Abstract
Background
Polymerase chain reaction (PCR) assays increase the rate of viral detection in clinical specimens, compared with conventional virologic methods. Studies suggest that PCR may detect virus nucleic acid (NA) that persists in the respiratory tract.
Methods
We analyzed virologic data from children having frequent upper respiratory infections (URI) who were followed in a longitudinal study. Nasopharyngeal secretions (NPS) were collected at URI onset and when acute otitis media (AOM) was diagnosed; virus studies were performed using conventional diagnostics and PCR. Repeated presence of adenovirus by PCR was further studied by sequencing and phylogenetic analysis.
Results
Of 581 URI episodes in 76 children, 510 viruses were detected. Of the viruses detected by PCR, 15% were those detected previously; repeated positives occurred most frequently with adenovirus. Sequencing results were available in 13 children with repeated adenovirus detection; four patterns of infection were identified (16 instances): 1) adenovirus of the same serotype and strain detected continuously (n=8 instances), 2) adenovirus of different serotypes detected during sequential URI episodes (n=3), 3) adenovirus of the same serotype but different strains detected during sequential URI episodes (n=3) and 4) adenovirus of the same serotype and strain detected intermittently (n=2).
Conclusions
Among children with frequent URIs, repeated positive PCR results for adenovirus NA may represent a new serotype/strain, or persistence of viral NA. Results must be interpreted with caution; clinical correlation and presence of other viruses are important. Further longitudinal studies of children during and after infection are required for better understanding of the clinical significance of positive PCR tests for adenovirus NA in the respiratory tract .
Keywords: upper respiratory tract infection, nasopharyngeal secretion, virus detection, PCR, adenovirus
INTRODUCTION
Respiratory viral infections are universal in young children and contribute to a high burden of morbidity and occasional mortality1-3. Polymerase chain reaction (PCR) assays reportedly have greater sensitivity than conventional viral assays,4,5 leading to an increased viral detection rate4,6,7. PCR has been used to provide detailed molecular identification of adenovirus in previous epidemiologic studies8,9. The prolonged presence of adenovirus nucleic acid (NA) in respiratory secretions following upper respiratory tract infection (URI) has not been studied, but prolonged adenovirus shedding has been described10. Adenovirus has been isolated from 6% of rectal samples and 0.6-3%of throat samples from healthy children11-13. In addition, adenovirus NA has been found in tonsil and adenoid tissue of young children undergoing routine tonsillectomy14. These studies suggest that, following initial infection, adenovirus NA can persist in the respiratory tract and be shed intermittently. Careful phylogenetic studies have not been performed to elucidate whether this phenomenon is caused by persistent infection with one adenovirus strain or caused by subsequent infections with different adenovirus strains.
We have previously shown in a prospective, longitudinal study of children with acute otitis media (AOM) complicating URI that children often test positive repeatedly by PCR for adenovirus15. This observation led us to perform a detailed phylogenetic analysis in a subset of children with repeated adenovirus infections. We herein describe a series of cases illustrating four distinct patterns associated with the repeated presence of adenovirus NA in nasopharyngeal secretions (NPS) during URI episodes.
MATERIALS AND METHODS
Study Design and Subjects
An analysis was performed using data from a prospective, longitudinal, cohort study of AOM complicating URI in children (January 2003 through March 2007)15 . Healthy children were enrolled at 6 to 35 months of age; each child was followed for one year to document URI and AOM occurrences. Data on demographic and otitis media risk factors were collected. Parents were asked to notify the study personnel as soon as the child developed symptoms of URI: nasal congestion, rhinorrhea, sore throat, cough, with or without fever. Children were seen by a study physician as soon as possible after URI onset, with a follow-up visit occurring 3-7 days later. Two additional home visits were performed at 2 and 3 weeks after URI onset. Children were also seen anytime parents reported AOM symptoms.
