Abstract
KCNQ1 channels are voltage-gated potassium channels that are widely expressed in various non-neuronal tissues, such as the heart, pancreas, and intestine. KCNE proteins are known as the auxiliary subunits for KCNQ1 channels. The effects and functions of the different KCNE proteins on KCNQ1 modulation are various; the KCNQ1–KCNE1 ion channel complex produces a slowly activating potassium channel that is crucial for heartbeat regulation, while the KCNE3 protein makes KCNQ1 channels constitutively active, which is important for K+ and Cl− transport in the intestine. The mechanisms by which KCNE proteins modulate KCNQ1 channels have long been studied and discussed; however, it is not well understood how different KCNE proteins exert considerably different effects on KCNQ1 channels. Here, we approached this point by taking advantage of the recently isolated Ci-KCNQ1, a KCNQ1 homologue from marine invertebrate Ciona intestinalis. We found that Ci-KCNQ1 alone could be expressed in Xenopus laevis oocytes and produced a voltage-dependent potassium current, but that Ci-KCNQ1 was not properly modulated by KCNE1 and totally unaffected by coexpression of KCNE3. By making chimeras of Ci-KCNQ1 and human KCNQ1, we determined several amino acid residues located in the pore region of human KCNQ1 involved in KCNE1 modulation. Interestingly, though, these amino acid residues of the pore region are not important for KCNE3 modulation, and we subsequently found that the S1 segment plays an important role in making KCNQ1 channels constitutively active by KCNE3. Our findings indicate that different KCNE proteins use different domains of KCNQ1 channels, and that may explain why different KCNE proteins give quite different outcomes by forming a complex with KCNQ1 channels.
INTRODUCTION
KCNQ1 is a voltage-gated potassium channel α subunit that is expressed in a wide variety of tissues, including the heart, kidney, pancreas, intestine, and inner ear. Although KCNQ1 alone can form functional ion channels on the plasma membrane, KCNQ1 is believed to form an ion channel complex with auxiliary subunit KCNE proteins (Barhanin et al., 1996; Sanguinetti et al., 1996; Schroeder et al., 2000). The stoichiometry of KCNQ1 and KCNE1 has long been thought to be 4:2 (two KCNE1 subunits bind to one KCNQ1 tetramer) (H. Chen et al., 2003; Morin and Kobertz, 2008), but some recent results including ours suggest that the stoichiometry may be variable and up to four KCNE1 subunits can bind to one KCNQ1 channel (Cui et al., 1994; Wang et al., 1998; Nakajo et al., 2010; Li et al., 2011; Nakajo and Kubo, 2011). There exist five KCNE genes (KCNE1–KCNE5) in the human genome. All KCNE proteins can change the channel properties of KCNQ1 at least in vitro, and all KCNE proteins are expressed in the human heart (Bendahhou et al., 2005; Lundquist et al., 2005). The interaction with KCNE1 enhances the KCNQ1 current amplitude approximately 10-fold and slows gating by nearly two orders of magnitude (Barhanin et al., 1996; Sanguinetti et al., 1996). Coexpression of KCNE3 turns the KCNQ1 channel into a constitutive active potassium channel (Schroeder et al., 2000). KCNE2, KCNE4, and KCNE5 have an inhibitory effect on KCNQ1 channels (McCrossan and Abbott, 2004). It is, however, not fully understood yet why distinct KCNE proteins cause very different outcomes upon association with KCNQ1 channels. It is also unclear why KCNQ1 is by far the most sensitive channel to KCNE proteins.
The molecular and structural mechanisms underlying the channel modulation by KCNE1 have been studied since the initial cloning of KCNE1 as “minK” (Takumi et al., 1988, 1991). In experiments using cysteine-scanning mutagenesis and monitoring of a channel block by cadmium, KCNE1 was suggested to be located near the conduction pathway (Wang et al., 1996; Tai and Goldstein, 1998; Kurokawa et al., 2001; Tapper and George, 2001). Melman et al. (2004) confirmed that KCNE1 actually binds to the pore region of KCNQ1 by coimmunoprecipitation. With the structural models of KCNQ1 based on the Kv1.2 crystal structure and the actual structure of KCNE1 solved by nuclear magnetic resonance, it has been speculated that KCNE1 is located between two adjacent voltage-sensing domains (Long et al., 2005a,b; Smith et al., 2007; Kang et al., 2008). Recent reports that KCNE1 affects the equilibrium of the voltage sensor position (Nakajo and Kubo, 2007; Rocheleau and Kobertz, 2008; Shamgar et al., 2008) and that KCNE1 can form a disulfide bond with the KCNQ1 voltage sensor domain (VSD) (Nakajo and Kubo, 2007; Rocheleau and Kobertz, 2008; Shamgar et al., 2008; Osteen et al., 2010) also support this hypothetical KCNE1 location. Melman et al. (2001, 2002) identified Thr58 and Leu59 on KCNE1 as critical amino acid residues for the modulation. Panaghie et al. (2006) subsequently identified Phe339 and Phe340 on the S6 segment of KCNQ1 as the partners for Thr58 and Leu59 on KCNE1. However, these two phenylalanine residues do not explain why other types of KCNQ channels are not modulated by KCNE1, because these phenylalanine residues are both conserved in all KCNQ subtypes.
Our recent effort to make a catalog of ion channels in ascidian Ciona intestinalis, a marine invertebrate chordate, gave us some insights into how the diversity of vertebrate ion channels has been expanded (Okamura et al., 2005). In most cases, ascidian has only prototypes for each ion channel subtype; for example, it has only one Kv4 channel, whereas humans have three types of Kv4 genes (Kv4.1–Kv4.3) (Nakajo et al., 2003; Okamura et al., 2005). In the human genome, five members of KCNQ channels have already been identified (Jentsch, 2000). We found only two types of KCNQ channels in C. intestinalis, Ci-KCNQ1 and Ci-KCNQ4/5 (Okamura et al., 2005; Hill et al., 2008). More interestingly, the Ciona genome contains no KCNE-like gene (Okamura et al., 2005).
In this study, we used the Ci-KCNQ1 gene to survey the unique and critical amino acid residues on human KCNQ1 for KCNE1 and KCNE3 modulation. Because the Ciona genome lacks KCNE-related genes, we expected that the Ci-KCNQ1 channel is not designed for KCNE modulation. We successfully identified some amino acid residues of the pore region important for KCNE1 modulation. On the other hand, those amino acid residues of the pore region are not important for KCNE3; instead, we found that phenylalanine residues on the S1 segment play a unique role in KCNE3 modulation. KCNE proteins target different parts of the KCNQ1 channel, and this may be the reason why the modulations by KCNE proteins are various. These domains or amino acid residues identified herein are not conserved in other types of KCNQ channels, and that may account for why only KCNQ1 channels are sensitive to KCNE proteins.
MATERIALS AND METHODS
Identification of the Ci-KCNQ1 gene, subcloning, mutagenesis, and RNA synthesis
The full-length complementary DNA for the C. intestinalis KCNQ1, Ci-KCNQ1 (available from GenBank/EMBL/DDBJ under accession no. FJ461775), was obtained by a combination of 3′ RACE and 5′ RACE (GeneRacer kit; Invitrogen) from a full-length cDNA pool derived from hatched Ciona larvae. The primers designed to amplify the RACE fragments were as follows: 5′-out, CAACGACGAGAATTGCTTCCATGAC; 5′-in, CGATTGTTGACAGGACACTGAA; 3′-out, TCCTTGAACGTCCAACTGGAGTCAA; 3′-in, CACGTCCTAGTATTCATGATGGTTCT. This gene transcript is expressed in the central and peripheral neurons of C. intestinalis tadpole larvae (Hill et al., 2008). Ci-KCNQ1, human KCNQ1 (GenBank accession no. AF000571; provided by T. Hoshi, University of Pennsylvania, Philadelphia, PA) and rat KCNE1 (GenBank accession no. NM_012973; provided by T. Takumi, Hiroshima University, Hiroshima, Japan) cDNAs were subcloned into the pGEMHE expression vector. Chimeras and point mutants were made by PCR with high fidelity polymerase KOD Plus (version 2; Toyobo) and subsequently confirmed by sequencing. cRNA was prepared from the linearized plasmid cDNA using a T7 RNA transcription kit (Invitrogen).
Preparation of Xenopus laevis oocytes
Xenopus oocytes were collected from frogs anesthetized in water containing 0.15% tricaine. After the final collection, the frogs were killed by decapitation. The isolated oocytes were treated with collagenase (2 mg/ml; type 1; Sigma-Aldrich) for 6 h to completely remove the follicular cell layer. Oocytes of similar size (stage V or VI) were injected with ∼50 nl of cRNA solution and incubated at 17°C in frog Ringer’s solution containing (in mM): 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.3 Ca(NO3)2, 0.41 CaCl2, and 0.82 MgSO4, pH 7.6, with 0.1% penicillin–streptomycin solution (Sigma-Aldrich). When coexpressing human KCNQ1/Ci-KCNQ1 and KCNE1/KCNE3, the ratio of the amount of respective RNAs was set at 10:1. All experiments conformed to the guidelines of the Animal Care Committee of the National Institute for Physiological Sciences.
