SIRT1 targets many transcription factors, such as p53, FOXO, E2F1, NF-κB, PGC-1α, LXR, and MyoD (3, 4). Previous studies have shown that SIRT1 is overexpressed and/or catalytically activated in varieties of human cancers and that SIRT1 overexpression blocks apoptosis and senescence, and promotes cell proliferation and angiogenesis (5–8). Inhibition of SIRT1 induces growth arrest and apoptosis in a variety of cancer cells (9, 10). Further, p53 is a target for SIRT1; SIRT1 binds and deacetylates p53 resulting in its inactivation (11). In turn, activated p53 downregulates SIRT1 translation via miR-34a (12). p53-null mice have increased levels of SIRT1 and several p53-null tumor cell lines show SIRT1 overexpression (13). These studies clearly show that SIRT1 functions as an oncogene. However, recent studies have suggested that SIRT1 may function as a tumor suppressor. SIRT1 suppresses intestinal tumorigenesis and colon cancer growth in a β-catenin-driven mouse model of colon cancer (14). Sirt1−/− mice show impaired DNA damage response, evidenced by genomic instability and tumorigenesis, and activation of SIRT1 protects against mutant BRCA1-associated breast cancer (15).
Estrogen (E2) influences many physiologic processes in mammals (16, 17). E2/ERα signaling plays an important role in the regulation of mammary gland development and function, and also contributes to the onset and progression of breast cancer. More than 70% of human breast cancers express ERα, and elevated levels of ERα in benign breast epithelium correlate with increased risk of breast cancer (18). In addition, studies have shown a positive correlation between ERα expression and age-dependent increase in cancer incidence and metastasis (19, 20).
SIRT1 is a histone deacetylase; it deacetylates several histone and non-histone proteins and thereby it inactivates tumor suppressor genes and other target proteins. ERα is one of the several targets of SIRT1. p300 acetylates ERα and the acetylation is reversed by SIRT1 (21). However, recent studies have shown that inhibition of SIRT1 suppresses ERα expression (22). Mammary gland-specific Sirt1 deletion interferes with E2-stimulated growth signaling in normal and malignant mammary epithelial cells (23). E2 prevents age-related bone loss by inducing SIRT1 expression in the bone marrow (24). In addition, E2 recruits ERα and SIRT1 at the NQO1 (an NRF2-dependent detoxifying enzyme) promoter to inhibit transcription (25). Since ERα and SIRT1 cooperate in the development of mammary tumorigenesis, a clear understanding of the interaction at the molecular level could potentially open up new therapeutic avenues for the treatment of breast cancer.
Materials and Methods
Cell lines, plasmids and transfection
The human normal mammary epithelial cell line HMEC was obtained from the Lonza, Walkersville, MD while the other two human normal mammary epithelial cell lines, MCF10A and MCF12A, were obtained from ATCC, Manassas, VA. HBL100, also a human normal mammary epithelial cell line, was kindly provided by Dr. S. Sukumar, Johns Hopkins University, Baltimore, MD. ER-positive human breast cancer cell lines (MCF7, T47D, ZR75.1, BT474, BT483, MDA-MB361, MDA-MB415) and the ER-negative human breast cancer cell lines (MDA-MB231, MDA-MB453, MDA-MB468 and HCC1937) were obtained from ATCC, Manassas, VA. The HMEC and MCF10A cells were grown in MEGM complete medium and MCF12A cells was grown in DMEM/F12 medium with 5% horse serum, 20 ng/ml human epidermal growth factor (EGF), 100 ng/ml cholera toxin, 0.01mg/ml bovine insulin and 500 ng/ml hydrocortisone. HBL100 cells was grown in McCoy 5A with 10% FBS. MCF7 and BT20 cells were grown in DMEM medium with 10% FBS. T47D, ZR75.1, BT474, BT485 and HCC1937 cells were grown in RPMI 1640 medium with 10% FBS. MDA-MB-231, -361, and MDA-MB-468 cells were grown in Leibovit’s L-15 medium with 10% FBS. MDA-MB415 cells was grown in Leibovit’s L-15 medium with 15% FBS and 0.01mg/ml insulin.
Plasmid constructs
Details are given in supplementary information.
Generation of SIRT1shRNA-expressing stable cell lines
Details are given in supplementary information.
Immunoprecipitation experiments
Immunoprecipitation (IP) was accomplished with the Universal Magnetic Co-IP kit. HMEC, MCF10A, MCF7, ZR75.1, MB231 and MB453 cells’ extracts were first incubated with protein A/G agarose beads. The cleared supernatants were incubated either with SIRT1-specific antibody or with ERα-specific antibody overnight before addition of protein A/G agarose beads. Normal rabbit IgG was used as control. After washing, immunoprecipitated materials were eluted and immunoblotted (IB) with human anti-SIRT1 and anti-ERα antibodies. For assessment of ERα acetylation, nuclear extracts were used for IP with an antibody specific for acetylated lysine, and the immunoprecipitates were used for immunoblotting with an ERα-specific antibody.
Immunofluorescence
Details are given in supplementary information.
Chromatin immunoprecipitation (ChIP) assays
ZR75.1 cells was transfected with expression constructs of ER family members or SIRT1-7. Chromatin immunoprecipitation (ChIP) assays were carried out using a ChIP assay kit (Millipore) using human SIRT1, ERα and mouse IgG antibodies. After ChIP, genomic DNA present in the immunoprecipitates was analyzed by PCR using the promoter-specific primers (Supplementary Table S1)
Immunoblot analysis
For immunoblot (IB) analysis, cell lysates were prepared by sonication of cells in cell lysis buffer with protease inhibitors. Protein samples were fractionated on SDS-PAGE gels and transferred to Protran nitrocellulose membrane (Whatman GmbH). Membranes were blocked with 5% non-fat dry milk and exposed to primary antibody at 4°C overnight followed by treatment with appropriate secondary antibody, conjugated to horseradish peroxidase at room temperature for 1 h, and developed by Enhanced Chemiluminescence SuperSignal Western System.
RT-PCR
SIRT1, ERα, p53, c-Myc, cyclin G2, cyclin G1, survivin and BMP7 mRNA expressions were determined by RT-PCR. Total RNA, isolated from ZR75.1-pLKO.1 and ZR75.1-SIRT1shRNA cells, was reverse transcribed using the GeneAmp RNA PCR kit (Applied Biosystems). PCR was performed on Veriti thermocycler (Applied Biosystems) using the human-specific primers (Supplementary Table S2). Representative images of triplicate experiments are shown.
SIRT1 expression in kidney, lung, liver, heart and colon was analyzed by RT-PCR in male and female mice (C57BL/6) at different ages (3, 6, and 9 months). We used three animals in each age group and SIRT1 expression was quantified by densitometry.
Lipid peroxidation, glutathione peroxidase and superoxide dismutase Assays
Commercial kits were used to assay the lipid peroxidation product malondialdehyde (MDA), glutathione peroxidase (Gpx) and superoxide dismutase (SOD) activities as per the manufacturer’s instructions. Details are given in supplementary information.
Cell cycle analysis
Cells were fixed in 50% ethanol, treated with 0.1% sodium citrate, 1 mg/mL RNase A, and 50 µg/mL propidium iodide, and subjected to fluorescence-activated cell sorting (FACS, Becton Dickinson) analysis.