Virologic Studies
NPS were collected during each initial URI visit and at subsequent follow-up visits only when AOM was diagnosed. Conventional viral diagnostics included tissue culture isolation and respiratory syncytial virus (RSV) antigen detection by enzyme immunoassay (EIA) during RSV season. PCR was performed only on samples collected during the initial URI episode for which culture/RSV-EIA were negative. Real time-PCR assay was used for detection of adenovirus, rhinovirus, enterovirus and coronavirus (OC43, 229E, and NL 63), and RT-PCR with electronic microarray detection (Nanochip® 400 system) was used for detection of RSV, parainfluenza types 1-3, and influenza A & B15-17. The PCR assays were performed at the Medical College of Wisconsin (Midwest Respiratory Virus Program).
Children having frequent symptomatic URI episodes (≥ 4 episodes in 6 months) in which viruses were detected by PCR were further studied. We first reviewed the longitudinal data for each virus and separated the data by whether they were ‘positive once’ or ‘repeatedly positive’ (see definitions in footnotes of table).
Adenovirus Sequencing and Phylogenetic Analysis
Cases with repeated positive PCR results for adenovirus were further studied for genetic analysis. Sequence analysis was performed using a protocol adopted from Lu and Erdman18. In brief, the hypervariable region of the adenovirus hexon gene was sequenced using a DNA Analyzer 3730 (Applied BioSystems, Foster City, CA). Sequence chromatograms were analyzed using the Staden package (preGap4 and Gap4)19 and the Seqman II program of DNAstar's Lasergene 6.0 package (DNAstar, Madison, WI). Chromatograms were manually verified and corrected when there were obvious inaccuracies. Adenovirus serotyping was performed using the “nucleotide BLAST” feature available on the National Center for Biotechnology Information's website. Following BLAST analysis, the sample was assigned a serotype based on that of the sequence showing the greatest homology to the sample (e.g. serotype 1, 2, 3, 4, etc; consistent with international adenovirus classification system).
After serotype assignment, sequences were aligned with Clustal W in the MEGA (molecular evolutionary genetics analysis) analytical package ( version 4.0)20, nucleotide comparison was performed using the MEGA analytical package (version 4.0)20 to compare the sequences of all viruses obtained from an individual child. The analyses were based on a 500 base pair (bp) region for which all samples had complete genetic information. It should be noted that sequence length varied slightly based on insertions or deletions in this region. We assumed that 98-100% similarity between two virus isolates is indicative of an identical strain
RESULTS
Study Population and Overall Virologic Data
Seventy-six of 294 (26%) children enrolled in the original study had ≥ 4 URI episodes in 6 months; 49% were male; 54% were Caucasian, 29% Black, 4% Asian, and 13% biracial. The mean age at enrollment was 15 months (range 6-33 months); the median age was 13.9 months. The mean follow up duration was 11.5 months. In these 76 children, 581 URI episodes (an average of 7 episodes per child-year) were evaluated. At least one respiratory virus was detected in 375 (65%) of the episodes. Of these 375 episodes- one virus was detected in 281 (75%); 2 viruses in 69 (18%), 3 viruses in 23 (6%), and 4 viruses in 2 (<1%). The number of viruses identified by each detection methods is shown in Table 1. Of the 510 viruses detected, adenovirus was the most common, followed by rhinovirus and enterovirus; collectively these 3 viruses represent 75% of all viruses detected (Table 1). Four-hundred and thirty-three (85%) viruses were detected once by any method, and 77 (15%) were viruses classified as ‘repeatedly positive’ by PCR as seen in Table 2. (See footnote for definitions). Repeated positive PCR occurred more frequently with adenovirus, rhinovirus and enterovirus combined (76 of 382 = 20%), than with other viruses (1 of 128 = 1%), p<0.001; chi square test. One child had parainfluenza 2 detected 20 days apart by PCR.
Table 1.
Viruses detected from nasopharyngeal secretions from 76 children by method of detectiona.
| Virus | Conventional assays (%) | PCR (%) | All methodsb (%) |
|---|---|---|---|
| Adenovirus | 21 (13) | 138 (87) | 159 (31) |
| Rhinovirus | 28 (23) | 93 (77) | 121(24) |
| Enterovirus | 18 (18) | 84 (82) | 102 (20) |
| Coronavirusc | 0 (0) | 41 (100) | 41(8) |
| Parainfluenza virusd | 17 (55) | 14 (45) | 31(6) |
| Respiratory syncytial viruse | 21 (81) | 5 (19) | 26 (5) |
| Herpes simplex virus | 4 (100) | 0 (0) | 4(1) |
| Influenzaf | 19 (73) | 7 (27) | 26(5) |
| Total | 128 | 382 | 510 |
510 viruses were detected by all methods from samples collected from 76 children (581 URI episodes). Conventional assays included tissue culture isolation and RSV-EIA (during RSV season).
