Abstract
The p19ARF tumor suppressor limits ribosome biogenesis and responds to hyperproliferative signals to activate the p53 checkpoint response. While its activation of p53 has been well characterized, ARF’s role in restraining nucleolar ribosome production is poorly understood. Here we report the use of a mass spectroscopic analysis to identify protein changes within the nucleoli of Arf-deficient mouse cells. Through this approach, we discovered that ARF limited the nucleolar localization of the RNA helicase DDX5 which promotes the synthesis and maturation of rRNA, ultimately increasing ribosome output and proliferation. ARF inhibited the interaction between DDX5 and nucleophosmin (NPM), preventing association of DDX5 with the rDNA promoter and nuclear pre-ribosomes. In addition, Arf-deficient cells transformed by oncogenic RasV12 were addicted to DDX5, since reduction of DDX5 was sufficient to impair RasV12-driven colony formation in soft agar and tumor growth in mice. Taken together, our findings indicate that DDX5 is a key p53-independent target of the ARF tumor suppressor and is a novel non-oncogene participant in ribosome biogenesis.
Keywords: ARF, DDX5, NPM, non-oncogene, ribosome biogenesis
Introduction
The role of ARF in regulating p53 is well established, but the mechanisms by which it exerts its p53-independent tumor suppressor function have yet to be fully characterized. A common theme in ARF’s p53-independent activity is its ability to regulate nucleolar ribosome biogenesis11,40, but mechanistic details of its involvement have remained elusive. Understanding the p53-independent functions of ARF in the nucleolus is an increasingly important focus in cancer biology.
The nucleolus is a dynamic organelle that assembles around ribosomal DNA (rDNA) repeats and is the cellular center for ribosome biogenesis. Characterization of the nucleolar proteome has revealed the broad spectrum of resident proteins (1). As nucleoli lack membranes, proteins freely diffuse into and out of nucleoli in response to varying conditions (2). Some of the most important residents of nucleoli are proteins that regulate ribosome production, including p19ARF (p14ARF in humans).
The canonical function of ARF is to activate p53 by binding and sequestering the p53 inhibitor Mdm2 (3–6). Arf-null mice develop spontaneous tumors consisting of predominantly fibrosarcomas and lymphomas (7, 8). However, ARF also possesses p53-independent roles that contribute to its growth-inhibitory function and suppression of tumorigenesis (9). For example, basal ARF maintains nucleolar structure and function (10) at least in part through its ability to interact with nucleophosmin (NPM) (11–15). The ability of ARF to regulate the nucleolar localization of Mdm2 (4) and the nuclear export of nucleophosmin (14) suggests that ARF may monitor nucleolar function by regulating the composition of the nucleolar proteome. To determine how the presence or absence of basal ARF affects nucleolar protein composition, we conducted a proteomic screen using isolated nucleoli from wild type and Arf−/− mouse embryonic fibroblasts (MEFs). Among the proteins enriched in nucleoli in the absence of Arf was DDX5, a DEAD-box protein also known as p68 RNA helicase.
The DEAD-box family of RNA helicases is defined by a conserved Asp-Glu-Ala-Asp motif that interacts with Mg2+ and is involved in ATP hydrolysis (16). DEAD-box proteins also contain several conserved motifs that have been shown to function in ATP binding, ATPase activity, and helicase activity (17). Many cellular functions of DEAD-box RNA helicases have been attributed to RNA duplex unwinding and ribonucleoprotein (RNP) complex remodeling (18). In yeast, several RNA helicases have been demonstrated to facilitate ribosome biogenesis (19), which involves both the processing of ribosomal RNA (rRNA) as well as its assembly into functional RNP complexes. Given that the cellular center for ribosome synthesis is the nucleolus, it is not surprising that many RNA helicases have been identified as components of the nucleolar proteome (1, 2).
The involvement of several known oncogenes and tumor suppressors in the regulation of protein synthesis underscores the importance of ribosomes and mRNA translational control in cancer (20). Thus, the ability of ARF to direct balanced RNA metabolism in the nucleolus could provide insights into how this major cellular axis might impact tumorigenesis. Apart from its classical function as a sensor of hyperproliferative signals (21–23), we now show that ARF limits non-oncogene-driven ribosome biogenesis to inhibit cellular transformation.
Materials and Methods
Cell culture and Reagents
Primary mouse embryonic fibroblasts were isolated and cultured as described (14). Rabbit anti-DDX5 (A300-523A) was purchased from Bethyl Laboratories. Mouse anti-NPM (cat no. 32-5200) was purchased from Zymed. Rat anti-p19ARF (ab26696) was purchased from Abcam. H-Ras, p21, and Gamma-tubulin antibodies were purchased from Santa Cruz Biotechnology.
Nucleolar Isolation
Nucleoli were isolated from 2×108 cells, essentially as described by Andersen and colleagues (1). Additional details for the nucleolar isolation protocol are included with the supplementary material.
Proteomic Analysis
Gel preparation, analysis and mass spectrometry were performed as previously described (24). Wild type nucleolar isolates were labeled with Cy3 and Arf−/− nucleolar isolates were labeled with Cy5. Samples were mixed and subjected to 2D SDS-PAGE. First-dimension isoelectric focusing was performed on immobilized pH gradient strips in an Ettan IPGphor system (GE Healthcare). Second dimension separation was performed on 10% isocratic SDS/PAGE gels (20 × 24cm). Imaging was performed using a Typhoon 9400 scanner (GE Healthcare) and Decyder DIA and BVA software (GE Healthcare) was used to quantify matched gel spots. Spots demonstrating >2-fold differences in intensity were isolated and identified by MALDI-TOF/TOF mass spectrometry.
Immunofluorescence
Cells were fixed with 4% paraformaldeyde in PBS for 10 minutes. Cells were permeabilized with 1% NP-40, blocked in 5% FBS, and stained with rabbit anti-DDX5 and mouse anti-NPM, followed by FITC-conjugated anti-mouse and RhodamineX-conjugated anti-rabbit (both from Jackson Immunoresearch). Samples were counterstained with DAPI and mounted with Vectashield (Vector Labs). Four independent MEF isolates were used to assess localization of DDX5. Images were acquired using a 100X oil immersion lens on a Zeiss LSM5 Pascal Vario Two UGB coupled to Axiovert 200 confocal microscope.