Colony formation assay
ZR75.1-pLKO.1 and ZR75.1-SIRT1shRNA cells were seeded in six-well plates (10,000 cells/ well) and grown in E2-free medium. After 24 h, cells were exposed to 10 nM 17β-estradiol for 2 weeks, changing the medium every 3 days. Cells were washed with PBS and fixed in 100% methanol for 30 min followed by staining with KaryoMax Giemsa stain for 1 h. The wells were washed with water and dried overnight at room temperature. Finally, cells were lysed with 1% SDS in 0.2 N NaOH for 5 min and the absorbance of the released dye was measured at 630 nm.
Mouse xenograft studies
Female athymic nude mice (6- to 8-wk old) were purchased from Taconic, Hudson, NY). Animals were housed in a pathogen-free isolation facility with a light/dark cycle of 12/12 h and fed with rodent chow and water ad libitum. We included 6 mice in each group. Seven days before tumor induction, mice were anesthetized and 3-mm pellets containing 17β-estradiol, 0.18 mg/21-day release (Innovative Research of America), was implanted subcutaneously in the animal’s back. The pellets provided a continuous release of E2 at serum concentrations of 150–250 pM, which is in the range of physiologic levels seen in mice during the estrous cycle. One week after surgery, ZR75.1-pLKO.1 and two clones of ZR75.1-SIRT1shRNA cells (1×107 cells in 100 µl PBS) were injected subcutaneously in the mammary fat pad. Tumor volume was quantified by measuring the length and width of the tumor every 7 days for 4 weeks using a caliper.
Collection of tissue samples from mouse
Tissues (kidney, lung, liver, heart and colon) were harvested from 3, 6 and 9 months-old male and female mice as per the Medical College of Georgia IACUC approved protocols. Tissue samples were immediately processed for the isolation RNA and protein samples.
Collection of ER-positive and ER-negative mammary tissues
The primary breast cancer specimens and corresponding normal control specimens, adjacent to the tumor, were obtained from the Medical College of Georgia Tumor Bank, which is operated jointly by the MCG Cancer Center and the Department of Pathology. The Tumor Bank has the approval from the Institutional Review Board and Human Assurance Committee for the collection of the tissues. The Tumor Bank has determined the tumor grade and the molecular signature of each of the cancer specimens in terms of expression of ER and progesterone receptor (PR) and also whether or not the tumor overexpresses HER2. We obtained 12 specimens and the corresponding normal controls from the Tumor Bank. In addition, we had 4 specimens from a commercial source. RNA was extracted from 12 ER-positive and 4 ER-negative normal and breast cancer tissues using the TRIZOL reagent (Invitrogen).
Institutional compliance
The animal experiments reported in this study were approved by the Medical College of Georgia IACUC and Biosafety Committees. Similarly, human breast cancer tissue and the surrounding normal tissues were obtained from the Medical College of Georgia tumor bank as per the approval of the Institutional Review Board and Human Assurance Committee.
Statistical analyses
Statistical analysis was done using one-way ANOVA followed by Bonferroni multiple comparison test. The software used was Graph Pad Prism, version 5.0. A p value <0.05 was considered statistically significant.
Results
Functional cooperation between E2/ERα and SIRT1
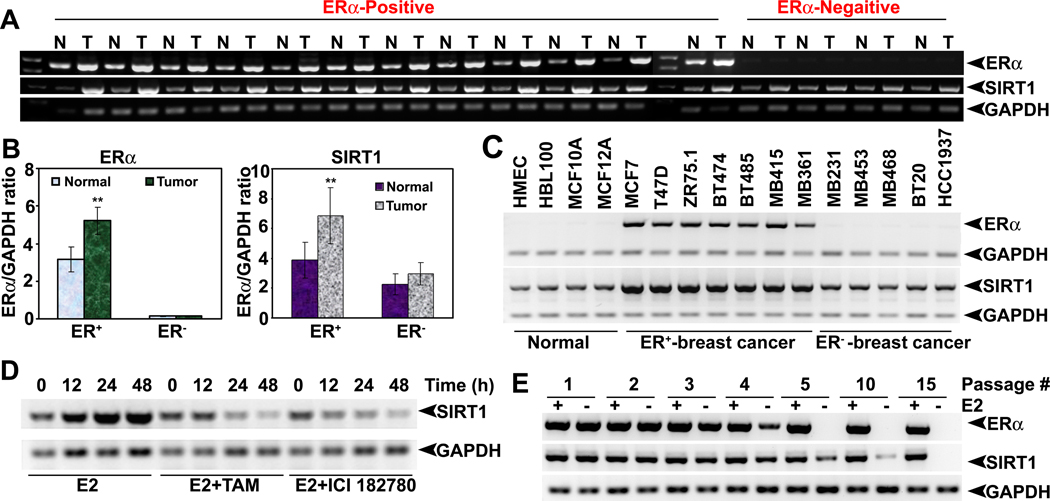
To understand the functional association between E2/ERα and SIRT1 in breast cancer, first we compared the expression levels of ERα and SIRT1 in ER-positive and ER-negative normal and breast tumor tissues as well as in nonmalignant and malignant mammary cell lines. We found a significant positive correlation between ERα and SIRT1 expressions (Fig. 1A, B and C). ERα-positive tumors and cancer cell lines expressed high levels of SIRT1 than ERα-negative tumors and cancer cell lines. Similarly, both ERα and SIRT1 proteins are overexpressed in breast tumor tissues compared to normal mammary tissues (Fig. S1). To confirm this positive association further, we treated ZR75.1 cells with E2 in the presence and absence of antiestrogens and monitored the expression levels of SIRT1. E2 significantly induced SIRT1 expression, and the effect was markedly blunted in the presence of antiestrogens (Fig. 1D). Similarly, when ZR75.1 cells were passaged several times in estrogen-free culture medium, ERα disappeared in these cells as did SIRT1 at passage numbers greater than 10 (Fig. 1E). These data showed that ligand-occupied ERα induces SIRT1 expression in mammary epithelial cells. This was supported further by the findings that SIRT1 levels were higher in various tissues in female mice than in male mice (Fig. S2). SIRT1 expression also correlated positively with E2 in various stages of mammary gland development (data not shown). All these results suggest that E2 plays a prominent role in regulation of SIRT1 expression.
Fig.1. Functional cooperation between E2-ERα and SIRT1.
(A) Expression of ERα and SIRT1 mRNAs in ER-positive (ER+) breast tumors tissues and adjacent normal tissues as well as in ER-negative (ER−) breast tumor tissues and corresponding normal tissues. (B) ERα and SIRT1 mRNA levels were quantified by densitometry. ** p<0.01. (C) SIRT1 mRNA expression in human normal, ER+ and ER− breast cancer cell lines. (D) SIRT1 mRNA expression in ZR75.1 cells, treated with or without E2 alone or in combination with TAM and ICI182740. (E) ERα and SIRT1 mRNA levels in ZR75.1 cells, cultured for several passages in regular and E2-free medium.