Column percent, percent of all viruses detected
Coronavirus OC43, 229E, NL-63
Types 1,2,3
RSV- was detected by culture and/or EIA; EIA positive only = 14
Influenza A and B
Table 2.
| Virus | Total Number of viruses detected | Positive Oncea (%) | Positive Repeatedlyb (%) |
|---|---|---|---|
| Adenovirus | 159 | 124 (78) | 35 (22) |
| Rhinovirus | 121 | 105 (87) | 16 (13) |
| Enterovirus | 102 | 77(75) | 25 (25) |
| Coronavirusc | 41 | 41(100) | 0(0) |
| Parainfluenzad | 31 | 30(97) | 1(3) |
| Respiratory syncytial virus | 26 | 26(100) | 0(0) |
| Herpes simplex virus | 4 | 4(100) | 0(0) |
| Influenzae | 26 | 26(100) | 0(0) |
| Total | 510 | 433 | 77 |
An individual virus detected once and did not reappear in the subsequent nasopharyngeal secretion either collected during AOM diagnosis or at the next new URI episode within 30 days interval. Presence of the same type of virus after a 30 day period was considered a new virus of the same type. Percent = % of total number of each virus detected
The same virus detected in consecutive samples without an intervening negative result and within 30 days of the previous sample
Coronavirus OC43, 229E, NL-63
Types 1,2,3;
Influenza A and B
Forty-one of the 76 (54%) children had more than one episode of adenovirus infection detected by PCR (125 samples); 66 of these samples could be retrieved for sequence analysis.
Adenovirus Sequencing Results and Phylogenetic Analysis
Of the 66 adenovirus samples, 36 yielded successful sequences that could be used for phylogenetic analysis; the remaining samples did not yield satisfactory results. Adenovirus serotypes 1, 2, 3, 4, 5 (Ad1-5) and 41 were identified using BLAST analysis. Several children had multiple positive results with adenoviruses of the same serotype. In most of the samples (81%), the virus sequences were identical (≥ 98% similarity) between episodes, possibly indicating a persistent infection with the same adenovirus strain and serotype. However, 21% of samples demonstrated cases in which a child had two or more positive results for the same adenovirus serotype, but with a larger number of nucleotide differences between the viruses (38-62% similarity), indicating infection with different strains. Phylogenetic analysis indicated that at least three different strains of Ad1 and two different strains of Ad2 were co-circulating during the study period.