Quantitative real-time PCR
Total RNA was isolated from WT and Arf −/− MEFs using Illustra RNAspin columns (GE Healthcare) according to manufacturer’s protocol. First strand cDNA synthesis and real-time PCR were as previously described (25).
[methyl-3H]-methionine labeling of rRNA
Equal numbers of MEFs were subjected to starvation in methionine-free media containing 10% dialyzed FBS for 15 minutes. Cells were treated with 50 μCi/mL [methyl-3H]-methionine and chased in complete media containing an excess of unlabeled methionine (10 μM) for the indicated times. Samples were lysed in RNASolv reagent (Omega Biotek) and extracted RNA was separated on agarose-formaldehyde gels and transferred to a Hybond XL membrane (GE Healthcare). The membrane was cross-linked and sprayed with En3Hance (Perkin-Elmer) prior to autoradiography. Band intensities were quantitated using ImageQuant TL (Amersham Biosciences).
Chromatin immunoprecipitation
Wild type and Arf−/− MEFs were cross-linked with formaldehyde and cell lysates were immunoprecipitated with the indicated antibodies at 4° C overnight. Samples were then washed with low salt, high salt, LiCl, and TE buffers, prior to elution. Crosslinks were reversed by addition of NaCl and samples were subjected to RNase A and proteinase K treatments. DNA was purified from samples using QIAquick PCR purification kits (QIAGEN). QPCR was performed as detailed above with primers sets specific to rDNA loci. Additional details for the ChIP protocol are provided in supplementary information.
rRNA immunoprecipitation
Arf−/− MEFs were starved as described above and labeled with [methyl-3H]-methionine for 4 hours. Cells were harvested, lysed, and subjected to immunoprecipitation and RNA extraction as previously described (25).
Ribosome Fractionation
Cells were treated with cycloheximide, collected, and fractionated by sucrose gradient centrifugation as previously described (25). Total protein was precipitated from individual fractions with trichloroacetic acid and analyzed by western blot.
Foci formation and proliferation assays
MEFs were plated in triplicate at the indicated concentrations and foci formation and proliferation assays were conducted as previously described (26).
Soft agar
Arf−/− MEFs were infected with shRNAs against luciferase or DDX5, prior to infection with either RasV12 or empty vector (pBabe). Cells were seeded onto soft agar at 104 cells per 6 cm2 dish and grown for 3 weeks. Cells were re-layered with soft agar on a weekly basis and visible colonies were counted after 3 weeks.
Tumorigenesis Assay
Arf−/− MEFs were infected with RasV12 and either shDDX5 or shSCR. Fibroblasts were trypsinized and resuspended in PBS at a concentration of 2×107 cells/mL. Athymic nude mice were injected subcutaneously with 2×106 cells along their left flank, with sample sizes of 5 mice per condition. Tumor size was monitored over an 18-day time course using calipers to measure the tumors in two dimensions. Tumor volume was calculated using the formula:
where height equals the smallest of the two measurements.
Additional Methods can be found in Supplementary Information
Results
p19ARF interferes with the nucleolar localization of DDX5 RNA helicase
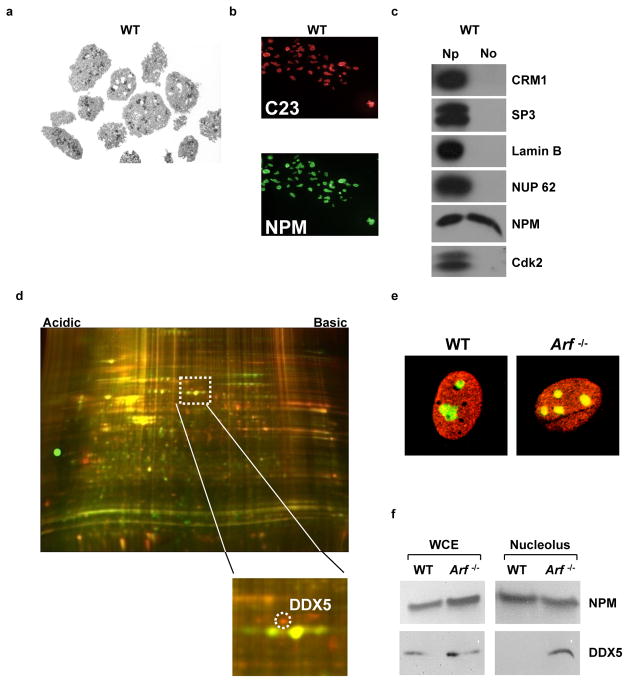
A proteomic screen was conducted to identify targets that displayed differential nucleolar localization in the presence or absence of basal ARF. Adapting a protocol from Andersen and colleagues (1), we isolated nucleoli from wild type and Arf−/− MEFs. Isolated nucleoli maintained in vivo morphology (Fig. 1A), were positive for nucleolar proteins by immunofluorescence (Fig. 1B), and were free of nucleoplasmic contaminants (Fig. 1C). Nucleolar isolates were subjected to comparative two dimensional differential gel electrophoresis (2D-DIGE) proteomic analysis. Twenty-six spots which showed differences greater than 2.5 standard deviations from the mean change were excised, and 19 were positively identified by mass spectroscopy (Supplementary Table 1). Among the differences between wild type and Arf−/− MEFs, enhanced nucleolar expression (10-fold) of DDX5 RNA helicase was observed in the absence of Arf (Fig. 1D). Immunofluorescence revealed enhanced nucleolar co-localization of DDX5 with NPM in Arf−/− MEFs (Fig. 1E). Biochemical fractionation confirmed the increased presence of DDX5 in Arf−/− nucleoli relative to wild-type nucleoli (Fig. 1F).