ERα binds to SIRT1 promoter and forms a complex in human breast cancer cells
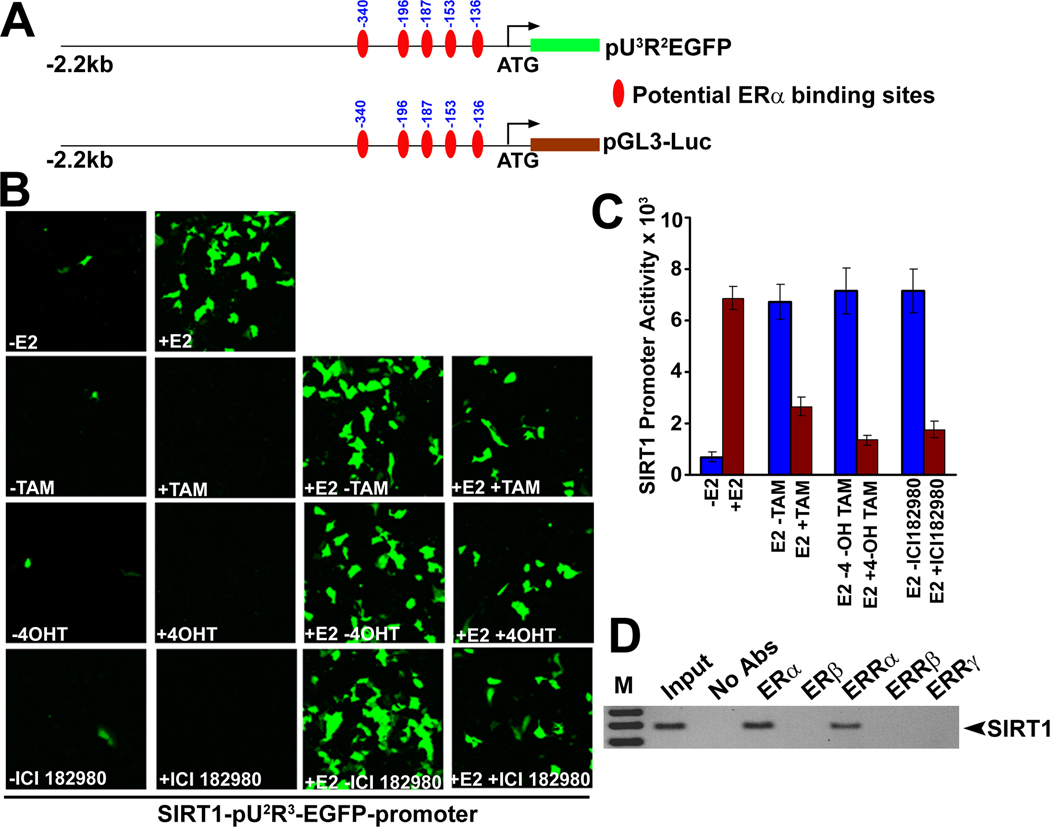
Unequivocal evidence for the induction of SIRT1 expression by E2/ERα came from the analysis of SIRT1 promoter activity. Analysis of the human SIRT1 promoter sequence revealed the presence of five putative ERα binding sequences in the upstream regulatory region (Fig. 2A). We cloned the 2.2 kb human SIRT1 promoter and subcloned it into pEGFP and luciferase (Luc) reporter vectors. These constructs were used for transfection into ERα-positive ZR75.1 cells. E2 treatment induced the SIRT1 promoter-specific EGFP fluorescence (Fig. 2B) as well as SIRT1 promoter-specific luciferase activity (Fig. 2C). Estrogen-induced EGFP fluorescence and luciferase activity were markedly blunted in the presence of antiestrogens (Fig. 2B and C). These experiments were complemented with ChIP assay. We expressed ER (ERα and ERβ) and ERR (ERRα, ERRβ and ERRγ) family members in ZR75.1 cells, performed immunoprecipitation with antibodies specific for these proteins, and examined the SIRT1 promoter in the immune complex by PCR. These studies showed that both ERα and ERRα interact with SIRT1 promoter and form a complex (Fig. 2D). These studies clearly demonstrate the transcriptional regulation of SIRT1 by E2 in mammary epithelial cells.
Fig.2. ERα binds to SIRT1 promoter and forms a complex.
(A) SIRT1-pGL3 and SIRT1-pU3R2EGFP reporter constructs and potential ERα binding sites. (B) ZR75.1 cells were transfected with SIRT1-pU3R2EGFP reporter construct and then cultured in E2-free medium for 24 h. Cells were then treated with and without 17β-estradiol (E2), tamoxifen (TAM), 4-hydroxy tamoxifen (4-OH TAM) and ICI182780 for 24 h and the expression of GFP was monitored by epifluorescence. (C) ZR75.1 cells were transfected with SIRT1-pGL3 reporter construct and then cultured in E2-free medium for 24 h. Cells were then treated with and without E2, TAM, 4-OH TAM and ICI182780 for 24 h and luciferase activity was measured using the cell lysates. (D) ZR75.1 cells were transfected with both ER (ERα and ERβ) and ERR (ERRα, ERRβ and ERRγ) family members and Chromatin immunoprecipitation (ChIP) assays were carried out using antibodies specific for these proteins. Genomic DNA present in the immunoprecipitates was examined using the SIRT1 promoter-specific primers by PCR.
Interaction of SIRT1 with ERα
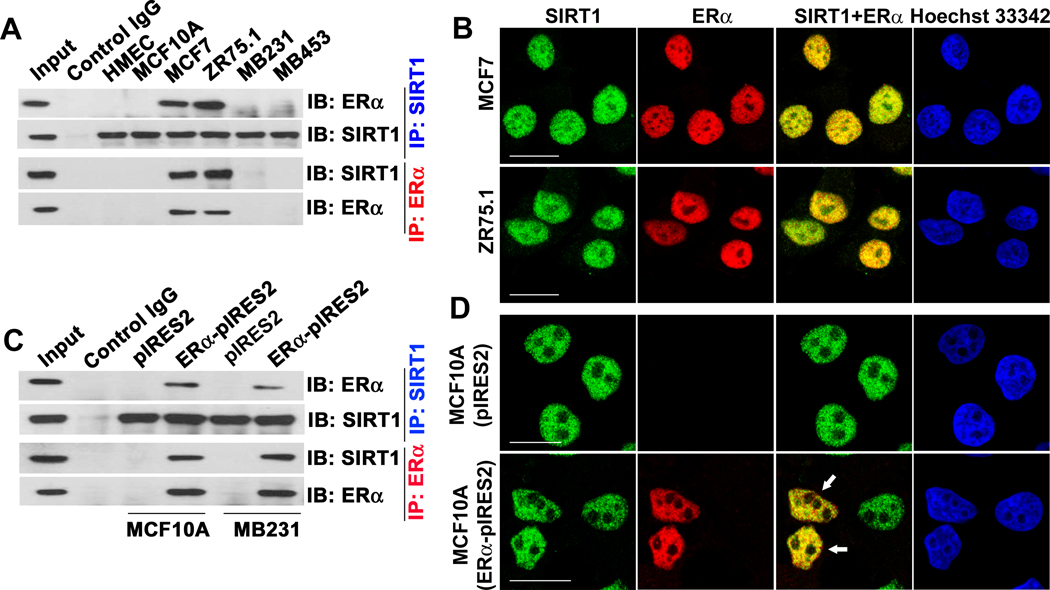
Then, we wanted to test whether ERα and SIRT1 physically interact and functionally colocalize with each other in mammary epithelial cells. We performed a series of co-immunoprecipitation assays using ERα-positive and ERα-negative cell lines. The interaction between SIRT1 and ERα was clearly evident in MCF7 and ZR75.1 cells that express both proteins (Fig. 3A). In these cells, both SIRT1 and ERα were colocalized in the nucleus (Fig. 3B). This interaction and nuclear colocalization was also confirmed in ERα-negative cells (MCF10A and MB231) following ectopic expression of ERα (Fig. 3C, D). There are two estrogen receptors, ERα and ERβ, and three estrogen receptor-related proteins, ERRα, ERRβ, and ERRγ; all these proteins share significant sequence homology (26). Except ERβ, all other members are associated with breast cancer initiation and progression (27). Therefore, we examined the interaction of SIRT1 with both ERs and ERRs in ZR75.1 cells that endogenously express all these proteins. These studies showed that SIRT1 interacts not only with ERα but also with ERRα (Fig. S3A and B). This was further confirmed in MCF7 cells as well as in two ER-negative human mammary epithelial cell lines, HMEC and HBL100 (data not shown). To test whether ERα specifically interacts with SIRT1 or also with other sirtuins, we expressed Flag-tagged SIRT1-7 in ZR75.1 cells, and examined the presence of ERα in immunoprecipitates with an anti-Flag antibody. The interaction was evident only with SIRT1 (Fig. S3C).