Case series and patterns of adenovirus infection
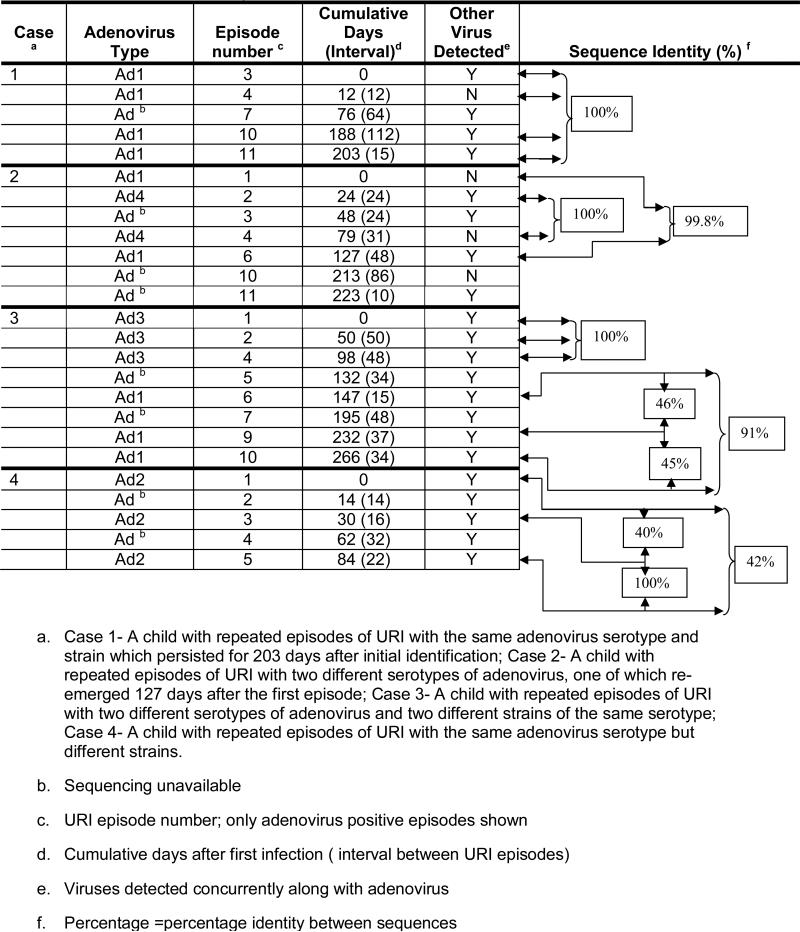
Thirty-six samples from 13 children were successfully sequenced and phylogenetically analyzed. Four cases below (Table 3) illustrate different patterns of repeatedly positive PCR for adenovirus. Of note, some children displayed more than one pattern.
Table 3.
Cases of children with persistent adenovirus detection
|
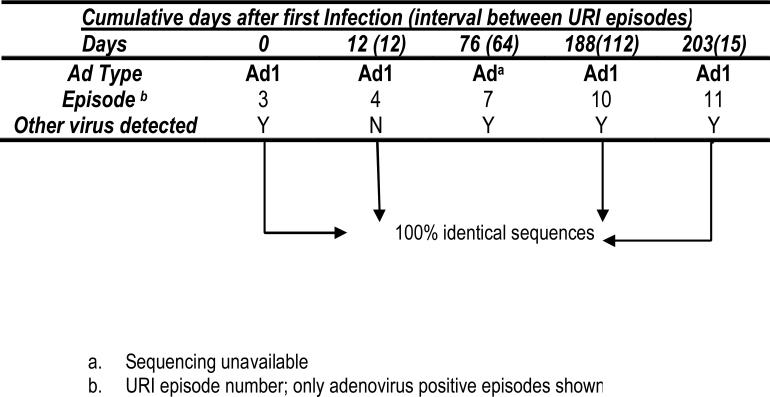
Case 1 (Figure 1a – website---) An 18-month old child had 12 URI episodes; 4 episodes of URI occurred with the same serotype and same strain of adenovirus over a 6 month period. The same strain of Ad1 was detected in URI episodes 3 (with rhinovirus), 4 (alone), 10 (with rhinovirus) and 11 (with coronavirus OC43/229E). Episode 7 was also positive for adenovirus (along with enterovirus and parainfluenza 2) but sequencing was unavailable. In two episodes, other respiratory viruses but not adenoviruses were detected (not shown): enterovirus and parainfluenza 2 in episode 6 and parainfluenza 3 in episode 12. Episodes 1, 2, 5, 8, and 9 (not shown) were negative for all viruses.
Figure 1a.
Case 1- A child with repeated episodes of URI with the same adenovirus serotype and strain which persisted for 203 days after initial identification.
This case illustrates the prolonged presence of Ad1 for 203 days after first detection.
Case 2 (Figure 1b) A 6-month old child who was followed through 11 URI episodes, had repeated adenovirus positive samples with two different serotypes-Ad1 and Ad4. In this child, Ad1 was detected in URI episode 1 and then re-emerged 127 days later during episode 6 along with rhinovirus. During the intervening period between episode 1 and episode 6, the child had 4 other virus positive episodes (episode 5 not shown). Viruses were detected in episodes 2 (Ad4, rhinovirus), 3 (adenovirus, Influenza A, parainfluenza 1 & 2, and rhinovirus), 4 (Ad4) and 5 (Influenza A). Sequencing was unavailable for the adenovirus detected in episode 3. Adenovirus was detected again in episodes 10 (by culture) and 11 (by PCR, along with rhinovirus) but sequencing was also not available for these episodes. Episodes 7, 8 and 9 (not shown) were negative for viruses.