Figure 1.
ARF maintains the nucleolar exclusion of DDX5. A, Nucleoli were isolated from WT and Arf−/− MEFs. Nucleolar morphology (shown for WT) was assessed by electron microscopy. B, Nucleoli (shown for WT) were analyzed by immunofluorescence microscopy for the nucleolar markers C23/nucleolin (red, Texas Red) and NPM (green, FITC). C, Immunoblotting of nucleolar and nucleoplasmic fractions was performed to determine purity (shown for WT). D, Proteins from isolated nucleoli were differentially labeled with Cy3 and Cy5 fluorophores and were subjected to 2-D DIGE. E, Localization of NPM and DDX5 in wild type and Arf−/− MEFs was determined by immunofluorescence. F, Western blotting of nucleolar lysates for NPM and DDX5 revealed a change in nucleolar DDX5 expression between genotypes.
To investigate whether nucleolar exclusion of DDX5 is mediated by ARF through its activation of p53, we treated Arf−/− MEFs with nutlin-3, a pharmacological inhibitor of Mdm2. Instead of stimulating DDX5 nucleolar exclusion, nucleolar localization of DDX5 persisted in the presence of nutlin-3 (Fig. S1). This demonstrates that p53 activation is not responsible for the ARF-dependent nucleolar exclusion of DDX5 observed in wild type MEFs, consistent with a p53-independent role for ARF in regulating DDX5 localization.
ARF regulates the association of DDX5 with rDNA, rRNA, and nuclear pre-ribosomes
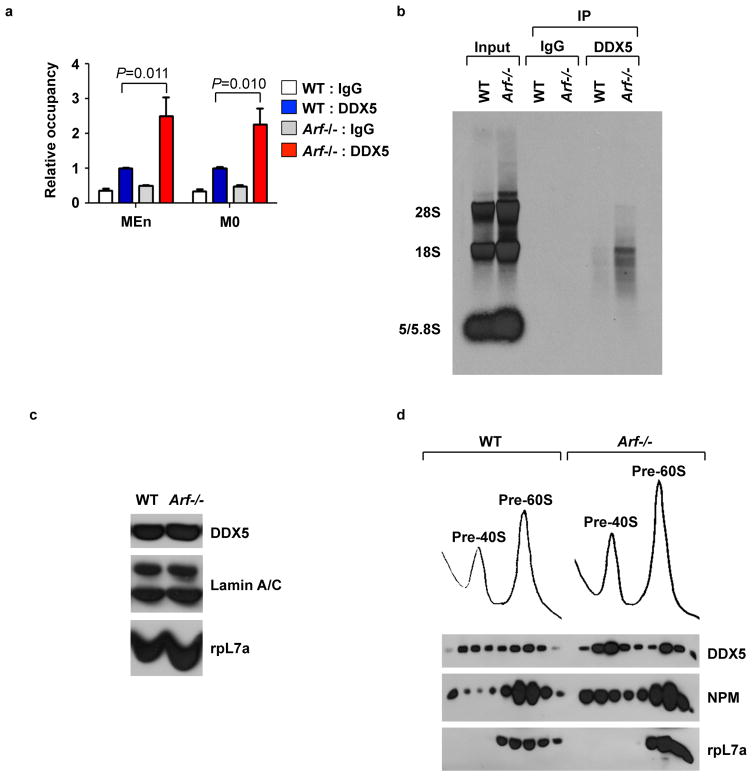
The nucleolar localization of DDX5, along with its function as an RNA helicase, suggested that DDX5 might be involved in the biogenesis of rRNA. The regulation of DDX5 localization by basal ARF led us to investigate whether ARF could control ribosome biogenesis through regulation of DDX5 function. Both p19ARF (mouse) and p14ARF (human) inhibit rRNA transcription (10, 27, 28), and DDX5 has been ascribed roles as a transcriptional regulator (17). However, it is unknown whether DDX5 regulates transcription at nucleolar rDNA loci. We conducted chromatin immunoprecipitation experiments to determine whether DDX5 associated with the rDNA promoter at two previously identified binding sites of the RNA polymerase I transcription factor UBF (29). ARF regulated DDX5 association with these sites, such that DDX5 occupancy at the rDNA promoter was over two-fold greater in Arf−/− MEFs compared to wild-type MEFs (Fig. 2A).
Figure 2.
ARF impairs association of DDX5 with nuclear pre-ribosomes. A, Wild type and Arf−/− MEFs were collected for chromatin immunoprecipitation using DDX5 or IgG control antibodies. QPCR with primers flanking two regions, MEn and M0, on the rDNA promoter was used to amplify DNA isolated from the immunoprecipitates. B, Wild type and Arf−/− MEFs were labeled with [methyl-3H]-methionine and DDX5 was immunoprecipitated from cell lysates. Radiolabeled RNA isolated from the DDX5 immunoprecipitate was visualized by autoradiography. C, Nuclear extract from wild type and Arf−/− MEFs was analyzed by western blot. D, Nuclear extracts from wild type and Arf−/− MEFs were subjected to sucrose density centrifugation. RNA absorbance was monitored at 254 nm as samples were fractionated and isolated proteins were analyzed by western blot.
Additionally, DDX5 has been shown to be involved in processing of the 5.8S rRNA (30) and the 28S rRNA from their respective rRNA precursors (31). By immunoprecipitation, we observed an interaction between DDX5 and the 28S and 18S rRNA species (Fig. 2B). This association with mature rRNA suggests that DDX5 could be involved at multiple stages in the production and assembly of ribosomes. In wild-type MEFs the interaction of DDX5 with rRNA was decreased relative to Arf−/− cells, suggesting that ARF can inhibit this association as well.
We hypothesized that ARF may interfere with the ability of DDX5 to stimulate ribosome biogenesis by impeding access of DDX5 to maturing pre-ribosomes. Nuclear lysates obtained from wild-type and Arf−/− MEFs were separated by sucrose gradient centrifugation. Enhanced association of DDX5 with the 40S and 60S pre-ribosomal fractions was observed in the Arf−/− nuclear lysates relative to the corresponding wild-type fractions (Fig. 2D). These changes were not due to altered expression since wild-type and Arf−/− MEFs expressed similar levels of DDX5 protein in both whole cell lysate (Fig. 1F) and nuclear extract (Fig. 2C).