Fig.3. SIRT1 interacts with ERα.
(A) SIRT1 was immunoprecipitated (IP) from ER− (HMEC, MCF10A, MB231 and MB453) and ER+ (MCF7 and ZR75.1) cells using SIRT1 or ERα antibodies. Immunoblotting (IB) was done with SIRT1 or ERα antibodies. (B) Colocalization of SIRT1 and ERα in MCF-7 and ZR75.1 cells. Hoechst staining was used to locate the nucleus. The scale bars represent 20 µm. (C) ER− MCF10A and MB231 cells were transfected with an expression construct of ERα. IP and IB were performed 48 h post-transfection with SIRT1 and ERα antibodies. (D) MCF10A cells were transfected with an ERα expression construct, and colocalization (arrows) was monitored with SIRT1 and ERα antibodies.
ERα ligands and catalytically active SIRT1 are required for the interaction between ERα and SIRT1
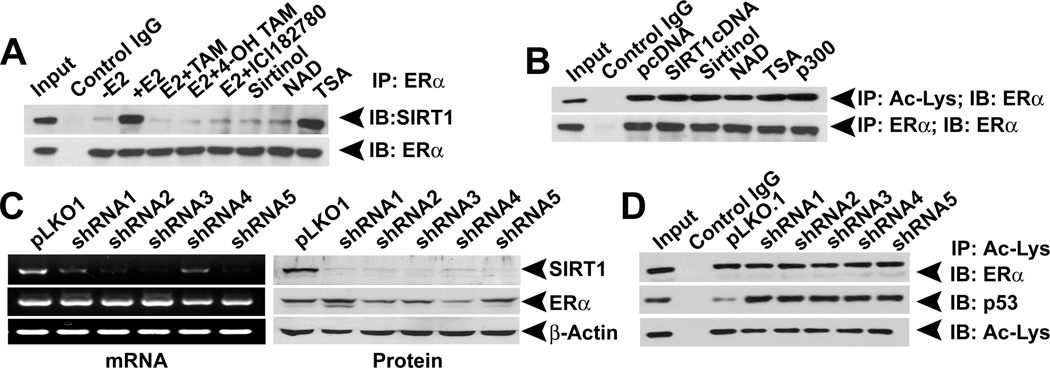
In the classical mechanism, E2 binds to ERα and the resultant complex interacts with the estrogen-response elements in E2 target genes, consequently promoting their transcription. To examine whether the interaction between ERα and SIRT1 requires E2 and whether functional disruption of E2/ERα complex with antiestorgens affects the interaction, we cultured ZR75.1 cells in E2-free medium and then treated the cells with E2 and antiestrogens. Immunoprecipitation experiments using these cell lysates showed that SIRT1-ERα interaction requires E2 and that functional disruption of E2/ERα complex dramatically reduces the interaction (Fig. 4A). Similarly, inhibition of the catalytic activity of SIRT1 with SIRT1 inhibitors also disrupted the interaction (Fig. 4A). In contrast, inhibition of type I and type II HDACs did not affect the interaction between ERα and SIRT1. This suggests that active ERα as well as catalytically active SIRT1 is required for the interaction. Further, the requirement of a catalytically active SIRT1 suggests that the interaction between ERα and SIRT1 may be regulated by SIRT1-mediated deacetylation of ERα. Therefore, we examined the acetylation status of ERα in the presence (SIRT1 overexpression) and absence (SIRT1 inhibitors) of SIRT1. Neither SIRT1 overexpression nor inhibition influenced the acetylation status of ERα (Fig. 4B), showing that SIRT1 is not involved in the deacetylation of ERα. To confirm this observation further, we used SIRT1shRNA in ZR75.1 cells (ERα-positive cells with endogenous SIRT1) and MDA-MB453 (ERα-negative cells with endogenous SIRT1) and monitored the protein expression and acetylation status of ERα (Fig. 4C, data not shown). Silencing of SIRT1 did not change either the expression or the acetylation status of ERα. As a positive control, we monitored the acetylation of p53, a known target for SIRT1. As expected, silencing of SIRT1 increased p53 acetylation (Fig. 4D). A recent study has shown that inhibition of SIRT1 suppresses ERα expression in breast cancer cells, suggesting that SIRT1 might be involved in the regulation of ERα expression (22). To confirm these observations, we treated ERα-positive breast cancer cells with SIRT1 inhibitors and analyzed ERα expression. Treatment of ERα-positive breast cancer cells with relatively high concentrations of sirtinol decreased ERα expression, and this process was accompanied with an increase in apoptosis (Fig. S4A and C). In contrast, another SIRT1 inhibitor, nicotinamide, neither affected ERα expression nor induced apoptosis in these three cell lines (Fig. S4B and D). These results suggest that sirtinol-associated ERα downregulation may not be directly related to SIRT1 inhibition.
Fig.4. SIRT1 interaction requires an active E2-ERα complex.
(A) ZR75.1 cells, treated with or without E2 (10 nM), E2 antagonists TAM (1µM), 4-OH TAM (1µM) and ICI182740 (1µM), SIRT1 inhibitors (NAD, 2 mM) and Sirtinol, 25 µM) or Type I and II HDAC inhibitor (TSA, 1 µM) were subjected to IP with an ERα antibody. The immunoprecipitates were used for IB with SIRT1 antibody. (B) ZR75.1 cells were transfected with pcDNA, SIRT1 and p300 expression constructs or treated with NAD+, Sirtinol and TSA. Nuclear extracts from these cells were used for IP with acetyl-lysine (Ac-Lys) antibody and the immunoprecipitates were immunoblotted with an ERα antibody. (C) ERα expression was analyzed in control and SIRT1 knockdown ZR75.1 cells. (D) Nuclear extracts from control and SIRT1 knockdown ZR75.1 cells were subjected to IP with Ac-Lys antibody. The immunoprecipitates were then used for immunoblotting with ERα and p53 antibodies.