Two-electrode voltage clamp
1–3 d after cRNA injection, K+ currents were recorded under two-electrode voltage clamp using an amplifier (OC725C; Warner Instruments) and pClamp10 software (Axon Instruments). Data from the amplifier were digitized at 10 kHz and filtered at 1 kHz. The microelectrodes were drawn from borosilicate glass capillaries (World Precision Instruments) to a resistance of 0.2 ∼ 0.5 MΩ when filled with 3 M K-acetate and 10 mM KCl. The bath solution (ND96) contained (in mM): 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES, pH 7.2. Oocytes were held at −90 mV, stepped up to various test voltages, and then to −30 mV to record tail currents. Tail current amplitudes were typically measured as the average value at 10–20 ms after the end of the test pulse. All experiments were performed at room temperature (25 ± 2°C).
Analysis of the voltage dependence
G-V relationships were plotted using tail current amplitudes obtained at −30 mV after 2-s prepulses of various membrane potentials. 2-s depolarization is not long enough to reach a new equilibrium, especially in slowly activating KCNQ1–KCNE1 current. However, it is not practical to wait until the current reaches its plateau; we took the tail current amplitude at 2 s as an apparent conductance “G.” Tail currents were fitted using pClamp10 software to a two-state Boltzmann equation: G = Gmin + (Gmax − Gmin)/(1 + e−zF(V−V1/2)/RT), where G is determined by the tail current amplitude, Gmax and Gmin are the maximum and minimum tail current amplitudes, z is the effective charge, V1/2 is the half-activation voltage, T is the temperature in degrees Kelvin, F is Faraday’s constant, and R is the Boltzmann constant. G/Gmax, which is plotted on the longitudinal axes in the presentations of the G-V curves, simply equals the normalized tail current amplitude. In some figures (Figs. 7 C and 9 B), the G/Gmax value becomes slightly lower than zero at negative membrane potentials, probably a result of some contamination of leak and endogenous chloride conductance.
Figure 7.
The V46F mutation with a human S5P connector changes Ci-KCNQ1 into a KCNE3-sensitive channel. (A) The constitutive activity index (G−80mV/G+20mV) with KCNE3 for the point mutants of the ciQ1hQ1(S2–S3,S5P) chimera. Asterisks indicate the significant increase in the level of G−80mV/G+20mV, with KCNE3 compared with that of ciQ1hQ1(S2–S3,S5P) with KCNE3 (Dunnett’s test). The locations of the mutations are indicated by yellow stars in the inset. (B) Representative current traces of ciQ1hQ1(S5P) V46F in the absence and presence of KCNE3. The membrane potential was stepped up from −100 to +60 mV and subsequently stepped to −30 mV for tail currents. (C) G-V relationships for Ci-KCNQ1 V46F and ciQ1hQ1(S5P) V46F in the absence (open symbols) and presence (closed symbols) of KCNE3. Plots are fitted with the Boltzmann equation.
Figure 9.
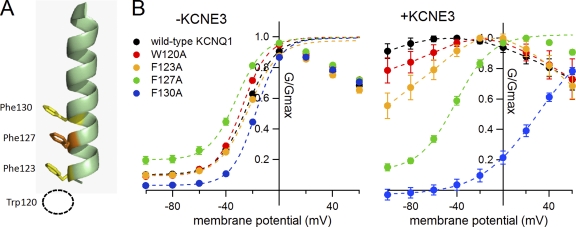
Phenylalanine residues on the S1 segment of KCNQ1 play a role in KCNE3 modulation. (A) Location of phenylalanine residues on the hKCNQ1 S1 segment. Trp120 is not on the S1 segment and therefore is not depicted. (B) G-V relationships for hQ1 F(W)-to-A mutants in the absence (left) and presence of KCNE3 (right). All but WT KCNQ1, W120A, and F123A with KCNE3 are fitted with the Boltzmann equation. For the WT KCNQ1, W120A, and F123A with KCNE3, plots were connected by dotted lines, as they could not be fitted with the Boltzmann equation.
Analysis of the constitutive activity of the KCNQ1–KCNE3 current
To evaluate the constitutive activity of KCNQ1 chimeras and mutants with KCNE3, we introduced a constitutive activity index (G−80mV/G+20mV), which is the ratio of the conductance at −80 and +20 mV (Figs. 6 and 7).
Figure 6.
The S1 segment, not the pore region of hKCNQ1, is important for the modulation by KCNE3. (A) Representative current traces of chimera ciQ1hQ1(S4–S6) in the absence and presence of KCNE3. The membrane potential was stepped up from −100 to +60 mV in 20-mV increments and subsequently stepped to −30 mV for tail currents. Insets are the expanded tail current traces after depolarization to +20, +40, and +60 mV. Expanded areas are indicated by red circles below. Bars in the insets indicate 0.1 s and 0.1 µA, respectively. The “hook” tail current, which reflects de-inactivation and is the characteristic feature of the homomeric KCNQ1 channel, was only seen in the homomeric ciQ1hQ1(S4–S6) channel, indicating that KCNE3 abolished inactivation. (B) G-V relationships for the ciQ1hQ1(S4–S6) in the absence (closed circles) and presence (blue triangles) of KCNE3. The G-V relationships of ciQ1hQ1(S4–S6) in the presence of KCNE1 (red dotted curve) is superimposed for comparison. (C) The constitutive activity indices (G−80mV/G+20mV) with KCNE3 for human KCNQ1 (hKCNQ1; left end), Ci-KCNQ1 (right end), and their chimeras are plotted. Asterisks indicate the significant reduction in the level of G−80mV/G+20mV, with KCNE3 compared with that of hKCNQ1 with KCNE3 (Dunnett’s test). The design for each chimera is depicted at the bottom of the bar graphs; red regions are from human KCNQ1, and blue regions are from Ci-KCNQ1.
Structural data and presentation
Structural data were presented using PyMOL software (DeLano Scientific).
Statistical analyses
The data are expressed as the mean ± SEM. Differences between means were evaluated using Dunnett’s test. All samples in Figs. 3 A, 4 A, 5 A, 6 C, and 7 A were compared with the leftmost sample (hKCNQ1 in Fig. 3 A, for example) in each panel. Values of P < 0.05 were considered significant. *, **, and *** denote values of P < 0.05, 0.01, and 0.001, respectively.
Figure 3.
Second half of the transmembrane region from hKCNQ1 is necessary for the modulation by KCNE1. (A) The half-maximal potential (V1/2) without (light gray bars) and with (dark gray bars) KCNE1 for human KCNQ1 (hKCNQ1; left end), Ci-KCNQ1 (right end), and their chimeras are plotted. Asterisks indicate the significant reduction in the level of V1/2 with KCNE1 compared with that of hKCNQ1 with KCNE1 (Dunnett’s test). The design for each chimera is depicted at the bottom of the bar graphs; red regions are from human KCNQ1, and blue regions are from Ci-KCNQ1. (B) Representative current traces of chimera ciQ1hQ1(S4–S6) in the absence and presence of KCNE1. The membrane potential was stepped from −100 to +60 mV in 20-mV increments and subsequently stepped to −30 mV for tail currents. (C) G-V relationships for ciQ1hQ1(S4–S6) in the absence and presence of KCNE1. Plots are fitted with the Boltzmann equation (red curves).
Figure 4.
Amino acid residues in the pore region of KCNQ1 have a large impact on the KCNE1 modulation. (A) V1/2 without (light gray bars) and with (dark gray bars) KCNE1 for point mutants of the ciQ1hQ1(S5P) chimera and Ci-KCNQ1 are plotted. Asterisks indicate the significant increase in the level of V1/2 with KCNE1 compared with those of ciQ1hQ1(S5P) or Ci-KCNQ1 with KCNE1 (Dunnett’s test). The locations of the mutations are indicated by yellow stars in the inset. (B) Representative current traces for the A191G/I258V double mutant of ciQ1hQ1(S5P) in the absence (left) and presence (right) of KCNE1. The membrane potential was stepped up from −100 to +40 mV in 10-mV increments for KCNE1-less current and from −100 to +60 mV in 20-mV increments for KCNE1 current. (C) G-V relationships for the point mutants in the absence (closed circles) and presence (open circles) of KCNE1.
Figure 5.