Figure 1b.
Case 2- A child with repeated episodes of URI with two different serotypes of adenovirus, one of which re-emerged 127 days after the first episode
This case illustrates the presence of two different adenovirus serotypes 24 days apart as well as demonstrating that the same serotype (and strain) can be detected intermittently for more than 4 months.
Case 3 (Figure 1c) A 24-month old child was infected with two different serotypes of adenovirus (Ad 3 and Ad1) as well as with two different strains of Ad1. This child had 8 of 11 URI episodes positive for adenovirus. Adenovirus was detected in episodes 1 (Ad3 with enterovirus) and 2 (Ad3 with enterovirus). Episode 3 (not shown) was positive for enterovirus only by PCR. Episode 4 was positive for Ad3 along with enterovirus. Episode 5 was also positive for enterovirus and adenovirus, but the adenovirus sequencing was unsuccessful due to dual peaks on chromatogram, possibly indicative of a dual infection with two adenovirus strains/serotypes, making it difficult to interpret. Adenovirus serotype 1 was detected along with other viruses in episodes 6 (enterovirus), 9 (rhinovirus) and 10 (coronavirus OC43/229E, enterovirus). However, while episodes 6 and 10 were positive for the same strain of Ad1, episode 9 was positive for a different strain of Ad1 (only 45-46% identical to the strain in episode 6 and 10). Episode 7 was positive for adenovirus by PCR (as well as for enterovirus and coronavirus OC43/229E) but sequencing was not available. Episode 8 (not shown) was positive for Influenza A only by culture.
Figure 1c.
Case 3- A child with repeated episodes of URI with two different serotypes of adenovirus and two different strains of the same serotype.
This case illustrates that adenoviruses of different serotypes can be detected during sequential URI episodes within a short period of time. In addition, adenoviruses of the same serotype but different strains can be detected during sequential URI episodes.
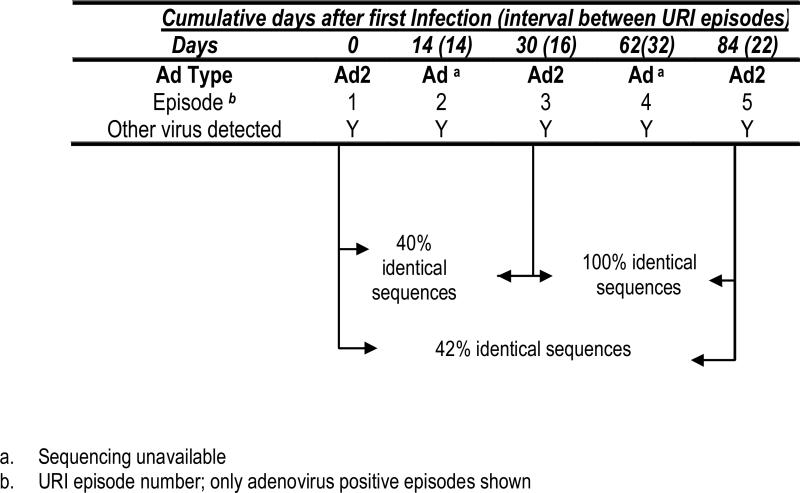
Case 4 (Figure 1d) A 7-month old child followed for 6 URI episodes was infected with two different strains of Ad2. This child had 5 episodes that were positive for adenovirus along with other respiratory viruses; episodes 1 (enterovirus), 2 (rhinovirus), 3 (rhinovirus), 4 (enterovirus and rhinovirus) and 5 (coronavirus OC43/229E, enterovirus). However, the strain of Ad2 found in episode 1 was only 40-42% identical to the strain of Ad2 found in episodes 3 and 5. Episode 6 (not shown) was positive for RSV by EIA.
Figure 1d.
Case 4- A child with repeated episodes of URI with the same adenovirus serotype but different strains.