SIRT1 is required for activation of antioxidant enzymes by E2-ERα in breast cancer cells
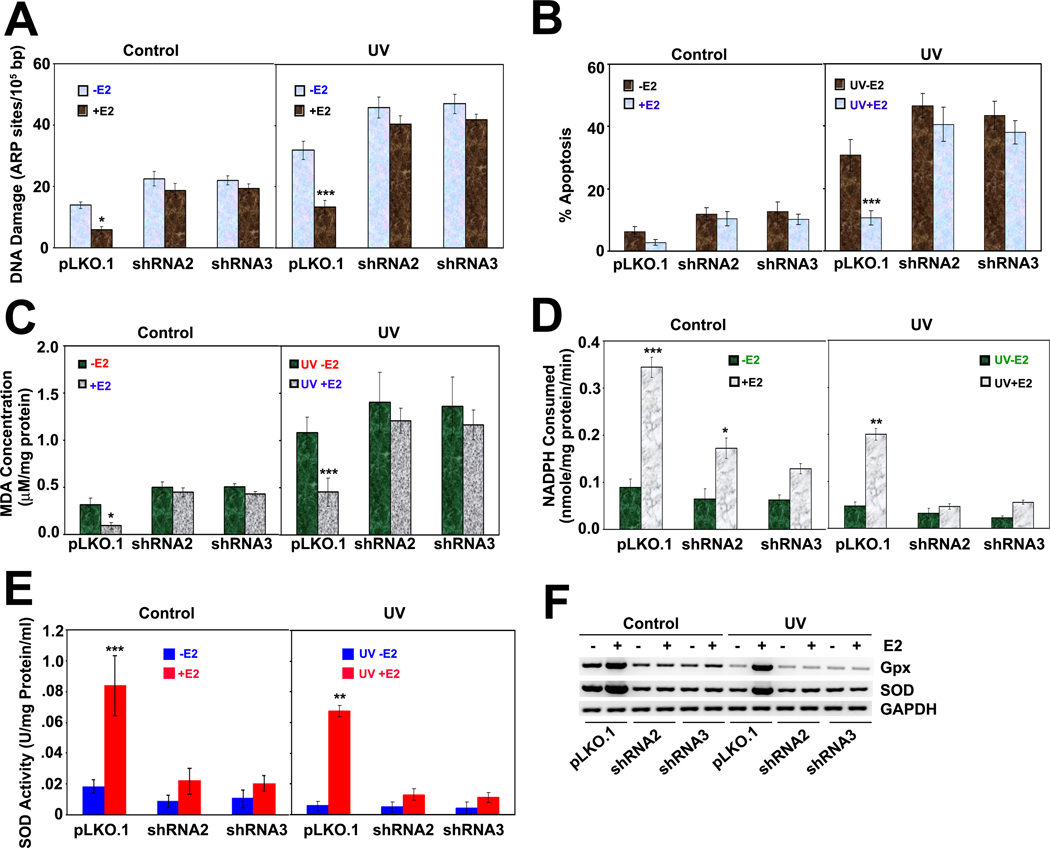
All the above results show that E2/ERα induces the transcriptional activation of SIRT1 and stabilizes SIRT1 expression by protein-protein interaction and that functional disruption of E2/ERα reduces SIRT1 expression. Thus, we wanted to find out the functional consequences of E2/ERα and SIRT1 interaction in breast cancer. It is known that E2 prevents reactive oxygen species (ROS) formation and thus protects cells from DNA damage. A well known example of E2-associated protection against ROS formation is that mitochondria from females produce approximately half the amount of ROS compared to males, and females express higher levels of antioxidative enzymes, especially Mn-SOD and Gpx, than males (28). Thus, females have a natural protection against ROS-induced cell damage. E2 also protects tumor cells from ROS-induced cell death by activating these enzymes. Further, recent studies have shown that overexpression of SIRT1 increases the transcription of SOD in human hepatoma cancer cell lines (29). This suggests that both E2/ERα and SIRT1 upregulate antioxidant enzymes to promote cell survival. Therefore, we examined the role of SIRT1 in the activation of the antioxidant machinery by E2. First we compared the extent of DNA damage, with or without UV radiation, in control (pLKO.1) and SIRT1-knockdown (SIRT1 shRNA) ZR75.1 cells in the presence and absence of E2. E2 prevented DNA damage in control cells to a significant extent, but did not do so in SIRT1 knockdown cells (Fig. 5A). The E2-mediated cellular defense was also reflected in the extent of cellular apoptosis (Fig. 5B), production of malondialdehyde (a surrogate marker for ROS) (Fig. 5C), and the expression and activities of the antioxidant enzymes Mn-SOD and Gpx (Fig. 5D, E and F); but the protective effect of E2 was evident only in control cells and not in SIRT1 knockdown cells. These studies showed that SIRT1 is required for the E2-mediated cellular defense mechanism in human breast cancer cells.
Fig.5. SIRT1 is required for activation of antioxidant enzymes by E2/ERα.
(A) DNA damage, with or without UV radiation, was quantified in control and SIRT1 knockdown ZR75.1 cells with or without E2 treatment. *** p<0.001; *p<0.05. (B) Cell cycle analysis was performed with cells described above, and the sub G0/1 cells were quantified using cell quest software. *** p<0.001. (C) The levels of malondialdehyde (MDA) were measured as a surrogate for lipid peroxidation in control and SIRT1 knockdown ZR75.1 cells with or without E2 treatment. *** p<0.001; *p<0.05. (D) Glutathione peroxidase (Gpx) and (E) superoxide dismutase (SOD) activities were measured in cell lysates. ***p<0.001; ** p<0.01; *p<0.05. (F) Gpx and SOD mRNA levels were analyzed in control and SIRT1 knockdown ZR75.1 cells.