Human KCNQ1 mutants confirm that Gly272, Val324, and Val334 of KCNQ1 are important for KCNE1 modulation. (A) V1/2 values without (light gray bars) and with (dark gray bars) KCNE1 for hKCNQ1 (hQ1) point mutants are plotted. Asterisks indicate the significant reduction in the level of V1/2 with KCNE1 compared with that of hKCNQ1 with KCNE1 (Dunnett’s test). (B) Representative current traces for hKCNQ1 G272A/V334I double mutant in the absence (left) and presence (right) of KCNE1. The membrane potential was stepped up from −100 to +60 mV in 20-mV increments. (C) G-V relationships for the hQ1 G272A/V334I mutant in the absence (open circles) and presence (closed red circles) of KCNE1. The G-V relationships of WT hKCNQ1 in the absence (black dotted curve) and presence (red dotted curve) of KCNE1 are superimposed.
RESULTS
KCNQ1 from C. intestinalis is not properly modulated by mammalian KCNE proteins
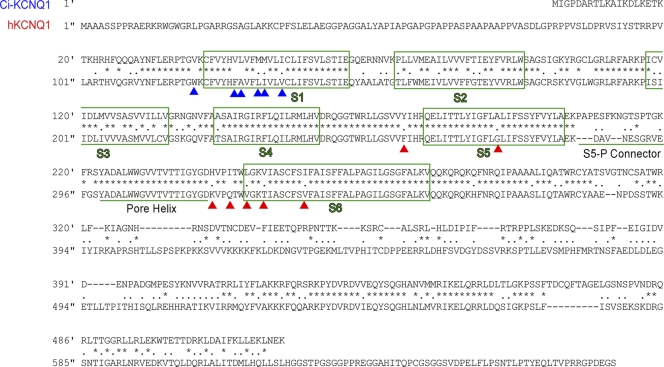
C. intestinalis has two KCNQ genes, Ci-KCNQ1 and Ci-KCNQ4/5, and no KCNE gene (Okamura et al., 2005; Hill et al., 2008). Ci-KCNQ1, a KCNQ1 orthologue encoding a 520–amino acid-long protein, has shorter N-terminal and C-terminal cytoplasmic regions than those of the human KCNQ1 channel (Fig. 1). The overall similarity of the amino acid sequence with human KCNQ1 (hKCNQ1) is 54%. The transmembrane region is relatively well conserved (75% homology), and the region from S4 to S6 segments (except the S5-P connector) is especially well conserved (84% homology between Ser 225 and Ala 383 of hKCNQ1).
Figure 1.
Amino acid sequences of Ci-KCNQ1 and human KCNQ1 (hKCNQ1) are aligned for comparison. Same and similar amino acid residues are marked with asterisks and dots, respectively. Transmembrane regions (S1–S6) are boxed in green. The S5-P connector and pore helix are underlined. Red and blue arrowheads indicate the mutated amino acid residues in this study.
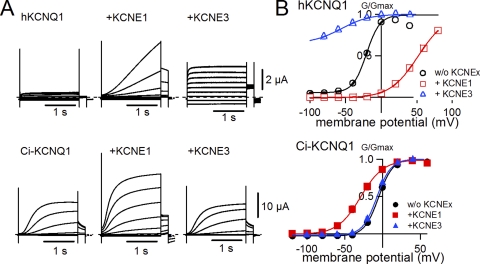
Because the genome of Ciona does not contain a KCNE gene, we hypothesized that the Ci-KCNQ1 channel might not be modulated by mammalian KCNE proteins. If that were the case, Ci-KCNQ1 would enable us to determine which part of KCNQ1 is important for the modulation induced by KCNE proteins. Ci-KCNQ1 was very well expressed in Xenopus oocytes; it required only 1 d after injection to be robustly expressed. In the absence of KCNE1, the current amplitude of Ci-KCNQ1 was much larger and the activation kinetics was slower than that of hKCNQ1 (Fig. 2 A). Rat KCNE1 actually modulated Ci-KCNQ1 current but in a different manner, as with hKCNQ1. The activation kinetics was not slowed as seen for hKCNQ1. The current amplitude was still enhanced by KCNE1, but it was merely a twofold increase and not comparable with the hKCNQ1 current, which was increased by more than 10 times when KCNE1 was coexpressed (Fig. 2 A). The most notable aspect of KCNE1 modulation against Ci-KCNQ1 was the shift of the G-V curve. Although KCNE1 shifted the G-V curve of hKCNQ1 >60 mV in the positive direction, KCNE1 failed to shift the G-V curve of Ci-KCNQ1 in a positive direction, and actually shifted in a negative direction (Fig. 2 B). Even more prominently, KCNE3, a different type of KCNE protein making the hKCNQ1 channel constitutively active, did not show any effect on the Ci-KCNQ1 current or its voltage dependence (Fig. 2, A and B). As we expected, Ci-KCNQ1 was not properly modulated by KCNE proteins. Therefore, we hypothesized that Ci-KCNQ1 lacked the subdomain or amino acid residues important for a “right kind” modulation by KCNE proteins.
Figure 2.
Ci-KCNQ1 is not properly modulated by KCNE1. (A) Representative current traces in the absence and presence of KCNE1 or KCNE3 for human KCNQ1 (hKCNQ1) and Ci-KCNQ1. The membrane potential was stepped up from −120 or −100 to 60 mV in 20-mV increments and subsequently stepped to −30 mV for tail currents. (B) G-V relationships for human KCNQ1 (top) and Ci-KCNQ1 (bottom) in the absence and presence of KCNE1 or KCNE3.
The pore region from human KCNQ1 is important for the positive G-V shift by KCNE1
To identify which part of hKCNQ1 is important for the shift of the voltage dependence by KCNE1, we made some chimeric mutants between hKCNQ1 and Ci-KCNQ1. We compared the shift of the half-maximal voltage (V1/2) of the voltage dependence with that of wild-type (WT) hKCNQ1 between these chimeric channels (Fig. 3 A and Table I). As shown in Fig. 1, the S4–S6 segments of human and Ciona KCNQ1 are particularly well conserved. Therefore, we first made two hKCNQ1-based chimeras, one having the C-terminal region of Ci-KCNQ1 (Fig. 3 A, hQ1ciQ1(C-term), second from the left), and the other having the N terminus through the S3 segment of Ci-KCNQ1 (Fig. 3 A, ciQ1(N-S3)hQ1(S4-C), third from the left). KCNE1 shifted the voltage dependence of these constructs in a similar manner to WT hKCNQ1. We next made a Ci-KCNQ1–based chimera having the S4–S6 segments from hKCNQ1 (ciQ1hQ1(S4–S6)), which also showed the large positive G-V shift (Fig. 3, A and B). This chimera, ciQ1hQ1(S4–S6), showed larger current and slower kinetics with the coexpression of KCNE1, similar to WT hKCNQ1 (Fig. 3 B). As in the case of WT Ci-KCNQ1, only 1 d was required for robust expression of the ciQ1hQ1(S4–S6) chimera, both in the absence and presence of KCNE1. The G-V of this chimera was shifted from −17.1 ± 0.7 mV to +44.1 ± 3.5 mV by KCNE1, comparable with hKCNQ1 (Fig. 3 C and Table I). Further deletion of the human KCNQ1 region diminished the shift of V1/2 by KCNE1. A ciQ1hQ1(S5P–S6) chimera lacking the S4 and S5 segments of hKCNQ1 and a ciQ1hQ1(S4–S5P) chimera lacking the S6 segment of hKCNQ1 showed a much smaller V1/2 shift, with KCNE1 of −8.3 ± 2.0 and −5.8 ± 4.7 mV, respectively (Fig. 3 A, fifth and sixth from the left). The ciQ1hQ1(S5P) chimera, which has only the S5-P connector region from hKCNQ1, showed a V1/2 value of −30.0 ± 1.1 mV, even with KCNE1 (Fig. 3 A, second from the right).
Table I.