DDX5 enhances the synthesis and processing of ribosomal RNA
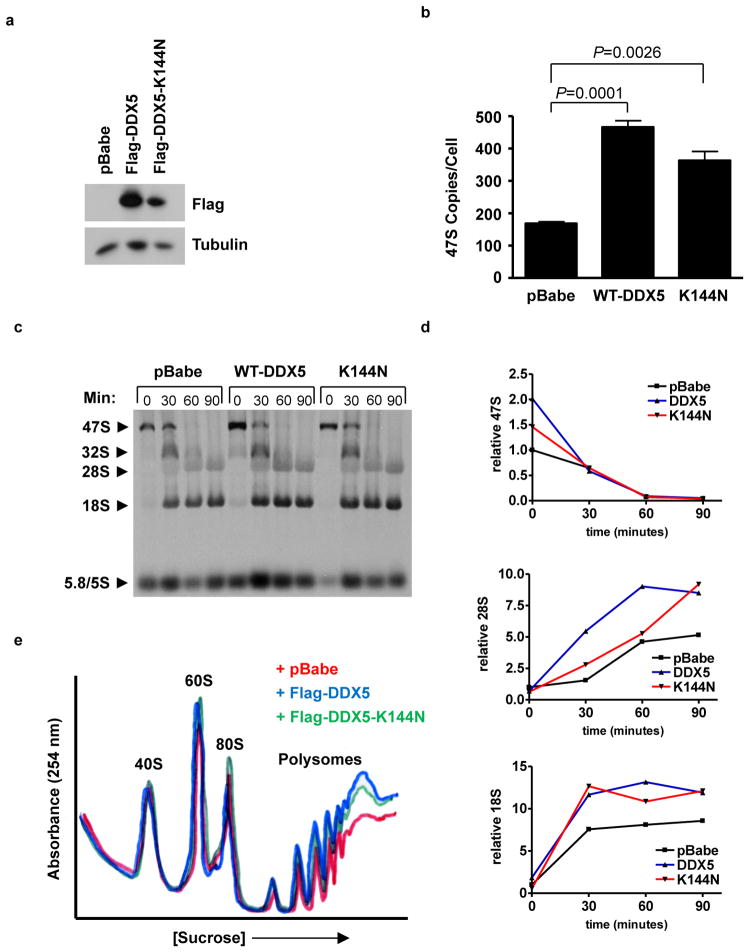
In order to determine whether DDX5 could accelerate ribosome biogenesis, wild-type MEFs were transduced with a Flag-epitope-tagged DDX5 or a mutant (K144N) deficient in ATP binding (Fig. 3A). The K144N mutation in the Walker A motif abrogates not only ATP binding, but also the ATPase and helicase activities of DDX5 (31). The earliest observed effect of DDX5 on ribosome biogenesis was at the level of 47S pre-rRNA transcription, where both Flag-DDX5 and Flag-DDX5-K144N increased the amount of 47S transcript per cell (Fig. 3B). The ability of DDX5 to regulate transcription of 47S pre-RNA concurred with its aforementioned association at the rDNA promoter. Monitoring the processing of the 47S pre-rRNA transcript by pulse-chase analysis, we discovered a more rapid accumulation of mature 28S and 18S rRNAs in cells expressing Flag-DDX5 or Flag-K144N versus vector-transduced cells (Figs. 3C and D). To determine whether the accelerated production of rRNA equated with increased protein synthesis, cytosolic fractions were collected for ribosome profile analysis. Both Flag-DDX5 and Flag-DDX5-K144N enhanced the amplitude of the actively translating polyribosome fraction (Fig. 3E), indicating that ectopic expression of Flag-DDX5 ultimately increases ribosome availability for translation, and that helicase activity is not required for this induction. These results indicate that DDX5 stimulates the production of functional ribosomes by increasing the total amount of mature rRNA.
Figure 3.
Overexpression of DDX5 promotes ribosome output. Wild-type MEFs were transduced with empty vector or Flag-DDX5 retroviruses. A, Flag immunoblot demonstrates expression of the retroviral fusion protein. B, Total RNA was analyzed by QPCR to determine copy number of the 47S pre-rRNA transcript. C, Cells were labeled with [methyl-3H]-methionine and chased for the indicated times. Total RNA was extracted, separated on an agarose gel, and transferred to membranes. Radiolabeled RNA was visualized by autoradiography. D, Relative band intensities were determined for rRNA in the processing assay. 47S, 28S, and 18S rRNAs were individually normalized to the pBabe sample at t=0 and tracked throughout the time course. E, Cytosolic extracts from 2.5 × 106 cells were separated by sucrose density gradient centrifugation. Ribosome profiles were obtained by measuring the absorbance of RNA at 254 nm.