SIRT1 is required for the suppression of p53 by E2-ERα
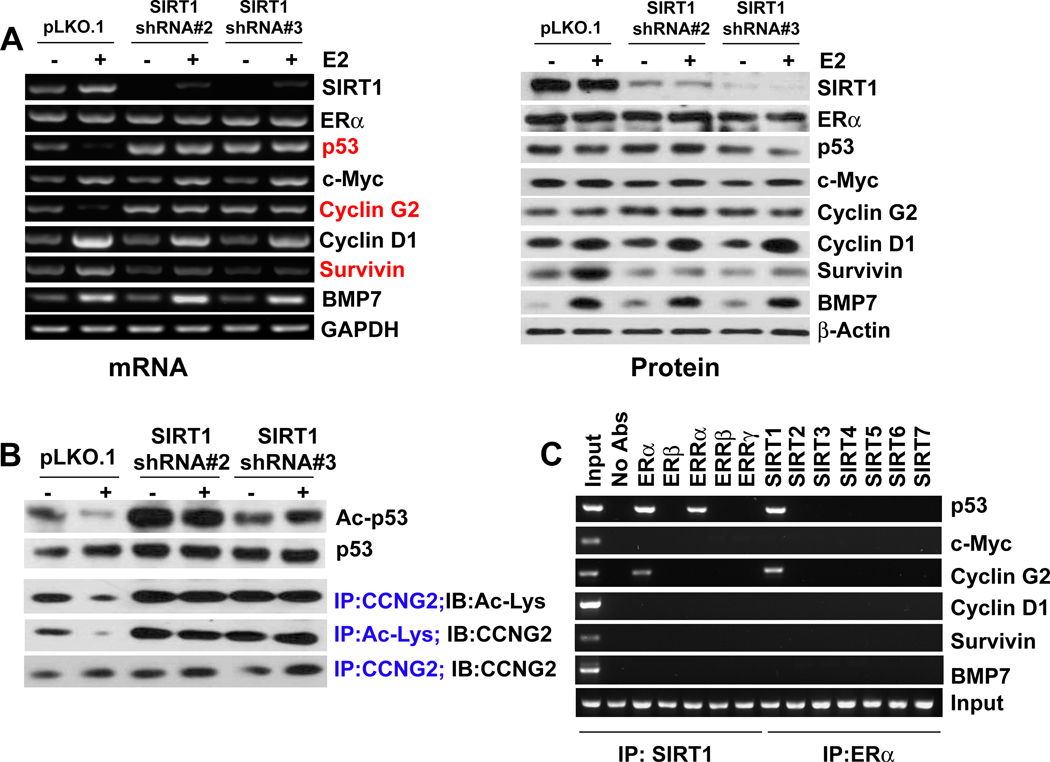
Studies have shown that ERα directly binds to p53 and represses its transcription and thus it interferes with p53-mediated cell cycle arrest and apoptosis (30). SIRT1 also binds and deacetylates p53 and inactivates its function. Thus, p53 is a common target for both ERα and SIRT1; therefore we determined whether SIRT1 is required for the inactivation of p53 by E2/ERα complex. We first monitored the transcriptional activity of E2/ERα by examining the expression levels of various E2/ERα target genes in control (pLKO.1) and in SIRT1 knockdown (shRNA) cell lines. The induction of c-Myc, cyclin D1 and BMP7 by E2/ERα was independent of SIRT1, but the downregulation of p53 and cyclin G2, and the induction of survivin by E2/ERα was dependent on SIRT1 (Fig. 6A). Because E2 treatment significantly reduced p53 mRNA expression in the pLKO.1 stable cell line and SIRT1 knockdown abolished that effect and thus, we wanted to examine the role E2 and SIRT1 in the transcriptional regulation of p53 at the promoter level. Genomic database analysis of p53 promoter suggested that it has 10 consensus binding sites for ERα (Fig. S5A). We transfected ZR75.1-pLKO.1 and ZR75.1-SIRT1shRNA cells with 2.4 kb and 356-bp p53 promoter-luciferase reporter plasmids (kindly provided by Dr. S. Sukumar, Johns Hopkins University) and treated the cells with E2 for 24 h. We observed that E2 treatment significantly suppressed the p53-promoter (2.4 kb)-dependent reporter activity only in ZR75.1-pLKO.1 cells (Fig. S5B). E2 treatment did not affect the shorter p53-promoter (356-bp) in these cells. Interestingly, SIRT1 knockdown itself transactivated the p53-promoter several-fold, but E2 treatment did not affect the p53-promoter-dependent reporter activity in these cells (Fig. S5B). These results suggest that SIRT1 is required for the transcriptional suppression of p53 by E2 in breast cancer cells.
Fig.6. SIRT1 is required for suppression of p53 by E2/ERα.
(A) The expression levels of E2 target genes (c-Myc, cyclin D1, BMP7, survivin, p53 and cyclin G2) were analyzed by RT-PCR and western blot in control and SIRT1 knockdown ZR75.1 cells with or without E2 treatment. The red highlights identify the genes whose regulation by E2 is mediated through SIRT1. (B) The acetylation status of p53 and cyclin G2 was analyzed in control and SIRT1 knockdown ZR75.1 cells with or without E2 treatment. (C) ChIP assay was performed to examine the binding of ERα/SIRT1 to E2 target genes.
The E2-induced p53 suppression in ZR75.1-pLKO.1 cells was not observed at the protein level even though the promoter activity showed suppression (Fig. 6B). This suggests that E2-mediated p53 inactivation is associated with post-translational modification and that SIRT1 might play role in this event. Therefore, we examined the role of SIRT1 in the acetylation of p53 and cyclin G2. In control cells, E2 caused deacetylation of p53 and cyclin G2, but this effect was abolished in SIRT1 knockdown cells (Fig. 6B). ChIP assays with ZR75.1 cells transfected with ER and ERR family members showed that immunoprecipitates with anti-SIRT1 antibody contained the promoters of p53 and cyclin G2 only in cells expressing ERα and ERRα (Fig. 6C). Similarly, ChIP assays with cells transfected with SIRT1-7 showed that immunoprecipitates with anti-ERα antibody contained the promoters of p53 and cyclin G2 only in cells expressing SIRT1. This was not the case for the other E2/ERα target genes c-Myc, cyclin D1, survivin, and BMP7. These results suggest that E2 recruits ERα and SIRT1 to suppress p53 and cyclin G2 expression in ER-positive breast cancer cells.