Parameters of chimeras and mutants for the KCNE1 study
| Chimera/mutant name | Without KCNE1 | With KCNE1 | ||||||
| n | I (μA) | V1/2 (mV) | z | n | I (μA) | V1/2 (mV) | z | |
| Human KCNQ1 | 29 | 0.4 ± 0.0 | −22.7 ± 0.7 | 2.8 ± 0.1 | 24 | 6.1 ± 0.7 | 35.2 ± 2.1 | 1.4 ± 0.0 |
| Ci-KCNQ1 | 15 | 3.9 ± 0.4 | −4.2 ± 0.9 | 3.1 ± 0.1 | 7 | 6.5 ± 1.1 | −29.4 ± 2.1 | 1.9 ± 0.1 |
| Chimeras | ||||||||
| hQ1ciQ1(C-term) | 5 | 0.8 ± 0.2 | −15.7 ± 0.8 | 2.7 ± 0.0 | 5 | 7.8 ± 1.5 | 43.8 ± 1.6 | 1.3 ± 0.0 |
| ciQ1(N-S3)hQ1(S4-C) | 6 | 1.0 ± 0.6 | −25.4 ± 1.1 | 2.4 ± 0.1 | 10 | 7.9 ± 1.5 | 29.8 ± 2.1 | 1.3 ± 0.0 |
| ciQ1hQ1(S4-S6) | 5 | 0.9 ± 0.2 | −17.1 ± 0.7 | 2.3 ± 0.0 | 5 | 8.0 ± 1.5 | 44.1 ± 3.5 | 1.5 ± 0.0 |
| ciQ1hQ1(S5P-S6) | 19 | 3.6 ± 0.3 | −12.1 ± 0.4 | 2.9 ± 0.0 | 18 | 11.0 ± 0.9 | −8.3 ± 2.0 | 1.7 ± 0.1 |
| ciQ1hQ1(S4-S5P) | 3 | 1.5 ± 1.0 | −18.1 ± 0.3 | 2.9 ± 0.2 | 6 | 7.6 ± 2.1 | −5.8 ± 4.7 | 1.7 ± 0.2 |
| ciQ1hQ1(S5P) | 13 | 4.2 ± 0.3 | −19.8 ± 0.3 | 3.0 ± 0.1 | 16 | 10.7 ± 1.1 | −30.0 ± 1.1 | 2.1 ± 0.1 |
| ciQ1hQ1(286-297) mutants | ||||||||
| Y175F | 8 | 5.0 ± 0.6 | −17.6 ± 0.5 | 3.2 ± 0.1 | 15 | 12.0 ± 0.9 | −21.0 ± 1.1 | 2.0 ± 0.1 |
| A191G | 18 | 2.6 ± 0.5 | −6.3 ± 1.4 | 2.8 ± 0.0 | 21 | 8.1 ± 0.6 | −2.2 ± 0.8 | 1.8 ± 0.0 |
| H242K/I245Q | 8 | 1.5 ± 0.2 | −16.0 ± 0.9 | 3.2 ± 0.1 | 8 | 5.3 ± 1.1 | −27.7 ± 2.9 | 1.9 ± 0.1 |
| L248V | 18 | 1.2 ± 0.1 | −17.1 ± 0.4 | 3.0 ± 0.0 | 18 | 5.2 ± 0.5 | 13.7 ± 2.5 | 1.4 ± 0.0 |
| V251T | 9 | 2.5 ± 0.3 | −21.6 ± 0.7 | 3.2 ± 0.1 | 9 | 7.0 ± 2.1 | −15.5 ± 2.7 | 1.7 ± 0.0 |
| I258V | 17 | 1.4 ± 0.1 | −19.9 ± 0.6 | 3.0 ± 0.1 | 17 | 5.9 ± 0.4 | −0.5 ± 2.5 | 1.5 ± 0.0 |
| A191G/L248V | 15 | 4.4 ± 0.5 | −9.5 ± 0.7 | 2.5 ± 0.1 | 19 | 10.6 ± 0.9 | 8.4 ± 2.9 | 1.7 ± 0.1 |
| A191G/I258V | 17 | 1.5 ± 0.2 | −17.3 ± 0.6 | 2.6 ± 0.1 | 17 | 6.9 ± 0.4 | 31.0 ± 2.6 | 1.3 ± 0.0 |
| L248V/I258V | 19 | 4.1 ± 0.6 | −16.0 ± 0.6 | 3.0 ± 0.1 | 19 | 15.6 ± 1.1 | 3.6 ± 1.1 | 1.6 ± 0.0 |
| A191G/L248V/I258V | 19 | 2.4 ± 0.5 | −9.6 ± 0.8 | 2.5 ± 0.0 | 19 | 14.4 ± 1.3 | 31.9 ± 2.4 | 1.3 ± 0.0 |
| Ci-KCNQ1 mutants | ||||||||
| A191G | 10 | 0.2 ± 0.0 | −1.4 ± 0.6 | 3.4 ± 0.2 | 10 | 2.3 ± 0.5 | −12.6 ± 1.5 | 2.5 ± 0.1 |
| L248V | 10 | 4.8 ± 0.6 | 1.2 ± 0.7 | 2.8 ± 0.1 | 10 | 5.9 ± 0.7 | −14.7 ± 1.3 | 2.4 ± 0.1 |
| I258V | 10 | 2.9 ± 0.4 | −6.8 ± 0.6 | 3.2 ± 0.1 | 10 | 6.7 ± 1.0 | −18.9 ± 1.4 | 2.6 ± 0.1 |
| hKCNQ1 mutants | ||||||||
| F256Y | 6 | 0.6 ± 0.1 | −27.0 ± 0.5 | 2.5 ± 0.1 | 6 | 11.5 ± 1.0 | 27.3 ± 1.7 | 1.4 ± 0.0 |
| G272A | 15 | 1.3 ± 0.2 | −19.6 ± 0.4 | 3.6 ± 0.1 | 18 | 8.8 ± 1.1 | 10.4 ± 2.5 | 1.5 ± 0.1 |
| V324L | 10 | 0.6 ± 0.1 | −29.5 ± 0.7 | 3.0 ± 0.1 | 10 | 12.9 ± 1.4 | 17.3 ± 1.9 | 1.6 ± 0.1 |
| T327V | 6 | 0.3 ± 0.1 | −28.5 ± 0.9 | 2.4 ± 0.0 | 6 | 8.6 ± 0.4 | 37.9 ± 2.0 | 1.4 ± 0.0 |
| V334I | 10 | 2.5 ± 0.2 | −18.8 ± 0.5 | 3.1 ± 0.1 | 10 | 16.8 ± 1.4 | 3.6 ± 2.5 | 1.5 ± 0.1 |
| G272A/I334L | 6 | 1.6 ± 0.2 | −17.4 ± 0.3 | 2.9 ± 0.1 | 6 | 15.6 ± 1.8 | −0.1 ± 1.8 | 1.0 ± 0.1 |
Number of experiments (n), average maximum tail current amplitudes (μA), and parameters deduced from the Boltzmann fitting for the chimeras and pore mutants of Ciona and human KCNQ1.
To identify which part or amino acid residues are responsible for the KCNE1 modulation, we further surveyed the S4–S6 region. Although the S5-P connector is the least conserved motif in the S4–S6 region, this motif itself is apparently not sufficient to induce the positive G-V shift, as shown in Fig. 3 A. There are only seven different amino acid residues in the S4–S6 segments of Ci-KCNQ1 other than the S5-P connector (Tyr175, Ala191, His242, Ile245, Leu248, Val251, and Ile258; see Fig. 1, red arrowheads). We introduced point mutations into these amino acid residues and found that A191G, L248V, and I258V rescued the G-V shift by KCNE1 to some extent (Fig. 4 and Table I). However, the point mutations we identified were less effective in the absence of the human S5-P connector (Fig. 4 A). We also examined double or triple mutations of these amino acids (Fig. 4 A) and found that double mutations of A191G and I258V with the human S5-P connector almost fully rescued the positive G-V shift by KCNE1 (Fig. 4, B and C).
To confirm that these amino acid residues are responsible for the KCNQ1 modulation by KCNE1, we introduced reverse point mutations into the corresponding amino acid residues of hKCNQ1 (Fig. 5 and Table I). Single mutations of G272A and V334I, which correspond to A191G and I258V in Ci-KCNQ1, respectively, showed a reduced voltage shift induced by KCNE1 coexpression (Fig. 5 A). The G-V shift of the double mutant of G272A/V334I induced by KCNE1 was significantly smaller than that of WT hKCNQ1 with KCNE1 (Fig. 5, A–C). Although other aspects of the current modulation by KCNE1, such as the current augmentation and the slow activation, were still observed, we concluded that these amino acids played an important role in the modulation of KCNQ1 gating, at least in terms of the voltage dependence.