This case illustrates that adenovirus of the same serotype but different strains can be detected during sequential URI episodes a month (30 days) apart.
Overall, our analysis of the sequential adenovirus-positive episodes from these 13 children illustrated four different patterns of repeatedly positive PCR for adenovirus (n=number of instances seen in these 13 children):
Pattern 1. Adenovirus of the same serotype and strain repeatedly and continuously detected in multiple episodes of URI, for more than 6 months (n =8); range of 48-203 days. (e.g. case 1)
Pattern 2. Adenovirus of different serotypes detected during sequential URI episodes within a short period of time (< 60 days) (n=3); range of 24-49 days. (e.g. cases 2 & 3)
Pattern 3. Adenovirus of the same serotype but different strains detected during sequential URI episodes at least a month apart (n=3); range of 30-85 days. (e.g. cases 3 & 4)
Pattern 4. Adenovirus of the same serotype and strain detected intermittently for more than 4 months (n =2); range 119-127 days. ( e.g. case 2)
Discussion
Using comprehensive viral diagnostic methods, we detected one or more respiratory viruses in 65% of URI episodes. PCR played a major role in increased detection of these viruses; PCR detected 87% of adenoviruses, as well as 82% of enteroviruses and 77% of rhinoviruses. Our data show a strong relationship between the diagnostic method and detection rate.
In keeping with other studies21-23, we used a 30-day time limit to define new versus repeated NA detection. We found that 15% of viruses had previously been detected in the same child within the 30 day time limit. We realize that this time period may not represent the actual repeated detection period. Prolonged presence of viral ribonucleic acids (RNA) of picornaviruses (rhinovirus and enterovirus) in respiratory secretions has also been described24,25.
We observed adenovirus of the same genetic strain detected repeatedly in multiple episodes of URI as the most common pattern seen among the children we studied. In one case we observed the same strain of adenovirus present for up to 203 days. Prolonged presence of adenovirus NA in respiratory secretions following URI has not been studied carefully but prolonged shedding of adenovirus has been described10. Adenovirus has been detected in tonsillar tissue26,27 and has been isolated from 0.6-3% of throat samples from healthy children11-13. Latency of adenovirus has been described in the lungs28,29 and reactivation of adenovirus has been described in immunosuppressed hosts30,31. While adenovirus NA continued to be detected in subsequent URI episodes, other respiratory viruses were also detected. The newly detected respiratory virus at the new URI onset is more likely the causative agent, especially when the virus is detected by the conventional assays. The role of other respiratory viruses in reactivation of latent adenovirus is still to be determined.
PCR is being used increasingly to serotype adenoviruses8,32-34. We used a hexon gene sequence typing method18 to identify the serotypes of the adenovirus samples obtained from the children we observed. The use of phylogenetic analysis enabled us to appreciate the existence of different strains within serotypes. We believe that the bp differences we observed were greater than could be explained by normal mutation. Our assumption that 98-100% similarity between virus samples is indicative of identical strains is as in keeping with Lereuz-ville et al35, who observed 100% similarity within isolates of the same strain of adenovirus in an epidemic. We demonstrated that repeated detection of adenovirus NA during symptomatic respiratory tract infection may be due to various patterns involving identical or different serotypes or strains. The presence of these different patterns of adenovirus infection poses a challenge as to how best to clinically interpret a positive adenovirus PCR result without additional phylogenetic analysis.
As the present study was a secondary data analysis, it is limited by the fact that the primary objective of the original prospective study15 was to determine the causative virus of URI and virus-specific characteristics of AOM complicating URI. We did not collect NPS samples from asymptomatic children; neither did we collect NPS on a regular basis. The samples were collected at the new URI onset and when the children subsequently developed AOM. The original study design did not include serologic studies, which would have required acute and convalescent blood drawings. Simultaneous adenovirus-specific serology may have helped with interpretation of repeated adenovirus NA detected. Even with the limitations mentioned here, our data clearly demonstrate various patterns of adenovirus infection and strongly suggest the possibility of prolonged presence of viral NAs after respiratory tract infection.
