Introduction
Although hereditary predisposition to cancer was known before 1900, it was only then, after the rediscovery of Gregor Mendel's once-ignored 19th century work, that hereditary predisposition to cancer could be rationalized. By then it was also known that the pattern of chromosomes in cancer cells is abnormal. The next contribution to understanding cancer genetics was made by Theodor Boveri, who proposed that some chromosomes might stimulate cell division and others might inhibit it, but his idea was long overlooked. Now we know that there are genes of both types.
In this review, we summarize the history of the study of the latter type of genes, tumour suppressor genes (TSGs), and the evidence supporting a role for both complete and partial tumour suppressor inactivation in the pathogenesis of cancer. We integrate the classical “two-hit” hypothesis of tumour suppression with a continuum model that accounts for subtle dosage effects of tumour suppressors, and we discuss other exceptions to the two-hit hypothesis such as the emerging concept of “obligate haploinsufficiency” in which partial loss of a TSG is more tumourigenic than complete loss. The continuum model highlights the importance of subtle regulation of TSG expression or activity such as regulation by miRNAs. Finally, we discuss the implications of this model on the diagnosis and therapy of cancer.
The “two-hit” hypothesis
The first evidence for a genetic abnormality as a cause of cancer came with the discovery, in 1960, of the abnormal “Philadelphia” chromosome in chronic myelogenous leukaemia cells1. Later, in 1973, it was discovered that this chromosome was a translocation between chromosomes 9 and 222 and in 1977, a translocation of chromosomes 15 and 17 was identified in acute promyelocytic leukaemia4. Eventually, the genes at the breakpoints of these translocations were cloned - BCR/ABL in 19833 and PML/RARα in 19915. Meanwhile, seminal work demonstrated that the cancer-causing gene within the avian sarcoma virus genome, v-src, was actually a co-opted and mutated version of a normal cellular gene termed a proto-oncogene, now called c-src6,7. These observation demonstrated that normal cellular genes, when mutated or altered, are able to cause cancer, and were followed by the identification of numerous other cellular oncogenes that are activated by mutation, chromosome translocation, or amplification. One of Boveri's predictions was proven to be correct.
In the 1980s, scientists verified Boveri's other prediction with the identification of a second class of genes involved in cancer, “tumour suppressor genes,” that inhibit cancer development and oppose oncogene function. In a normal cell, a physiological balance between tumour suppressors and oncogenes maintains homeostasis and allows carefully regulated cell proliferation without unrestrained malignant tumour growth. Somatic cell fusion experiments pointed to the existence of such TSGs, because fusion of a normal cell with a malignant cell could revert the malignant cell to a normal phenotype8. One paradox that puzzled cancer researchers was that cancer susceptibility syndromes usually display a dominant mode of inheritance whereas the hypothesized “tumour suppressor genes” appeared to function in a recessive manner in the in vitro cell fusion experiments.
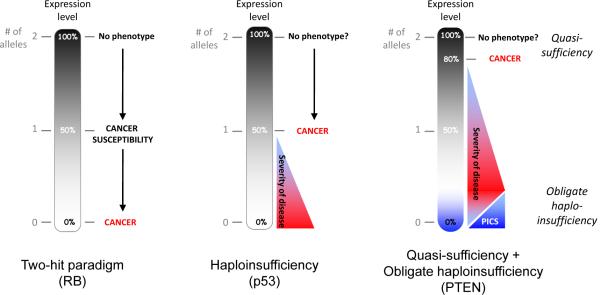
The resolution of this problem was provided by the analysis of a tumour of children, retinoblastoma (RB), which is sometimes present even at birth9. Statistical modelling indicated that hereditary cases likely developed after only one somatic mutational event. It was hypothesized that that one mutant allele is inherited and the other is generated somatically during growth of the developing eye. Because of the high chance of the second “hit” occurring, almost all individuals with the inherited first hit (the mutant allele) will develop retinoblastoma, and therefore the cancer susceptibility phenotype is inherited in a dominant manner (Figure 1). In contrast, tumour initiation requires both hits, and therefore tumourigenesis is recessive. A similar mechanism could be extended to sporadic retinoblastoma, the only difference being that in the sporadic cases both the first and second hits occur in somatic tissue. At the time of the original analysis, it wasn't known if these two hits occurred in the same or distinct genes, but it was clear that the search for the secondary mutation would start with the disease gene itself.
Figure 1. Paradigms of tumour suppression.
A black gradient represents a continuum of expression that is also related to the number of alleles present (gray numbering). Left, the two-hit paradigm as exemplified by the tumour suppressor, RB. Loss of one allele induces cancer susceptibility; loss of two alleles induces cancer. Middle, classic haploinsufficiency. Loss of one allele is sufficient for induction of cancer. Right, Quasi-sufficiency and obligate haploinsufficiency. Quasi-sufficiency refers to the phenomenon whereby tumour suppression is impaired after subtle expression downregulation without loss of even one allele. Obligate haploinsufficiency occurs when TSG haploinsufficiency is more tumourigenic than complete loss of the TSG, usually due to the activation of fail-safe mechanisms following complete loss of TSG expression.