The S1 segment of hKCNQ1 is critical for making KCNQ1 channels constitutively active by KCNE3
As we have shown in the previous section, and also according to previous immunoprecipitation work (Melman et al., 2004), the pore region of KCNQ1 seems to be an important part for the binding and modulation of KCNE proteins. However, KCNE3 failed to make the ciQ1hQ1(S4–S6) chimera constitutively active (Fig. 6, A and B), indicating that another region is required for the modulation by KCNE3. The ratio of the conductance between −80 and +20 mV (G−80mV/G+20mV), which we used for assessing the constitutively active component, is merely 0.01 ± 0.00 for ciQ1hQ1(S4–S6), whereas the WT hKCNQ1 with KCNE3 showed a G−80mV/G+20mV of 0.73 ± 0.02 (Fig. 6 C). Interestingly, KCNE3 was able to partly modulate ciQ1hQ1(S4–S6) in a similar manner as KCNE1 modulates KCNQ1: KCNE3 changed the activation kinetics of ciQ1hQ1(S4–S6) and slightly shifted its G-V relationship (Fig. 6, A and B). In addition, KCNE3 abolished inactivation seen in the homomeric KCNQ1 channel (Pusch et al., 1998; Tristani-Firouzi and Sanguinetti, 1998), which was also seen in homomeric ciQ1hQ1(S4–S6) (see “hook” tail current, the characteristic feature of homomeric KCNQ1 channel, in Fig. 6 A, insets). These observations suggest that KCNE3 could bind and modulate ciQ1hQ1(S4–S6) but failed to make the channel constitutively active. We subsequently explored which part of hKCNQ1 is important for the constitutive activity caused by KCNE3 modulation. hQ1ciQ1(S4-C), which has the first half (N-terminal region through S3 segment) of hKCNQ1 and the second half (S4 segment and the rest) of Ci-KCNQ1, showed a significant constitutively active component (0.18 ± 0.03; Fig. 6 C, third graph from the left), indicating that the first half of hKCNQ1 might be important for the constitutive opening. By further substituting the Ciona S5-P connector with the human S5-P connector, which assists the proper modulation of KCNE1 (see Fig. 4), the value of G−80mV/G+20mV became 1.10 ± 0.04, even larger than that of the WT (Fig. 6 C, fourth graph from the left). Exchanging the N-terminal region through the S1 segment with Ci-KCNQ1 resulted in a reduction in G−80mV/G+20mV, indicating that the S1 segment might play a significant role in the KCNE3 modulation (Fig. 6 C, fifth graph from the left). Six amino acid residues (Val39, Val46, Leu47, Met50, Met51, and Ile54) are different in the S1 segment of Ci-KCNQ1 compared with that of hKCNQ1 (see Fig. 1, blue arrowheads). By introducing point mutations on the ciQ1hQ1(S2–S3,S5P) chimera, we found that the V46F mutation significantly revived the KCNE3-sensitive KCNQ1 channel phenotype, whereas all other point mutants failed (Fig. 7, A and B). V46F alone was not enough to make Ci-KCNQ1 KCNE3-sensitive, and thus the S5-P connector from hKCNQ1 was still necessary (Fig. 7 C), although the human S5-P connector alone could not make Ci-KCNQ1 KCNE3-sensitive, as shown in Fig. 6 C.
We also examined whether Phe127 of hKCNQ1, which corresponds to Val46 in Ci-KCNQ1, has a significant impact on the modulation by KCNE3. As shown in Fig. 8, the F127V hKCNQ1 mutant somewhat impaired the ability of KCNE3 to modulate KCNQ1 channels; F127V/KCNE3 was a partly voltage-dependent channel retaining some constitutively active component (Fig. 8 B, blue triangles). Importantly, the mutation had almost no impact on the KCNQ1 channel property in the absence of KCNE proteins and in the presence of KCNE1 (Fig. 8 B). These results confirm that Phe127 plays a significant role only in the presence of KCNE3. By using this intermediate phenotype of F127V with KCNE3, we also tested whether the F127V/KCNE3 channel could be closed by hyperpolarization and subsequently activated normally. A series of 2-s depolarizing step pulses from two different holding potentials of −80 and −120 mV for 8 s was applied to oocytes expressing F127V and KCNE3 (Fig. 8, C and D). F127V/KCNE3 channels were closed by the holding potential of −120 mV and behaved as if they were normal voltage-gated potassium channels (Fig. 8 C, right). This observation strongly suggests that KCNE3 modifies the voltage dependence via the S1 segment and subsequently stabilizes the open state of KCNQ1 channels.
Figure 8.
The human KCNQ1 F127V mutant confirms that the S1 segment of KCNQ1 is important for the KCNE3 modulation. (A) Representative current traces for the hKCNQ1 F127V mutant in the absence (left) and presence (right) of KCNE3. The membrane potential was stepped up from −100 to +60 mV in 20-mV increments. (B) G-V relationships for the hQ1 F127V mutant in the absence (closed circles) and presence of KCNE1 (red squares) or KCNE3 (blue triangles). The G-V relationships of WT hKCNQ1 in the absence (black dotted curve) and presence of KCNE1 (red dotted curve) or KCNE3 (blue dotted curve) are superimposed. (C) Representative current traces for the hKCNQ1 F127V mutant in the presence of KCNE3 from the same oocyte at different holding potentials (−80 and −120 mV). 2-s depolarizing pulses were applied at every 10 s. (D) G-V relationships for the hQ1 F127V mutant with KCNE3 at a holding potential of −80 mV (open circles) and −120 mV (closed circles).
Phenylalanine residues on the S1 segment are important for KCNE3 modulation
Phe127 is located in the bottom half of the S1 segment and is exposed to the lipid side according to the structural models of the KCNQ1 channel (Smith et al., 2007). Interestingly, three phenylalanine residues (Phe123, Phe127, and Phe130) are aligned on the S1 helix (Fig. 9 A). We next examined whether these phenylalanine residues or aromatic rings are necessary for the constitutive activity induced by KCNE3. In addition to these three phenylalanine residues, we also tested Trp120, which could be aligned just under Phe123 if it is on the S1 segment. We substituted alanine residues for these aromatic amino acids to remove the aromatic (or indole) rings. In the absence of KCNE3, these F(W) to A mutants did not show any significant changes compared with WT hKCNQ1 (Fig. 9 B, left). In the presence of KCNE3, on the other hand, the constitutive activity was completely lost in F130A and significantly weakened in F127A and to some extent in F123A (Fig. 9 B, right). W120A had the least effect in the presence of KCNE3.
We next examined double mutations on Phe127 and Phe130 to further confirm the importance of these phenylalanine residues. We introduced alanine, valine, and leucine for the substitution of the phenylalanine residues. All introduced amino acid residues are hydrophobic with different sizes and lack aromatic rings. All the double mutants showed little effects on the KCNQ1 current without KCNE proteins (Fig. 10, black traces and plots, and Table II). The positive shift induced by KCNE1 was not impaired by the mutations, although F127A/F130A and F127V/F130V mutants showed a further positive shift of the G-V curve compared with those of hKCNQ1 WT and F127L/F130L mutant (Fig. 10, red traces and plots, and Table II). Most importantly, the current of the phenylalanine double mutants did not show any constitutive activity at all with KCNE3 coexpression (Fig. 10, blue traces and plots, and Table II). We therefore concluded that the phenylalanine residues on the S1 segment of hKCNQ1, especially Phe127 and Phe130, play a pivotal role exclusively in the induction of the constitutive activity in the presence of KCNE3.
Figure 10.
KCNE3 sensitivities are lost in the S1 phenylalanine double mutants. (A) Representative current traces of the hKCNQ1 F127L/F130L mutant in the absence (black) and presence of KCNE1 (red) or KCNE3 (blue) are shown. The membrane potential was stepped up from −100 to +60 mV in 20-mV increments. (B) G-V relationships for the human KCNQ1 WT and phenylalanine double mutants in the absence (black) and presence of KCNE1 (red) or KCNE3 (blue). All plots are fitted with the Boltzmann equation except the hKCNQ1 WT with KCNE3, which could not be fitted with it.
Table II.
Parameters of double phenylalanine-substituted mutants
| Mutant name | Without KCNE1 | With KCNE1 | With KCNE3 | |||||||||
| n | I (μA) | V1/2 (mV) | z | n | I (μA) | V1/2 (mV) | z | n | I (μA) | V1/2 (mV) | z | |
| Human KCNQ1 | 6 | 0.5 ± 0.0 | −19.6 ± 0.2 | 2.1 ± 0.1 | 6 | 14.7 ± 1.3 | 30.9 ± 2.5 | 1.6 ± 0.1 | 6 | 1.6 ± 0.2 | NA | NA |
| F127A/F130A | 6 | 0.3 ± 0.0 | −12.1 ± 1.2 | 2.2 ± 0.1 | 6 | 4.9 ± 0.4 | 84.6 ± 1.8 | 2.2 ± 0.1 | 6 | 1.3 ± 0.2 | −14.9 ± 1.6 | 1.4 ± 0.0 |
| F127V/F127V | 6 | 0.5 ± 0.1 | −6.3 ± 1.0 | 2.6 ± 0.1 | 6 | 9.0 ± 0.7 | 81.0 ± 1.8 | 1.9 ± 0.1 | 6 | 1.5 ± 0.2 | −15.5 ± 0.9 | 1.3 ± 0.0 |
| F127L/F130L | 6 | 0.6 ± 0.1 | −6.6 ± 0.4 | 2.3 ± 0.0 | 6 | 10.6 ± 1.1 | 37.8 ± 1.2 | 1.4 ± 0.1 | 6 | 1.0 ± 0.3 | 9.9 ± 2.9 | 1.5 ± 0.1 |
Number of experiments (n), average maximum tail current amplitudes (µA), and parameters deduced from the Boltzmann fitting for the S1 mutants of human KCNQ1. Data were taken from the same batch of oocytes, therefore, the data set for WT human KCNQ1 is different from the data set of Table I. NA, not applicable.