In summary, PCR assays significantly increased detection of respiratory viruses in NPS specimens compared with virus culture. For adenovirus, repeated or prolonged presence of viral NA's may be due to the same strain with continuous or intermittent detection, or different serotypes/strains during symptomatic URI. Because positive PCR for adenovirus without phylogenetic analysis is subject to various interpretations, caution should be employed in clinical interpretation of the results obtained. Future longitudinal studies documenting the ability to detect viral NA during symptomatic and asymptomatic periods, as well as implementing quantitative PCR, might help elucidate the role of positive PCR results during respiratory tract infections.
Acknowledgement
This work was supported by National Institutes of Health grant R01 DC005841. The study was conducted at the General Clinical Research Center at the University of Texas Medical Branch, which is funded by National Center for Research Resources (National Institutes of Health, US Public Health Service) grant M01 RR 00073.
Reference List
- 1.Cherry JD, Nieves DJ. The Common Cold. In: Feigin R, Cherry J, Demmler-Harrison GJ, Kaplan S, editors. Textbook of Pediatric Infectious Diseases. 6th ed. Saunders Elsevier; Philadelphia, PA: 2009. pp. 138–46. [Google Scholar]
- 2.Kendall EJ. Acute respiratory infections in the population. J R Soc Med. 1985;78:282–290. doi: 10.1177/014107688507800403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kvaerner KJ, Nafstad P, Jaakkola JJ. Upper respiratory morbidity in preschool children: a cross-sectional study. Arch Otolaryngol Head Neck Surg. 2000;126:1201–1206. doi: 10.1001/archotol.126.10.1201. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg GA, Erdman DD, Edwards KM, et al. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J Infect Dis. 2004;189:706–710. doi: 10.1086/381456. [DOI] [PubMed] [Google Scholar]
- 5.Freymuth F, Vabret A, Cuvillon-Nimal D, et al. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol. 2006;78:1498–1504. doi: 10.1002/jmv.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Elden LJ, van Kraaij MG, Nijhuis M, et al. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis. 2002;34:177–183. doi: 10.1086/338238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrickson KJ. Cost-effective use of rapid diagnostic techniques in the treatment and prevention of viral respiratory infections. Pediatr Ann. 2005;34:24–31. doi: 10.3928/0090-4481-20050101-08. [DOI] [PubMed] [Google Scholar]
- 8.Gray GC, McCarthy T, Lebeck MG, et al. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004-2006. Clin Infect Dis. 2007;45:1120–1131. doi: 10.1086/522188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang SY, Lee CN, Lin PH, et al. A community-derived outbreak of adenovirus type 3 in children in Taiwan between 2004 and 2005. J Med Virol. 2008;80:102–112. doi: 10.1002/jmv.21045. [DOI] [PubMed] [Google Scholar]
- 10.Brandt CD, Kim HW, Vargosko AJ, et al. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. Am J Epidemiol. 1969;90:484–500. doi: 10.1093/oxfordjournals.aje.a121094. [DOI] [PubMed] [Google Scholar]
- 11.Brandt CD, Kim HW, Jeffries BC, et al. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. II. Variation in adenovirus infections by year and season. Am J Epidemiol. 1972;95:218–227. doi: 10.1093/oxfordjournals.aje.a121389. [DOI] [PubMed] [Google Scholar]
- 12.Maletzky AJ, Cooney MK, Luce R, Kenny GE, Grayston JT. Epidemiology of viral and mycoplasmal agents associated with childhood lower respiratory illness in a civilian population. J Pediatr. 1971;78:407–414. doi: 10.1016/s0022-3476(71)80219-6. [DOI] [PubMed] [Google Scholar]
- 13.Edwards KM, Thompson J, Paolini J, Wright PF. Adenovirus infections in young children. Pediatrics. 1985;76:420–424. [PubMed] [Google Scholar]