Loss of SIRT1 is associated with elimination of E2-induced cell survival and mammary tumorigenesis
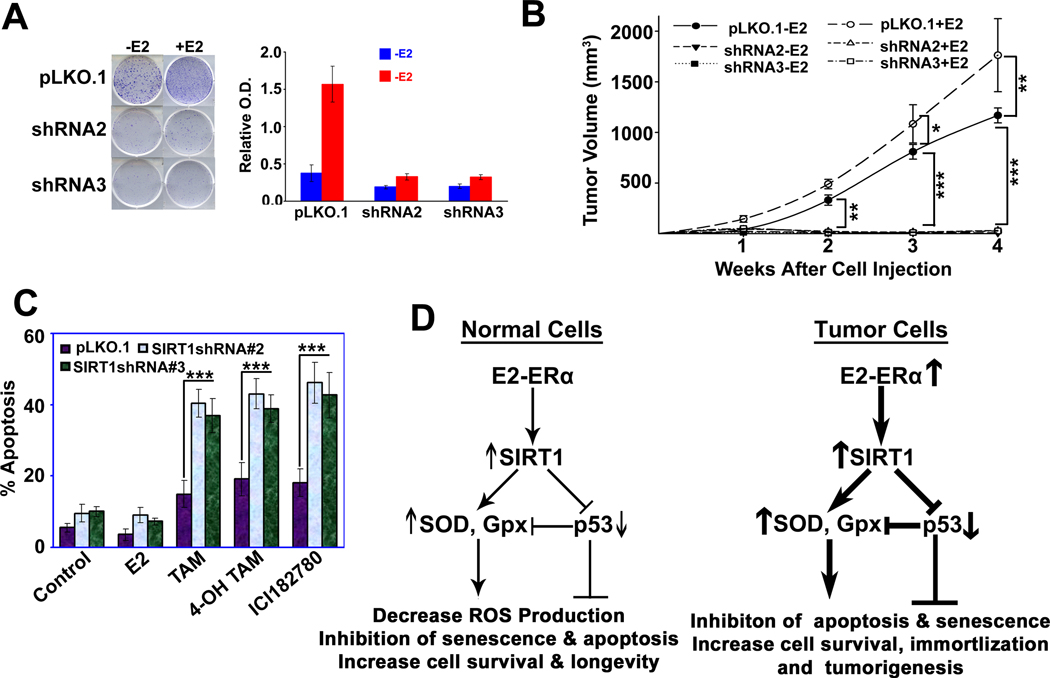
Since E2 promotes the development and progression of many hormone-dependent cancers, especially breast cancer, and SIRT1 also promotes several human malignancies by deacetylating tumor suppressor genes, we wanted to test whether SIRT1 plays a role in E2-induced mammary tumorigenesis. To determine the role of SIRT1 in E2-mediated promotion of cell survival and breast cancer growth, we first monitored the ability of E2 to promote colony formation in vitro in control cells and SIRT1 knockdown cells. E2 was able to induce colony formation only in control cells but not in SIRT1-depleted cells (Fig. 7A). This phenomenon was confirmed in vivo in mouse xenografts. Xenografts with control ZR75.1 cells grew in a time-dependent manner, and the growth was promoted by E2 (Fig. 7B). In contrast, SIRT1-depleted ZR75.1 cells failed to grow in xenografts with or without E2 (Fig. 7B), showing that SIRT1 is absolutely required for the promotion of tumor growth by E2. To confirm the functional cooperation between ERα and SIRT1 in the promotion of the growth of xenografted breast cancer cells, we treated the three different human breast cancer cell lines (MCF7, T47D and ZR75.1) with Tamoxifen (an antiestrogen) and Sirtinol and nicotinamide (SIRT1 inhibitors) alone or in combination. The combination treatment produced a greater magnitude of apoptosis in these cells than the individual treatment (Fig. S6). This was true even for the Tamoxifen-resistant cell line MCF7-TAM(R) (Fig. S7A). Interestingly, the MCF7-TAM(R) cells had higher levels of SIRT1 expression than the parent MCF7 cells (Fig. S7B). We confirmed this further with SIRT1 knockdown in ZR75.1 cells where antiestrogens caused more apoptosis in SIRT1 knockdown cells than in control cells (Fig. 7C). Based on these results, we propose a model for the interaction of ERα and SIRT1 and a molecular mechanism involved in the promotion of breast cancer by the ERα-SIRT1 complex (Fig. 7D).
Fig.7. Loss of SIRT1 is associated with elimination of E2-induced cell survival and mammary tumorigenesis.
(A) Control and SIRT1 knockdown ZR75.1 cells were treated with or without E2 for 2 weeks and the resulting colonies were quantified with Giemsa staining. (B) Seven days before tumor induction, female athymic nude mice (6 animals per group; 3 groups) were anesthetized and 3-mm pellets containing E2, 0.18 mg/21-day release, were implanted subcutaneously in the animal’s back. Another 18 animals were used as a control, without E2 treatment. One week after pellet implantation, ZR75.1-pLKO.1 and ZR75.1-SIRT1shRNA (two independent shRNA clones shRNA#2 and shRNA#3) cells (1×107 cells in 100 µl PBS) were injected s.c. in the mammary fat pad of all animals. Tumor volume was used as a measure of tumor growth at 1, 2, 3 and 4 weeks after cell injections. We compared the tumor volume of pLKO.1 and SIRT1shRNA-induced tumor in the presence and absence of E2 and the statistical significance was calculated as ***p<0.001; ** p<0.01; *p<0.05*p<0.05 in each time point (C) Control and SIRT1 knockdown ZR75.1 cells were treated with E2, TAM, 4-OH TAM and ICI182780 for 48 h. Apoptotic cell death was analyzed by FACS. We compared the apoptotic cell death in control pLKO.1 and SIRT1shRNA cells treated with E2, TAM, 4-OH TAM and the statistical significance was calculated as ***p<0.001 (D). Proposed mechanism of E2-ERα and SIRT1 complex in regulation of tumor cell immortalization and tumor development.
Discussion
The association of estrogen and its receptor (ERα) in development and progression of many hormone-dependent cancers, including breast cancer, has been well established. In the absence of estrogen, ERα is inactivated by the binding of a heat shock protein (HSP90) within the nucleus. Upon estrogen binding, the receptor undergoes conformational change so that HSP90 is released, enabling ERα to activate gene transcription. ERα functions as a transcriptional activator by binding to the estrogen response element in the target gene promoter (16, 17). ERα also functions as a transcription repressor upon binding to certain antagonists, such as tamoxifen, as well as to various corepressors and histone deacetylases (31). Thus, depending on the binding partner, ERα functions as either transcriptional activator or repressor of the target gene. Our findings demonstrate for the first time that SIRT1 is the binding partner of ERα in mammary epithelial cells and that the ERα-SIRT1 complex functions as a transcriptional activator of Mn-SOD and Gpx and as a transcriptional repressor of p53 and cyclin G2. In addition to its role as a binding partner of SIRT1, ERα also regulates the expression of SIRT1 through transcription as well as by protein-protein interaction. Our results also suggest that SIRT1 could be a downstream target of E2 in mammary epithelium.
In our study, we found a direct physical interaction between ERα and SIRT1. This interaction requires a catalytically active SIRT1. However, SIRT1 does not affect the acetylation status of ERα. Furthermore, knockdown of SIRT1 in ER-negative breast cancer cells is not able reactivate ERα expression. However, HDAC1 and HDAC3 deacetylate ERα and knockdown of these HDACs reactivates ERα expression (Fig. S8). This suggests that SIRT1 does not play a role in deacetylation of ERα. E2/ERα induces SIRT1 expression by directly binding to SIRT1 promoter. We conclude that E2/ERα signaling activates SIRT1 and that SIRT1 is the molecular transducer of E2-induced oncogenic signaling in mammary epithelial cells. Further, we also found a direct interaction between SIRT1 and ERRα in human breast cancer, and ER-negative breast cancer cells still have high expression of SIRT1 compared to normal mammary epithelial cells. In addition, we found higher levels of ERRα expression in ER-negative breast cancer cells than in normal and ER-positive breast cancer cells (unpublished data). These observations suggest that SIRT1 and ERRα might play a crucial role in regulation of survival signaling in ER-negative breast cancer. The findings in the literature that ERRα is critical for the growth of ER-negative breast cancer and that SIRT1 directly interacts with ERRα and enhances its transcriptional function (32, 33) support our data and conclusions.
E2 increases cell survival and oncogenic transformation via activation of anti-oxidative enzymes, MAPK, PI3K and telomerase, and inhibition of p53. Likewise, SIRT1 also increases cell survival and extents cellular longevity by preventing ROS formation, regulating MAPK and inhibiting p53 function. Our results demonstrate that E2 efficiently protects breast cancer cells from oxidative and radiation-induced DNA damage by inducing SOD and Gpx activity. However, E2-associated cellular protection and antioxidant enzyme induction obligatorily depends on functional SIRT1. Thus, both E2/ERα and SIRT1 target a common molecular signaling pathway to protect cells from oxidative and radiation-induced DNA damage. Our results show that ERα and SIRT1 interact with each other and promote cell survival and tumor growth in breast cancer cells.