DISCUSSION
We used Ci-KCNQ1, whose current amplitude and kinetics were not effectively modulated by KCNE1 and totally insensitive to KCNE3, to survey the important amino acid residues in each KCNE modulation. Although only nonconserved amino acid residues between the Ciona gene and the human gene could be discovered by this strategy, we successfully identified several amino acid residues in the pore region that are important only for KCNE1 modulation and several phenylalanine residues on the S1 segment that are required for KCNE3 modulation. These findings give us some structural perspectives on the mechanisms of KCNQ1 modulation by KCNE proteins, as discussed below.
Structural implications given by the ineffective modulation of Ci-KCNQ1 by KCNE1
KCNE1 binds to the pore region (S5–S6 segments) of KCNQ1 (Melman et al., 2004), probably by penetrating into the gap of two adjacent voltage-sensing domains (Nakajo and Kubo, 2007; Xu et al., 2008; Chung et al., 2009). KCNE1 can also bind to various types of voltage-gated potassium channels, including KCNQ4, Kv1.5, Kv2.1, and Kv4.3 (McCrossan and Abbott, 2004; Melman et al., 2004; Radicke et al., 2006, 2009; McCrossan et al., 2009), but KCNQ1 is thought to be the main target for KCNE1 because of its drastic effects on the kinetics and voltage dependence. So which parts or amino acid residues of KCNQ1 are responsible for the modulation by KCNE1? The pore region of the KCNQ1 channel has long been considered to be the main binding site for KCNE1 protein, according to previous electrophysiological and biochemical studies (Kurokawa et al., 2001; Tapper and George, 2001; Melman et al., 2004; Panaghie et al., 2006; Kang et al., 2008). However, a growing list of recent studies concerning the VSD suggests that the VSD of the KCNQ1 channel plays a significant role in KCNE1 modulation (Nakajo and Kubo, 2007; Panaghie and Abbott, 2007; Rocheleau and Kobertz, 2008; Shamgar et al., 2008; Osteen et al., 2010; Wu et al., 2010a,b). In addition to the above studies, interaction between the cytosolic domains of KCNQ1 and KCNE1 has been suggested (Rocheleau et al., 2006; Wiener et al., 2008; Chen et al., 2009; Haitin et al., 2009; Lvov et al., 2010). The interaction at the extracellular region has also been suggested (Nakajo and Kubo, 2007; Xu et al., 2008; Chung et al., 2009; Wang et al., 2011). In the present study, we basically concentrated on the transmembrane region and especially on the pore region for KCNE1 modulation. Because we used Ci-KCNQ1 to identify important domains and amino acid residues in KCNE modulation, we could only address domains or amino acids that are not conserved between human and Ciona. For example, as the S4 segment is almost perfectly conserved, it is impossible to identify important amino acid residues in the S4 segment, if any. Therefore, our current study does not deny the importance of other regions in KCNE modulation.
As for the pore domain, Panaghie et al. (2006) have applied a tryptophan scanning through the second half of the S6 segment (from Ala336 to Gly345) and identified that two phenylalanine residues at 339 and 340 are responsible for binding with Leu59 and Thr58 of KCNE1, which are two out of the three residues known as the “activation triplet” in KCNE1 (Melman et al., 2001, 2002). However, the same phenylalanine residues are conserved in all KCNQ family members, including KCNQ4, which can be coimmunoprecipitated with KCNE1 but not modulated by KCNE1 (Melman et al., 2004). These residues may be the contacting sites for KCNE1 but do not explain why only KCNQ1 current is dramatically modulated by KCNE1. In this study, we successfully identified three amino acid residues (Gly272 in S5, and Val324 and Val334 in S6) involved in the KCNE1 modulation and not conserved among KCNQ family members (Fig. 11). Two residues on the S6 segment (Val324 and Val334) are exposed to the membrane lipid side (Fig. 12), and these mutants showed little effect on the KCNQ1 current in the absence of KCNE1 (Fig. 5), suggesting that these two amino acid residues can be the interaction sites for KCNE1. Because all valine, leucine, and isoleucine residues have nonpolar hydrophobic side chains, the residues involved in the interaction between KCNQ1 and KCNE1 may be intolerant to even subtle mutations, especially on Val334, which is highly conserved among vertebrate KCNQ1 channels (Fig. 11). The location of Val334 is in the middle of the S6 segment. Cys331, located one turn above Val334 on the α-helical S6 segment, can form a disulfide bond with engineered F54C or G55C mutant residues in KCNE1 (Tapper and George, 2001). Together with the previous report suggesting that Phe-339 and Phe-340 are in close proximity to Leu59 and Thr58 (Panaghie et al., 2006), Val334 may interact with the amino acids between Phe54 and Leu59 of KCNE1. Val324, on the other hand, is located on the extracellular end of the S6 segment. An engineered V324C in KCNQ1 can form a disulfide bond with an engineered L42C in KCNE1 and yields constitutively open channels (Chung et al., 2009). Leu42 of KCNE1 is located at the end of the extracellular domain and probably interacts with Val324 of KCNQ1 in the extracellular space. Interestingly, the amino acid residues Lys326 and Thr327, which were recently identified to be critical for the inhibition by the inhibitory subunit KCNE4 (Vanoye et al., 2009), are located in the region between Val324 and Val334. According to the data of Vanoye et al. (2009), Val324 and Val334 do not play a significant role in the inhibition by KCNE4. Conversely, T327V has a smaller impact on the modulation induced by KCNE1 (Fig. 5 A). The upper part of the S6 segment may be a common contacting site for all KCNE family members; however, each KCNE subunit may use different amino acid residues to modulate KCNQ1, as Vanoye et al. (2009) claimed.
Figure 11.
Sequence comparisons among different species and other KCNQ channels. KCNQ1 amino acid sequences of part of the S1 segment (122–135), S5 segment (269–275), S5-P connector (286–298), and the upper half of the S6 segment (324–334) from various species are aligned. Other types of KCNQ channels (KCNQ2-5) from humans are also aligned. Conserved Phe123, Phe127, Phe130, Gly272, Val324, and Val334 are colored in red. Conserved amino acid residues are highlighted in bold.
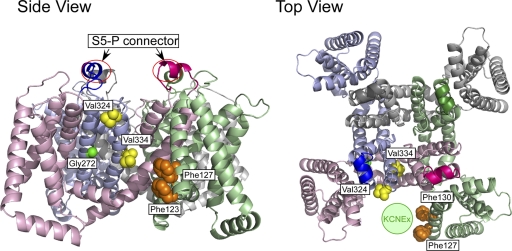
Figure 12.
Locations of important amino acid residues determined in this study and a putative binding site of the KCNE protein. The structural model for the open state of human KCNQ1 (Smith et al., 2007) is used to locate Phe123 (hidden below Phe127 in the top view), Phe127 and Phe130 (hidden behind Phe127 in the side view) on the S1 segment (orange), Gly272 on the S5 segment (green), and Val324 and Val334 on the S6 segment (yellow). S5-P connectors are circled in the side view (left). All four α subunits are shown, and each subunit is in a different color. The hypothetical location for the KCNE protein (“KCNEx”) is indicated as a light green circle in the top view (right).
The situation for the G272A mutant in the KCNQ1 S5 segment may be slightly different. Because glycine has the smallest side chain, just a hydrogen atom, the glycine residue might not directly contribute to the binding of KCNE1. Instead, glycine might give the S5 segment some flexibility. There is another glycine residue on the S5 segment at position 269. Although Gly269 is conserved among all KCNQ family members, Gly272 is seen only in KCNQ1 (Fig. 11). This extra glycine residue might confer extra flexibility to the KCNQ1 S5 segment, and that may contribute to the modulability of KCNQ1 by KCNE1. According to the KCNQ1 structural model (Smith et al., 2007), Gly272 sits right next to Val334 (Fig. 12); therefore, these two sites might work synergistically.