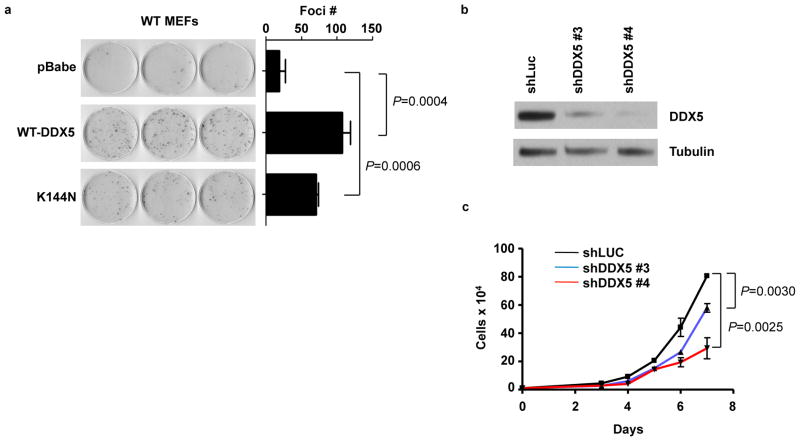
DDX5 stimulates proliferation in mouse embryonic fibroblasts
The ability of DDX5 to stimulate rRNA synthesis suggested that it might also be critical for growth and proliferation. The enhanced ribosome biogenesis caused by DDX5 overexpression corresponds to an increased proliferative capacity as evidenced by the ability of Flag-DDX5 and Flag-DDX5-K144N to stimulate foci formation in wild-type MEFs (Fig. 4A). Furthermore, using two different shRNA constructs, we demonstrated that knockdown of DDX5 reduced proliferation of Arf −/− MEFs in a dose-dependent manner (Figs. 4B and C). The dependency on DDX5 for unrestricted growth was not exclusive to Arf−/− MEFs, as foci formation in p53−/− MEFs was impaired by shRNAs targeting DDX5 (Fig S2A and B). DDX5 has been linked to p53 function in several reports, either as a transcriptional co-activator (32), or as a partner of p53 in microRNA processing (33). While these relationships suggest that DDX5 could inhibit growth through its interactions with p53, our data point to the opposite conclusion, specifically that the dominant role of DDX5 is not growth inhibition as would be inferred from the aforementioned studies, but rather growth stimulation.
Figure 4.
DDX5 stimulates growth and proliferation.. A, Wild type MEFs expressing empty vector, Flag-DDX5 or Flag K144N were plated at 103 cells per dish and grown for 12 days. Foci were fixed in methanol and stained with Giemsa. B, Arf−/− MEFs were infected with lentiviral shRNAs targeting luciferase (control) or DDX5. Western blot demonstrates the efficacy of DDX5 knockdown. C, Cells were plated in triplicate at a density of 104 per well in a 6-well plate for a proliferation assay and counted over a 7-day time course.
Knockdown of DDX5 phenocopies the p53-independent functions of ARF on ribosome output
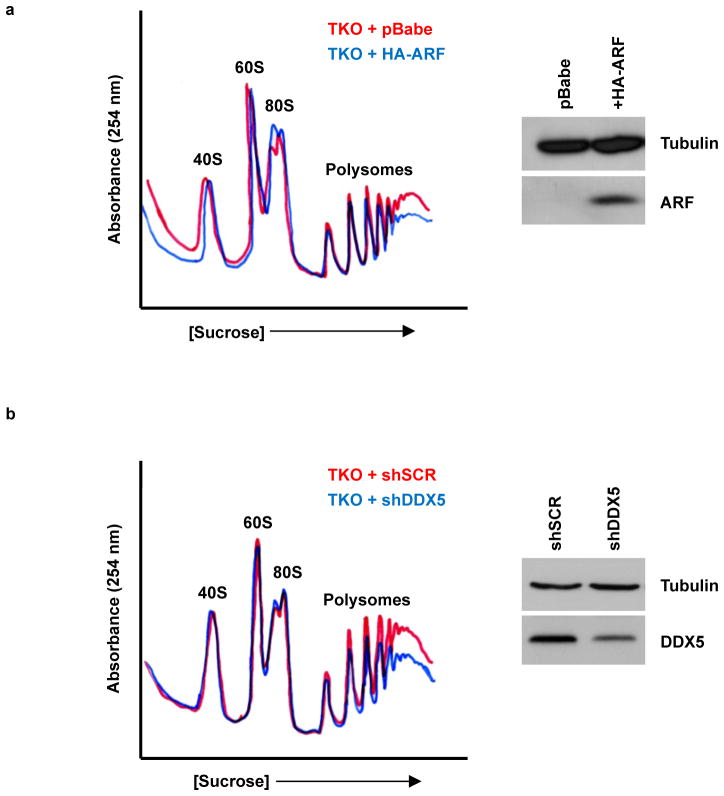
DDX5 stimulates ribosome production, whereas ARF inhibits ribosome biogenesis at several stages: 47S transcription, rRNA processing, and rRNA export (10, 28, 34). Ultimately, the effects of Arf loss are exhibited by the enhanced ribosome profiles of Arf−/− MEFs relative to wild-type MEFs (10). It was unclear, however, whether these effects of ARF on the cellular ribosome profile were truly p53-independent. To characterize the p53-independent functions of ARF on ribosome biogenesis, we utilized TKO (p53−/−; Mdm2−/−; Arf −/−) MEFs, in which the entire ARF-Mdm2-p53 axis has been removed (9). By adding ARF back into TKO MEFs we investigated growth-inhibitory effects of ARF that are completely independent of p53. HA-ARF expression reduced cytosolic ribosomes, most notably in the actively translating polyribosome fraction (Fig. 5A), demonstrating a p53-independent role for ARF in the regulation of ribosome output. Knockdown of DDX5 in TKO MEFs mimicked the effects of ARF overexpression on cytosolic ribosome content (Fig. 5B), causing a notable decrease in polyribosome peak amplitude. Thus, a DDX5 loss-of-function is equivalent to a p53-independent ARF gain-of-function on ribosome output.
Figure 5.
ARF overexpression and DDX5 knockdown each reduce cytosolic polyribosomes in a p53-independent manner. Cytosolic extracts from TKO (Arf/p53/Mdm2−/−) MEFs expressing A, HA-ARF or B, shDDX5 were loaded onto a sucrose density gradient and samples were monitored for the absorbance of RNA at 254 nm. (Side panels) Expression of ARF and DDX5 were assessed by western blot.
ARF inhibits the interaction between DDX5 and NPM
We previously identified an interaction between NPM and DDX5 while probing for NPM binding partners (25). Like DDX5, NPM is a multifunctional protein with key roles at multiple stages of ribosome biogenesis. NPM associates with the rDNA locus (35), regulating transcription and processing of the rRNA (11). Further, NPM functions as a nuclear export chaperone for ribosomes (25), a function that is antagonized by ARF (14). Interestingly, early embryonic lethality is a phenotype of both Npm1−/− and Ddx5−/− mice (12, 30, 36). We hypothesized that ARF impaired DDX5 function through regulation of its interaction with NPM.