- 14.Garnett CT, Erdman D, Xu W, Gooding LR. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol. 2002;76:10608–10616. doi: 10.1128/JVI.76.21.10608-10616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chonmaitree T, Revai K, Grady JJ, et al. Viral upper respiratory tract infection and otitis media complication in young children. Clin Infect Dis. 2008;46:815–823. doi: 10.1086/528685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan J, Henrickson KJ, Savatski LL. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex). Clin Infect Dis. 1998;26:1397–1402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 17.Henrickson KJ, Kraft AJ, Canter D, Shaw J. Comparison of electronic microarray to enzyme hybridization assay for multiplex reverse-transcriptase PCR detection of common respiratory viruses in children. Clinical microbiology newsletter. 2007;29:113–119. doi: 10.1016/j.clinmicnews.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol. 2006;151:1587–1602. doi: 10.1007/s00705-005-0722-7. [DOI] [PubMed] [Google Scholar]
- 19.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 21.Nokso-Koivisto J, Pitkaranta A, Blomqvist S, et al. Viral etiology of frequently recurring respiratory tract infections in children. Clin Infect Dis. 2002;35:540–546. doi: 10.1086/341773. [DOI] [PubMed] [Google Scholar]
- 22.Wald ER, Guerra N, Byers C. Upper respiratory tract infections in young children: duration of and frequency of complications. Pediatrics. 1991;87:129–133. [PubMed] [Google Scholar]
- 23.Winther B, Alper CM, Mandel EM, Doyle WJ, Hendley JO. Temporal relationships between colds, upper respiratory viruses detected by polymerase chain reaction, and otitis media in young children followed through a typical cold season. Pediatrics. 2007;119:1069–1075. doi: 10.1542/peds.2006-3294. [DOI] [PubMed] [Google Scholar]
- 24.Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 25.Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: Association with symptomatic illness and effect of season. J Med Virol. 2006;78:644–650. doi: 10.1002/jmv.20588. [DOI] [PubMed] [Google Scholar]
- 26.Garnett CT, Talekar G, Mahr JA, et al. Latent species C adenoviruses in human tonsil tissues. J Virol. 2009;83:2417–2428. doi: 10.1128/JVI.02392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann R, Genersch E, Eggers HJ. Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 1987;7:93–97. doi: 10.1016/0168-1702(87)90060-8. [DOI] [PubMed] [Google Scholar]
- 28.Retamales I, Elliott WM, Meshi B, et al. Amplification of inflammation in emphysema and its association with latent adenoviral infection. Am J Respir Crit Care Med. 2001;164:469–473. doi: 10.1164/ajrccm.164.3.2007149. [DOI] [PubMed] [Google Scholar]
- 29.Matsuse T, Hayashi S, Kuwano K, Keunecke H, Jefferies WA, Hogg JC. Latent adenoviral infection in the pathogenesis of chronic airways obstruction. Am Rev Respir Dis. 1992;146:177–184. doi: 10.1164/ajrccm/146.1.177. [DOI] [PubMed] [Google Scholar]
- 30.Hierholzer JC. Adenoviruses in the immunocompromised host. Clin Microbiol Rev. 1992;5:262–274. doi: 10.1128/cmr.5.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ison MG, Hayden FG. Viral infections in immunocompromised patients: what's new with respiratory viruses? Curr Opin Infect Dis. 2002;15:355–367. doi: 10.1097/00001432-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Pring-Akerblom P, Trijssenaar FE, Adrian T, Hoyer H. Multiplex polymerase chain reaction for subgenus-specific detection of human adenoviruses in clinical samples. J Med Virol. 1999;58:87–92. [PubMed] [Google Scholar]
- 33.Pring-Akerblom P, Adrian T. Type- and group-specific polymerase chain reaction for adenovirus detection. Research in Virology. 1994;145:25–35. doi: 10.1016/s0923-2516(07)80004-5. [DOI] [PubMed] [Google Scholar]
- 34.Wong S, Pabbaraju K, Pang XL, Lee BE, Fox JD. Detection of a broad range of human adenoviruses in respiratory tract samples using a sensitive multiplex real-time PCR assay. J Med Virol. 2008;80:856–865. doi: 10.1002/jmv.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leruez-Ville M, Chardin-Ouachee M, Neven B, et al. Description of an adenovirus A31 outbreak in a paediatric haematology unit. Bone Marrow Transplant. 2006;38:23–28. doi: 10.1038/sj.bmt.1705389. [DOI] [PubMed] [Google Scholar]