The role of the S1 segment in KCNE modulation
In the presence of KCNE3, which makes KCNQ1 channels constitutively active, the VSD seems to be fixed in the upstate or other ion-conducting state (Nakajo and Kubo, 2007; Rocheleau and Kobertz, 2008). In this study, we identified that the S1 segment, rather than the pore region, is important for sensitivity to KCNE3. Although there have been no reports yet indicating that KCNE3 directly interacts with the S1 segment of KCNQ1, there is some evidence that the S1 segment is in close proximity with KCNE1 (Xu et al., 2008; Chung et al., 2009; Wang et al., 2011). The cysteine residue introduced on the C-terminal end (extracellular side) of the S1 segment can form a disulfide bond with another introduced cysteine residue on the extracellular region of the KCNE1 protein (Xu et al., 2008; Chung et al., 2009; Wang et al., 2011). As KCNE1 is thought to be located between two adjacent VSDs, one KCNE1 subunit can interact with three different KCNQ1 subunits via the pore domain, S4 segment, and S1 segment (Fig. 12, top view). Because the S5-P connector is also required to convert Ci-KCNQ1 into a KCNE3-sensitive channel (Fig. 7), the pore region is a likely binding site for KCNE3 as well. KCNE3 is probably located between two VSDs as KCNE1 is and might be close to the pore region and S1 segment of KCNQ1 subunits (Fig. 12). We therefore assume that KCNE3 directly interacts with the S1 segment and modifies the equilibrium of VSD. Interestingly, two gain of function mutations have been found in the upper part of the S1 segment (S140G and V141M), which are associated with atrial fibrillation (Y.H. Chen et al., 2003; Hong et al., 2005). These mutations slow the deactivation of the KCNQ1 channel in the presence of KCNE1; indicating KCNE1 can affect the VSD movement upon depolarization through the S1 segment (Restier et al., 2008). Fig. 8 implies that KCNQ1/KCNE3 can be closed by strong hyperpolarization; therefore, KCNE3 might stabilize the open state by a similar mechanism of the atrial fibrillation mutants.
The phenylalanine residues (Phe123, Phe127, and Phe130) herein identified on the S1 segment of hKCNQ1 may be direct interaction sites for KCNE3. KCNE1 might also make contact with these phenylalanine residues because F127A/F130A and F127V/F130V double mutants showed a further positive shift of the G-V curve in the presence of KCNE1 (Fig. 10). However, the aromatic rings may not be important for the KCNE1 modulation because F127L/F130L showed normal G-V shift with KCNE1 (Fig. 10). All three residues seem to be exposed to the lipid side according to the open-state model of KCNQ1 (Smith et al., 2007) (see Fig. 12), meaning that these amino acid residues are accessible for KCNE3. This speculation is supported by the observation that the phenylalanine mutations showed almost no impact on the KCNQ1 current in the absence of KCNE proteins (Figs. 8–10). It is still not known which part of KCNE3 is important for the binding with any part of the KCNQ1 channel. Future work to identify the contacting site on the KCNE3 side is necessary for understanding mechanisms of the modulation of the KCNQ1 channel by KCNE3.
The role of the S5-P connector
Another important finding of this study is that the extracellular S5-P connector of hKCNQ1 is indispensable for the modulation by KCNE proteins, although the connector itself does not have a sufficient ability to induce a positive shift of the voltage dependence (see Figs. 3, 4, 6, and 7). According to the previous immunoprecipitation experiments, the pore region of hKCNQ1 is enough to coimmunoprecipitate KCNE1 (Melman et al., 2004). The S5-P connector of hKCNQ1 may be involved in the binding of KCNE proteins. However, it is apparent that binding is not enough to induce the complete modulation of KCNQ1, including the G-V shift, according to the results obtained with KCNQ4 (Melman et al., 2004) and our current study of Ci-KCNQ1. Proper arrangement may be required for the modulation for KCNE proteins, and the S5-P connector and probably the intracellular region of KCNQ1 might help to put KCNE proteins in a proper position to induce the modulation.
Three amino acid residues in the S5-P connector have been reported as mutation sites in human LQT1 patients (E290K, G292D, and R293C) (Tester et al., 2005; Smith et al., 2007). These mutations might have a problem for the KCNE protein binding and thus might not be modulated properly by KCNE proteins.
Acknowledgments
We thank Y. Asai for the preparation of oocytes and other technical assistance.
This work was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (KAKENHI 20790184 and 22790223 to K. Nakajo). This work was also supported by General Collaborative Project by the National Institute for Physiological Sciences.
Angus C. Nairn served as editor.
Footnotes
Abbreviations used in this paper:
- VSD
- voltage sensor domain
- WT
- wild type
References
- Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. 1996. KVLQT1 and lsK (minK) proteins associate to form the IKs cardiac potassium current. Nature. 384:78–80 10.1038/384078a0 [DOI] [PubMed] [Google Scholar]
- Bendahhou S., Marionneau C., Haurogne K., Larroque M.M., Derand R., Szuts V., Escande D., Demolombe S., Barhanin J. 2005. In vitro molecular interactions and distribution of KCNE family with KCNQ1 in the human heart. Cardiovasc. Res. 67:529–538 10.1016/j.cardiores.2005.02.014 [DOI] [PubMed] [Google Scholar]
- Chen H., Kim L.A., Rajan S., Xu S., Goldstein S.A. 2003. Charybdotoxin binding in the IKs pore demonstrates two MinK subunits in each channel complex. Neuron. 40:15–23 10.1016/S0896-6273(03)00570-1 [DOI] [PubMed] [Google Scholar]
- Chen J., Zheng R., Melman Y.F., McDonald T.V. 2009. Functional interactions between KCNE1 C-terminus and the KCNQ1 channel. PLoS ONE. 4:e5143 10.1371/journal.pone.0005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Xu S.J., Bendahhou S., Wang X.L., Wang Y., Xu W.Y., Jin H.W., Sun H., Su X.Y., Zhuang Q.N., et al. 2003. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 299:251–254 10.1126/science.1077771 [DOI] [PubMed] [Google Scholar]
- Chung D.Y., Chan P.J., Bankston J.R., Yang L., Liu G., Marx S.O., Karlin A., Kass R.S. 2009. Location of KCNE1 relative to KCNQ1 in the IKS potassium channel by disulfide cross-linking of substituted cysteines. Proc. Natl. Acad. Sci. USA. 106:743–748 10.1073/pnas.0811897106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Kline R.P., Pennefather P., Cohen I.S. 1994. Gating of IsK expressed in Xenopus oocytes depends on the amount of mRNA injected. J. Gen. Physiol. 104:87–105 10.1085/jgp.104.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitin Y., Wiener R., Shaham D., Peretz A., Cohen E.B., Shamgar L., Pongs O., Hirsch J.A., Attali B. 2009. Intracellular domains interactions and gated motions of IKS potassium channel subunits. EMBO J. 28:1994–2005 10.1038/emboj.2009.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A.S., Nishino A., Nakajo K., Zhang G., Fineman J.R., Selzer M.E., Okamura Y., Cooper E.C. 2008. Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet. 4:e1000317 10.1371/journal.pgen.1000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K., Bjerregaard P., Gussak I., Brugada R. 2005. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J. Cardiovasc. Electrophysiol. 16:394–396 10.1046/j.1540-8167.2005.40621.x [DOI] [PubMed] [Google Scholar]
- Jentsch T.J. 2000. Neuronal KCNQ potassium channels: physiology and role in disease. Nat. Rev. Neurosci. 1:21–30 10.1038/35036198 [DOI] [PubMed] [Google Scholar]
- Kang C., Tian C., Sönnichsen F.D., Smith J.A., Meiler J., George A.L., Jr, Vanoye C.G., Kim H.J., Sanders C.R. 2008. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 47:7999–8006 10.1021/bi800875q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa J., Motoike H.K., Kass R.S. 2001. TEA+-sensitive KCNQ1 constructs reveal pore-independent access to KCNE1 in assembled IKs channels. J. Gen. Physiol. 117:43–52 10.1085/jgp.117.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zaydman M.A., Wu D., Shi J., Guan M., Virgin-Downey B., Cui J. 2011. KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc. Natl. Acad. Sci. USA. 108:9095–9100 10.1073/pnas.1100872108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005a. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005b. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 309:903–908 10.1126/science.1116270 [DOI] [PubMed] [Google Scholar]
- Lundquist A.L., Manderfield L.J., Vanoye C.G., Rogers C.S., Donahue B.S., Chang P.A., Drinkwater D.C., Murray K.T., George A.L., Jr 2005. Expression of multiple KCNE genes in human heart may enable variable modulation of IKs. J. Mol. Cell. Cardiol. 38:277–287 10.1016/j.yjmcc.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Lvov A., Gage S.D., Berrios V.M., Kobertz W.R. 2010. Identification of a protein–protein interaction between KCNE1 and the activation gate machinery of KCNQ1. J. Gen. Physiol. 135:607–618 10.1085/jgp.200910386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrossan Z.A., Abbott G.W. 2004. The MinK-related peptides. Neuropharmacology. 47:787–821 10.1016/j.neuropharm.2004.06.018 [DOI] [PubMed] [Google Scholar]