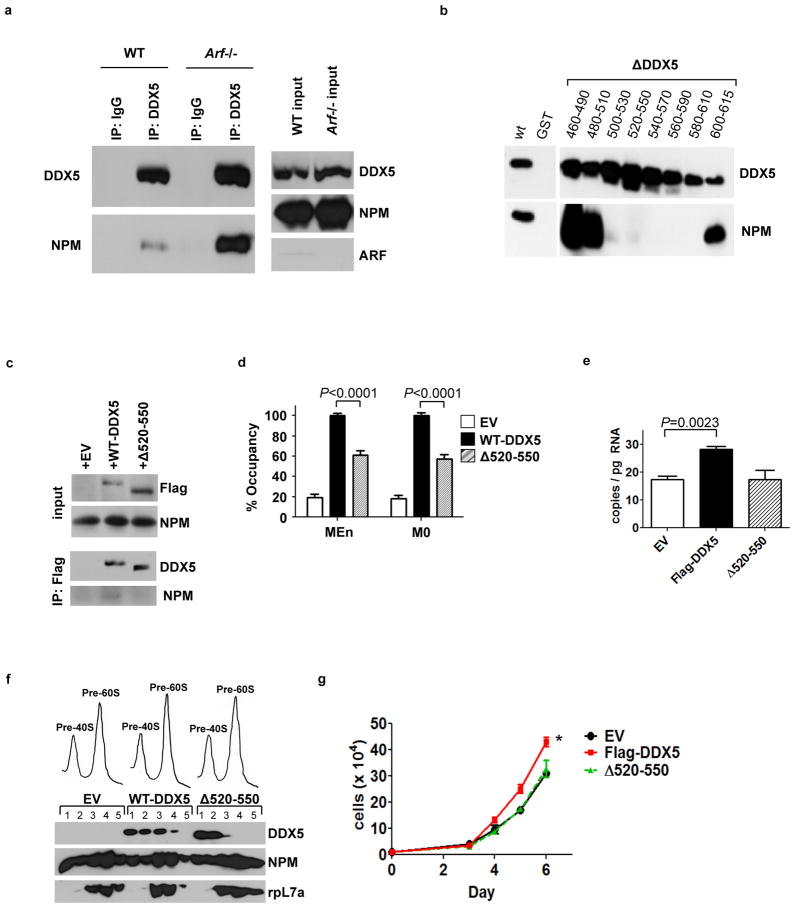
Given the ability of ARF to regulate both proteins individually, we tested whether ARF effected the interaction between DDX5 and NPM. Comparison of WT and Arf−/− MEF lysates by co-immunoprecipitation revealed that ARF significantly reduced the interaction of DDX5 with NPM (Fig. 6A). We then sought to determine the NPM-binding domain on DDX5 to assess whether this interaction was critical for the growth-stimulatory abilities of DDX5. Little has been reported on the proteins that interact with DDX5 through its C-terminal domain. Given the possibility that core domain mutations might directly impair conserved features that are critical in the DEAD-box helicase family and complicate any interpretations of its overall importance, we instead focused on mutations in the C-terminus. A panel of overlapping C-terminal deletion mutations was introduced to DDX5 in a GST-fusion protein expression vector. In vitro immunoprecipitation reactions using His-tagged NPM and GST-DDX5 or its mutants mapped an NPM interaction motif to residues 500–610 at the C-terminus of DDX5 (Fig. 6B). For further experiments, we chose a smaller mutant within this domain, DDX5Δ520–550. While ectopically expressed Flag-DDX5 interacted with endogenous NPM in Arf−/− MEFs, the Δ520–550 mutant displayed no visible interaction (Fig. 6C). Flag-DDX5-Δ520–550 also had reduced occupancy of the rDNA promoter compared to wild-type Flag-DDX5 (Fig. 6D), and did not stimulate 47S pre-rRNA transcription (Fig. 6E). Further, while Flag-DDX5 associated with nuclear pre-ribosomal fractions containing the 40S and 60S ribosomal subunits, Flag-DDX5-Δ520–550 was almost completely absent from the 60S fractions containing the large ribosomal protein rpL7a (Fig. 6F). Finally, in transduced Arf−/− MEFs, Flag-DDX5-Δ520–550 expression did not affect proliferation compared to the empty vector control, whereas Flag-DDX5 expression enhanced proliferation (Fig. 6G). Thus, it appears that DDX5 cooperates with NPM, through a direct interaction that is antagonized by ARF, to stimulate rRNA synthesis and proliferation.
Figure 6.
The ARF-regulated interaction between DDX5 and NPM is required for the growth-stimulatory effects of DDX5. A, The interaction between DDX5 and NPM was compared in lysates from wild type (WT) and Arf−/− MEFs by co-immunoprecipitation with antibodies against DDX5. B, A panel of GST-Flag-DDX5 mutants was subjected to GST-pull-down following incubation with His-NPM proteins and immunoblotting was performed with antibodies recognizing DDX5 and NPM. C, Arf−/− MEFs expressing Flag-DDX5 or Flag-DDX5- 520–550 were subjected to immunoprecipitation with an antibody against the Flag epitope. D, Arf−/− MEFs were subjected to chromatin immunoprecipitation reactions with an antibody against the Flag epitope. DNA recovered from the reactions was subjected to QPCR using primers to two different areas, MEn and M0, within the rDNA promoter. E, QPCR was performed for the 47S pre-rRNA from total RNA isolated from wild type MEFs. F, Nuclei from Arf−/− MEFs expressing Flag-DDX5 or Flag-DDX5- 520–550 were subjected to sucrose gradient centrifugation. Expression of rpL7a or Flag-tagged proteins in the isolated fractions was determined by western blot. G, Arf−/− MEFs were plated in triplicate at 20,000 cells per well for a proliferation assay and counted daily over a time course. *, P=0.0058.