Our studies also demonstrate that ERα binds to p53 promoter and suppresses its expression and that the process is obligatorily dependent on SIRT1. Thus, ERα-SIRT1 complex functions as a suppressor of p53 gene in breast cancer cells. Further, E2 treatment results in deacetylation of p53, and SIRT1 knockdown abolishes this effect, suggesting that SIRT1 is necessary for the ability of ERα to suppress p53 signaling in breast cancer. Similarly, E2/ERα directly binds to cyclin G2 promoter and suppresses its expression (34). Our studies show for the first time that SIRT1 is needed for this effect.
E2/ERα complex is linked to the development and promotion of breast cancer. There is also strong evidence that SIRT1 is involved in oncogenic signaling in mammary epithelial cells. First, SIRT1 knockout mice exhibit p53 hyperacetylation and increased radiation-induced apoptosis consistent with SIRT1 inhibition of p53 function (35). This raises the possibility that SIRT1 can facilitate tumor growth by antagonizing the p53 function. Second, DBC1 (deleted in breast cancer) inhibits SIRT1 in human mammary epithelial cells, and repression of SIRT1 by DBC1 hyperacetylates and activates p53 (6, 36). Third, HIC1 (hypermethylated in cancer) binds to SIRT1 promoter and represses its transcription; at the same time, HIC1 promotes p53 expression (7). Since HIC1 is silenced in many types of tumors, including breast cancer, this results in upregulation of SIRT1, thus promoting tumorigenesis. Further, upregulation of SIRT1 has been observed in various cancers (5–8). All these findings show that SIRT1 is a tumor promoter. Our studies demonstrate unequivocally that SIRT1 plays a critical role as a tumor promoter in ER-positive breast cancer. In normal cells, the ERα-SIRT1 complex mediates several beneficial functions such as the protection of the cells from the ROS-induced DNA damage, maintainance of genomic integrity and telomerase function, and inactivation of apoptotic signaling. The same functions promote the survival and growth of tumor cells. Furthermore, SIRT1 also seems to play a crucial role in breast cancer cells in the development of resistance to anti-estrogen therapy. These findings suggest that appropriate combination therapies targeting both ERα and SIRT1 could offer a novel and effective strategy for treatment of breast cancer.
Supplementary Material
Acknowledgements
We would like to thank Dr. Saraswati Sukumar, John Hopkins University, Baltimore, MD. We also thank the Medical College of Georgia Flow Cytometry and Sequencing Cores for FACS and sequencing analyses. We also thank the Medical College of Georgia Tumor Bank for providing the human mammary tissue samples.
Financial Support: This work was supported by grants from National Institute of Health (R01 CA131402) and Department of Defense (BC074289).
Footnotes
All authors declare no conflict of interest.
REFERENCES
- 1.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificient seven’, function, metabolism and longevity. Ann Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 2.Haigis MC, Guarente LP. Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 3.Feige JN, Auwerx J. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol. 2008;20:303–309. doi: 10.1016/j.ceb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 6.Kim JE, Chen J, Lou Z. DBC1 is negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 7.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisi J. Suppressing cancer: the importance of being senescent. Science. 2005;309:886–887. doi: 10.1126/science.1116801. [DOI] [PubMed] [Google Scholar]
- 10.Ota H, Tokunage E, Chang K, Hikasa M, Lijima K, Eto M, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MPAK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 11.Vaziri H, Dessain SK, Ng Eaton E, Imai S, Frye RA, Pandita TK, et al. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 12.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 14.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Zhang, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deroo BJ, Korach KS. Estrogen receptors and human diseases. J Clini Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke RB, Anderson E, Howell A. Steroid receptors in human breast cancer. Trends Endocrinol Metab. 2004;15:316–323. doi: 10.1016/j.tem.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39:655–660. doi: 10.1038/ng2006. [DOI] [PubMed] [Google Scholar]
- 19.Cooper JA, Rohan TE, Cant EL, Horsfall DJ, Tilley WD. Risk factors for breast cancer by oestrogen receptor status: a population-based case-control study. Br J Cancer. 1989;59:119–125. doi: 10.1038/bjc.1989.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazi AA, Jones JM, Koos RD. Chromatin immunoprecipitation analysis of gene expression in the rat uterus in vivo: Estrogen-induced recruitment of both estrogen receptorα and hypoxia-inducible factor 1 to the vascular endothelial growth factor promoter. Mol Endocrinol. 2005;19:2006–2019. doi: 10.1210/me.2004-0388. [DOI] [PubMed] [Google Scholar]
- 21.Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. Acetylation of estrogen receptor α by p300 at lysine 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol Endocrinol. 2006;20:1479–1493. doi: 10.1210/me.2005-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao Y, Li H, Gu Y, Davidson NE, Zhou Q. Inhibition of SIRT1 deacetylase suppresses estrogen receptor signaling. Carcinogenesis. 2010;31:382–387. doi: 10.1093/carcin/bgp308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Rajendran GK, Liu N, Ware C, Rubin BO, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1–R12. doi: 10.1186/bcr1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elpaz A, Rivas D, Duque G. Effect of estrogens on bone marrow adipogenesis and Sirt1 in aging C57BL/6J mice. Biogerontol. 2009;10:744–755. doi: 10.1007/s10522-009-9221-7. [DOI] [PubMed] [Google Scholar]
- 25.Yao Y, Brodie AMH, Davidson NE, Kensler TW, Zhou Q. Inhibition of estrogen signaling activates the NRF2 pathway in breast cancer. Breast Cancer Res Treat. 2010;124:585–591. doi: 10.1007/s10549-010-1023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laudet V, Hanni C, Coll J, Catzeflis F, Stehelin D. Evolution of the nuclear receptor gene super family. EMBO J. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein RA, McDonnell DP. Estrogen-related receptor alpha as a therapeutic target in cancer. Endocr Relat Cancer. 2006 Suppl.1:S25–S32. doi: 10.1677/erc.1.01292. [DOI] [PubMed] [Google Scholar]
- 28.Vina J, Consuelo BS, Gambini J, Sastre J, Pallardo FV. Why females live longer than males? Importance of the upregulation of longevity-associated genes by oestrogenic compounds. FEBS Letts. 2005;579:2541–2545. doi: 10.1016/j.febslet.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 29.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Konduri SD, Bansal S, Nayak BK, Rajasekaran SZ, Karuppayil SM, et al. Estrogen receptor-α binds p53 tumor suppressor protein directly and represses the function. J Biol Chem. 2006;281:9837–9840. doi: 10.1074/jbc.C600001200. [DOI] [PubMed] [Google Scholar]
- 31.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: Evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 32.Stein RA, Chang CY, Kazmin DA, Way J, Schroeder T, Wergin M, et al. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 2008;68:8805–8812. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguère V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha. Mol Endocrinol. 2010;24:1349–1358. doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem. 2006;281:16271–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- 35.Cheng HL, Mostolavsky R, Saito S, Manis JP, Gu Y, Patel P, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci. USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.