- McCrossan Z.A., Roepke T.K., Lewis A., Panaghie G., Abbott G.W. 2009. Regulation of the Kv2.1 potassium channel by MinK and MiRP1. J. Membr. Biol. 228:1–14 10.1007/s00232-009-9154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman Y.F., Domènech A., de la Luna S., McDonald T.V. 2001. Structural determinants of KvLQT1 control by the KCNE family of proteins. J. Biol. Chem. 276:6439–6444 10.1074/jbc.M010713200 [DOI] [PubMed] [Google Scholar]
- Melman Y.F., Krumerman A., McDonald T.V. 2002. A single transmembrane site in the KCNE-encoded proteins controls the specificity of KvLQT1 channel gating. J. Biol. Chem. 277:25187–25194 10.1074/jbc.M200564200 [DOI] [PubMed] [Google Scholar]
- Melman Y.F., Um S.Y., Krumerman A., Kagan A., McDonald T.V. 2004. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 42:927–937 10.1016/j.neuron.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Morin T.J., Kobertz W.R. 2008. Counting membrane-embedded KCNE beta-subunits in functioning K+ channel complexes. Proc. Natl. Acad. Sci. USA. 105:1478–1482 10.1073/pnas.0710366105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajo K., Kubo Y. 2007. KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel. J. Gen. Physiol. 130:269–281 10.1085/jgp.200709805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajo K., Kubo Y. 2011. Nano-environmental changes by KCNE proteins modify KCNQ channel function. Channels (Austin). 5:397–401 [DOI] [PubMed] [Google Scholar]
- Nakajo K., Katsuyama Y., Ono F., Ohtsuka Y., Okamura Y. 2003. Primary structure, functional characterization and developmental expression of the ascidian Kv4-class potassium channel. Neurosci. Res. 45:59–70 10.1016/S0168-0102(02)00193-1 [DOI] [PubMed] [Google Scholar]
- Nakajo K., Ulbrich M.H., Kubo Y., Isacoff E.Y. 2010. Stoichiometry of the KCNQ1-KCNE1 ion channel complex. Proc. Natl. Acad. Sci. USA. 107:18862–18867 10.1073/pnas.1010354107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y., Nishino A., Murata Y., Nakajo K., Iwasaki H., Ohtsuka Y., Tanaka-Kunishima M., Takahashi N., Hara Y., Yoshida T., et al. 2005. Comprehensive analysis of the ascidian genome reveals novel insights into the molecular evolution of ion channel genes. Physiol. Genomics. 22:269–282 10.1152/physiolgenomics.00229.2004 [DOI] [PubMed] [Google Scholar]
- Osteen J.D., Gonzalez C., Sampson K.J., Iyer V., Rebolledo S., Larsson H.P., Kass R.S. 2010. KCNE1 alters the voltage sensor movements necessary to open the KCNQ1 channel gate. Proc. Natl. Acad. Sci. USA. 107:22710–22715 10.1073/pnas.1016300108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaghie G., Abbott G.W. 2007. The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J. Gen. Physiol. 129:121–133 10.1085/jgp.200609612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaghie G., Tai K.K., Abbott G.W. 2006. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J. Physiol. 570:455–467 10.1113/jphysiol.2005.100644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch M., Magrassi R., Wollnik B., Conti F. 1998. Activation and inactivation of homomeric KvLQT1 potassium channels. Biophys. J. 75:785–792 10.1016/S0006-3495(98)77568-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicke S., Cotella D., Graf E.M., Banse U., Jost N., Varró A., Tseng G.N., Ravens U., Wettwer E. 2006. Functional modulation of the transient outward current Ito by KCNE beta-subunits and regional distribution in human non-failing and failing hearts. Cardiovasc. Res. 71:695–703 10.1016/j.cardiores.2006.06.017 [DOI] [PubMed] [Google Scholar]
- Radicke S., Cotella D., Sblattero D., Ravens U., Santoro C., Wettwer E. 2009. The transmembrane beta-subunits KCNE1, KCNE2, and DPP6 modify pharmacological effects of the antiarrhythmic agent tedisamil on the transient outward current Ito. Naunyn Schmiedebergs Arch. Pharmacol. 379:617–626 10.1007/s00210-008-0389-1 [DOI] [PubMed] [Google Scholar]
- Restier L., Cheng L., Sanguinetti M.C. 2008. Mechanisms by which atrial fibrillation-associated mutations in the S1 domain of KCNQ1 slow deactivation of IKs channels. J. Physiol. 586:4179–4191 10.1113/jphysiol.2008.157511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau J.M., Kobertz W.R. 2008. KCNE peptides differently affect voltage sensor equilibrium and equilibration rates in KCNQ1 K+ channels. J. Gen. Physiol. 131:59–68 10.1085/jgp.200709816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau J.M., Gage S.D., Kobertz W.R. 2006. Secondary structure of a KCNE cytoplasmic domain. J. Gen. Physiol. 128:721–729 10.1085/jgp.200609657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M.C., Curran M.E., Zou A., Shen J., Spector P.S., Atkinson D.L., Keating M.T. 1996. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 384:80–83 10.1038/384080a0 [DOI] [PubMed] [Google Scholar]
- Schroeder B.C., Waldegger S., Fehr S., Bleich M., Warth R., Greger R., Jentsch T.J. 2000. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 403:196–199 10.1038/35003200 [DOI] [PubMed] [Google Scholar]
- Shamgar L., Haitin Y., Yisharel I., Malka E., Schottelndreier H., Peretz A., Paas Y., Attali B. 2008. KCNE1 constrains the voltage sensor of Kv7.1 K+ channels. PLoS One. 3:e1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.A., Vanoye C.G., George A.L., Jr, Meiler J., Sanders C.R. 2007. Structural models for the KCNQ1 voltage-gated potassium channel. Biochemistry. 46:14141–14152 10.1021/bi701597s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai K.K., Goldstein S.A. 1998. The conduction pore of a cardiac potassium channel. Nature. 391:605–608 10.1038/35416 [DOI] [PubMed] [Google Scholar]
- Takumi T., Ohkubo H., Nakanishi S. 1988. Cloning of a membrane protein that induces a slow voltage-gated potassium current. Science. 242:1042–1045 10.1126/science.3194754 [DOI] [PubMed] [Google Scholar]
- Takumi T., Moriyoshi K., Aramori I., Ishii T., Oiki S., Okada Y., Ohkubo H., Nakanishi S. 1991. Alteration of channel activities and gating by mutations of slow ISK potassium channel. J. Biol. Chem. 266:22192–22198 [PubMed] [Google Scholar]
- Tapper A.R., George A.L., Jr 2001. Location and orientation of minK within the IKs potassium channel complex. J. Biol. Chem. 276:38249–38254 [DOI] [PubMed] [Google Scholar]
- Tester D.J., Will M.L., Haglund C.M., Ackerman M.J. 2005. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2:507–517 10.1016/j.hrthm.2005.01.020 [DOI] [PubMed] [Google Scholar]
- Tristani-Firouzi M., Sanguinetti M.C. 1998. Voltage-dependent inactivation of the human K+ channel KvLQT1 is eliminated by association with minimal K+ channel (minK) subunits. J. Physiol. 510:37–45 10.1111/j.1469-7793.1998.037bz.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoye C.G., Welch R.C., Daniels M.A., Manderfield L.J., Tapper A.R., Sanders C.R., George A.L., Jr 2009. Distinct subdomains of the KCNQ1 S6 segment determine channel modulation by different KCNE subunits. J. Gen. Physiol. 134:207–217 10.1085/jgp.200910234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.W., Tai K.K., Goldstein S.A. 1996. MinK residues line a potassium channel pore. Neuron. 16:571–577 10.1016/S0896-6273(00)80076-8 [DOI] [PubMed] [Google Scholar]
- Wang W., Xia J., Kass R.S. 1998. MinK-KvLQT1 fusion proteins, evidence for multiple stoichiometries of the assembled IsK channel. J. Biol. Chem. 273:34069–34074 10.1074/jbc.273.51.34069 [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Jiang M., Xu X.L., Hsu K.L., Zhang M., Tseng G.N. 2011. Gating-related molecular motions in the extracellular domain of the IKs channel: implications for IKs channelopathy. J. Membr. Biol. 239:137–156 10.1007/s00232-010-9333-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener R., Haitin Y., Shamgar L., Fernández-Alonso M.C., Martos A., Chomsky-Hecht O., Rivas G., Attali B., Hirsch J.A. 2008. The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction. J. Biol. Chem. 283:5815–5830 10.1074/jbc.M707541200 [DOI] [PubMed] [Google Scholar]
- Wu D., Delaloye K., Zaydman M.A., Nekouzadeh A., Rudy Y., Cui J. 2010a. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J. Gen. Physiol. 135:595–606 10.1085/jgp.201010408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Pan H., Delaloye K., Cui J. 2010b. KCNE1 remodels the voltage sensor of Kv7.1 to modulate channel function. Biophys. J. 99:3599–3608 10.1016/j.bpj.2010.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Jiang M., Hsu K.L., Zhang M., Tseng G.N. 2008. KCNQ1 and KCNE1 in the IKs channel complex make state-dependent contacts in their extracellular domains. J. Gen. Physiol. 131:589–603 10.1085/jgp.200809976 [DOI] [PMC free article] [PubMed] [Google Scholar]