RasV12-induced transformation of Arf−/− MEFs requires DDX5
Transduction of wild-type MEFs with oncogenic RasV12 results in ARF induction and growth arrest (21). Conversely, transduction of RasV12 transforms Arf−/− MEFs, as determined by colony formation in soft agar. To determine whether DDX5 meets the criteria of a classic oncogene, wild-type MEFs expressing Flag-DDX5 alone or in combination with RasV12, were plated in soft agar to evaluate anchorage-independent growth. While RasV12-transduced Arf−/− MEFs plated in parallel formed robust colonies, wild-type MEFs expressing Flag-DDX5 and RasV12 did not form colonies (Supplementary Fig. S3A). Further, unlike RasV12, Flag-DDX5 was unable to stimulate transformation of TKO MEFs (Supplementary Fig. S3B). This suggests that DDX5 is not an oncogene, as it cannot, in combination with Arf loss and p53 loss, or RasV12 overexpression, drive transformation.
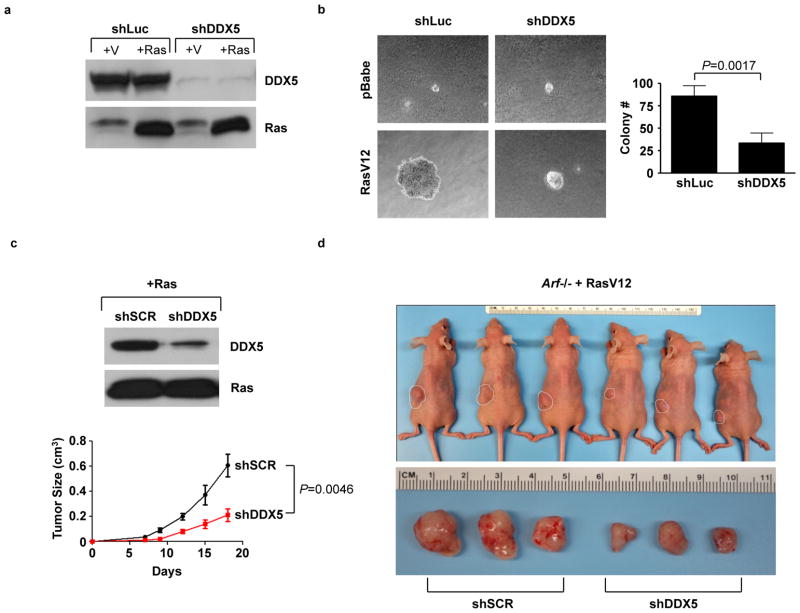
Despite not being sufficient to transform cells, it remained possible that DDX5 was necessary for transformation. To determine whether DDX5 is required for oncogenic transformation in the absence of Arf, we transduced Arf−/− MEFs with shRNA against DDX5 followed by ectopic expression of RasV12 (Fig. 7A). Knockdown of DDX5 impaired the ability of RasV12 to stimulate colony formation and anchorage-independent growth (Fig. 7B), suggesting that transformation of MEFs by RasV12 requires the cooperation of DDX5.
Figure 7.
Non-oncogene addiction to DDX5 in transformed Arf−/− MEFs. A, Arf−/− MEFs were infected with shRNAs against luciferase or DDX5. Cells were then infected with either oncogenic RasV12 or empty vector (pBabe) and expression was confirmed by western blot. B, Cells were seeded onto soft agar and grown for 3 weeks. C, Arf−/− MEFs were infected with RasV12, then subsequently infected with shDDX5 or scrambled shRNA. 2×106 cells were used for subcutaneous injection into nude mice. Tumor burden was monitored over an 18-day time course. At the endpoint, mice were sacrificed and tumors were excised and photographed. Images of three representative mice and their tumors (the 3 mice with tumor volumes closest to the median) are displayed in D.
To determine whether Ras-transformed fibroblasts could form tumors in vivo, Arf−/− MEFs transduced with RasV12 and shDDX5 or a scrambled shRNA were subcutaneously inoculated into the flanks of nude mice. RasV12-induced tumor growth in nude mice was reduced by knockdown of DDX5 (Figs. 7C and D). The dependence on DDX5 for the growth of these Arf-null tumors suggests that DDX5 may function as a non-oncogene by sustaining the levels of ribosome production required by transformed cells to maintain their accelerated proliferation rates.
Discussion
The role of ARF in regulating p53 is well established, but the mechanisms by which it exerts its p53-independent tumor suppressor function have yet to be fully characterized. Our group and others have recently demonstrated the regulation of translation by ARF, but mechanistic details of its involvement are limited. Both mouse and human ARF interact with nucleolar proteins involved in ribosome biogenesis as well as ribosomal components themselves (11, 37). Furthermore, ectopic expression of human p14ARF decreases polyribosomes in a p53-independent manner (37). ARF has recently been linked to ribosome biogenesis through its regulation of TTF1 (28) and its ability to inhibit ribosome export via its nucleolar interaction with NPM (14, 38). Here we have shown that ARF can control the protein composition of the nucleolus, the central organelle in ribosome biogenesis. Our observation that ARF can regulate DDX5 RNA helicase, provides a mechanistic explanation for the inhibitory effects of ARF on 47S rRNA transcription and processing (34).
Our data suggest that most of the endogenous pool of DDX5 may be excluded from nucleoli and inactive in ribosome biogenesis until a cellular perturbation stimulates this activity. Consistent with this model, upon loss of Arf a substantial increase in nucleolar DDX5 was observed, accompanied by tremendous gains in ribosome production. Surprisingly, both DDX5 and the helicase-dead DDX5 mutant (K144N) were able to stimulate 47S transcription and cellular ribosome output. The ability of DDX5-K144N to increase 47S pre-rRNA transcription is consistent with reports that helicase activity may be dispensable for the activities of DDX5 as a transcriptional co-regulator (32, 39). NPM was important for DDX5 to associate with the rDNA promoter and to facilitate 47S pre-rRNA transcription. The DDX5 NPM-binding mutant was also unable to associate with the nuclear 60S pre-ribosomal fraction or enhance proliferation, further underscoring the link between the effects of DDX5 on ribosome biogenesis with those on growth and proliferation. Clearly, the formation of DDX5-NPM complexes, enhanced in the absence of Arf, is necessary for the nucleolar gain-of-function activity reported here for DDX5.
Our results provide a new perspective for understanding the tumor suppressor function of ARF, which has classically been thought of as a checkpoint sensor of hyperproliferative signals. The data presented here suggest that an equally important mechanism by which ARF functions as a tumor suppressor is to limit ribosome output as a defense against oncogene activation and the attendant enhanced cellular protein requirements. Therefore, in the absence of Arf, DDX5 becomes a requisite non-oncogene effector that promotes an increased translational output in accord with the higher demand for protein production required upon oncogene activation. The ability of ectopic DDX5 expression to stimulate ribosome biogenesis and growth further proves the central role of DDX5 in regulating this translational output.
Our data showing the growth-stimulatory functions of DDX5 in ribosome biogenesis provides a strong rationale to explain the link between DDX5 and cancer. Although still in its infancy, most non-oncogenes are thought of as critical regulators of cellular stress responses and that their expression provides cancer cells the means to tolerate multiple stresses (40). It is unclear how DDX5 and ribosome biogenesis fit into this stress tolerance model. Rather, DDX5 may represent a class of non-oncogenes whose activities are unleashed in the absence of crucial tumor suppressors. In this setting, the role of the DDX5 non-oncogene is to make a required cellular process, such as ribosome biogenesis, more efficient or prolific in preparation for the tremendous protein synthesis demands following malignant transformation. It remains to be determined whether DDX5 will be an efficacious target in the treatment of cancer; however our results validate its importance in supplying the sustained ribosome output required for oncogenic transformation. In summary, DDX5 participation in ribosome biogenesis is negatively regulated by ARF, which inhibits the DDX5-NPM interaction, suggesting a dynamic interplay through which ARF and DDX5 duel for nucleolar growth control.
Supplementary Material
Acknowledgments
The authors would like to thank the members of the Weber laboratory for their advice and technical assistance and Greg Longmore, Alan Diehl, Charles Sherr and Martine Roussel for insightful comments and reagents. A.J.S. was supported by Komen for the Cure KG091234. This work was supported by NIH grant CA120436, American Cancer Society grant RSG-08-286-01-GMC, and an Era of Hope Scholar Award in Breast Cancer Research (BC007304) to J.D.W.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 2.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 3.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292–7. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–6. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 5.Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–23. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–34. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 7.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–59. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 8.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–22. [PubMed] [Google Scholar]
- 9.Weber JD, Jeffers JR, Rehg JE, Randle DH, Lozano G, Roussel MF, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14:2358–65. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apicelli AJ, Maggi LB, Jr, Hirbe AC, Miceli AP, Olanich ME, Schulte-Winkeler CL, et al. A non-tumor suppressor role for basal p19ARF in maintaining nucleolar structure and function. Mol Cell Biol. 2008;28:1068–80. doi: 10.1128/MCB.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–96. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, et al. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–86. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, et al. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25:1258–71. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–38. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enomoto T, Lindstrom MS, Jin A, Ke H, Zhang Y. Essential role of the B23/NPM core domain in regulating ARF binding and B23 stability. J Biol Chem. 2006;281:18463–72. doi: 10.1074/jbc.M602788200. [DOI] [PubMed] [Google Scholar]
- 16.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–41. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 17.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–15. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–63. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–52. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–92. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 21.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–6. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 22.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–33. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Stanchina E, McCurrach ME, Zindy F, Shieh SY, Ferbeyre G, Samuelson AV, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–42. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bredemeyer AJ, Lewis RM, Malone JP, Davis AE, Gross J, Townsend RR, et al. A proteomic approach for the discovery of protease substrates. Proc Natl Acad Sci U S A. 2004;101:11785–90. doi: 10.1073/pnas.0402353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maggi LB, Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol Cell Biol. 2008;28:7050–65. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelletier CL, Maggi LB, Jr, Brady SN, Scheidenhelm DK, Gutmann DH, Weber JD. TSC1 sets the rate of ribosome export and protein synthesis through nucleophosmin translation. Cancer Res. 2007;67:1609–17. doi: 10.1158/0008-5472.CAN-06-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene. 2006;25:7577–86. doi: 10.1038/sj.onc.1209743. [DOI] [PubMed] [Google Scholar]
- 28.Lessard F, Morin F, Ivanchuk S, Langlois F, Stefanovsky V, Rutka J, et al. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol Cell. 2010;38:539–50. doi: 10.1016/j.molcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 29.O’Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol. 2002;22:657–68. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 31.Jalal C, Uhlmann-Schiffler H, Stahl H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 2007;35:3590–601. doi: 10.1093/nar/gkm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, et al. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. Embo J. 2005;24:543–53. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 34.Sugimoto M, Kuo ML, Roussel MF, Sherr CJ. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell. 2003;11:415–24. doi: 10.1016/s1097-2765(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 35.Murano K, Okuwaki M, Hisaoka M, Nagata K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol Cell Biol. 2008;28:3114–26. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–53. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 37.Rizos H, McKenzie HA, Ayub AL, Woodruff S, Becker TM, Scurr LL, et al. Physical and functional interaction of the p14ARF tumor suppressor with ribosomes. J Biol Chem. 2006;281:38080–8. doi: 10.1074/jbc.M609405200. [DOI] [PubMed] [Google Scholar]
- 38.Yu Y, Maggi LB, Jr, Brady SN, Apicelli AJ, Dai MS, Lu H, et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol Cell Biol. 2006;26:3798–809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen ED, Niu L, Caretti G, Nicol SM, Teplyuk N, Stein GS, et al. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. J Cell Biochem. 2008;103:1438–51. doi: 10.1002/jcb.21526. [DOI] [PubMed] [Google Scholar]
- 40.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.