The success of the two-hit model came with the identification of deletion of a locus on chromosome 13 in RB tumours and the subsequent cloning of the RB1 gene as mutated in familial retinoblastoma10–17. This discovery was made possible by the development of restriction fragment length polymorphism (RFLP) mapping technology. Using this approach, it was shown that many families with familial retinoblastoma did indeed harbour mutations or deletions of RB1 in the germline (the first hit), and tumours from RB patients nearly always contained mutation or loss of the other RB1 allele10–17 (the second hit). This latter event most often results from somatic mitotic recombination, while in a minority of cases, the normal allele acquires a distinct mutation. The role of recessive TSGs in dominantly inherited cancer susceptibility syndromes could then be understood, and Boveri's second prediction was substantiated.
These findings and others established the two-hit hypothesis as a model that not only explained hereditary cancer susceptibility but also provided a proof-of-principle that TSGs could be identified by the study of chromosomal deletions and genetic linkage in hereditary cases of a cancer and subsequent analysis of the second allele of the locus. Such a strategy was critical to the ultimate identification of TP53 (commonly called p53) as a TSG18 and was used successfully in the identification of other TSGs such as APC19, BRCA120, and BRCA221. Now the advent of high-throughput sequencing and genome-wide copy number profiling is allowing further systematic identification of regions of mutation and deletion in tumour genomes22,23.
It should be noted that it was initially hypothesized that two hits would be sufficient for the initiation of some tumours such as paediatric cancers, but additional mutations would be necessary for others. It is now thought that non-hereditary cancers require at approximately four or more distinct mutation events that result in perturbation of critical cellular signalling pathways such as PI3K, p53, or RB24. Malignancy is thought to result from an iterative process of somatic mutation followed by clonal expansion25. In the context of hereditary cancer syndromes, full inactivation of the involved disease gene may be the rate-limiting step for tumour initiation, but other events may occur, particularly to promote or enhance tumour progression. In this respect, the “two-hit hypothesis” should be applied directly to the TSG, with two hits representing the number of events necessary for inactivation of the TSG, but not necessarily for tumourigenesis. Haploinsufficiency, as we discuss below, must also be interpreted in this context; in many cases, a gene might be haploinsufficient for a specific cellular function, but TSG haploinsufficiency may promote tumour formation only in the context of other genetic or environmental insults.
Haploinsufficiency and quasi-sufficiency
The two-hit hypothesis clearly explained the differences in tumour number and age of onset of RB in hereditary vs. sporadic cases of RB. Moreover, it led to the eventual cloning of RB1 as the first bona fide human TSG. The hypothesis has been useful in cloning genes for hereditary cancers and for their non-hereditary counterparts. However, this success has led to a questionable dogma in the field that all important tumour suppressors should behave according to the “two-hit” model.
Investigators sought to apply the principles learned from the two-hit model to sporadic cancers in order to identify novel TSGs. Many recurring regions of chromosomal deletion have been identified in sporadic cancers, suggestive of the presence of a TSG. Unfortunately, it has become apparent that not all regions of consistent loss are accompanied by obvious aberrations on the other allele, raising the question of whether these deletions may represent “passenger” events not relevant to carcinogenesis. Indeed, very few of the identified regions of chromosomal losses have yielded clear TSGs in those loci that conform to the two-hit hypothesis. This finding has led many to conclude that the relevant tumour suppressors in those regions have not been identified or do not exist, when in fact many genes in those loci are known to have tumour suppressive properties in vitro or in vivo.
Alternatively, these regions could harbour bona fide TSGs but may represent regions/genes where single copy mutation or loss plays a role in tumourigenesis. These single copy events may be even be selected for during tumourigenesis instead of biallelic TSG loss. One possibility is that the initial lesion, when reduced to homozygosity, may lead to cell death or senescence. The lethality could be due to homozygosity of the disease gene itself or of other distinct genes included in the initial “hit,” since recombination is a principal “second hit” in tumourigenesis26. Such a scenario would result in an “obligate haploinsufficiency” in which selection pressure during tumourigenesis favours partial, but not complete, loss of the TSG.
A second possibility is that the single copy mutation of a TSG functions as a dominant-negative towards the wild-type gene/protein. After mutation of one allele, the mutant protein product interferes with the normal wild-type protein produced from the remaining wild-type allele. Because the complete normal function of the TSG is already impaired with only one hit, there is no selection pressure by the tumour for loss or mutation of the wild-type allele.
A third possibility is that a gene or genes in the region of consistent deletion exhibits haploinsufficiency for tumour suppressor function and single-copy loss of the TSG is sufficient for aberrant TSG function and promotion of cancer (Figure 1). In contrast with classical TSGs that are insensitive to large reductions in expression or activity, the function of haploinsufficient TSGs is impaired by a 50% partial reduction in expression or activity, or sometimes by subtler changes, as in a “quasi-insufficiency” of TSG function that we discuss below.
The role of haploinsufficiency in cancer has been met with scepticism, despite the well-established role of haploinsufficiency in numerous developmental disorders such as, for instance, aniridia (caused by haploinsufficiency for PAX6) and Grieg syndrome (caused by haploinsufficiency of GLI3)27. Just as increased dosage of genes can result in developmental syndromes, such as Down syndrome, increased dosage or activity of oncogenes is an established genetic mechanism of malignancy28, exemplified by the role of MYC amplification or RAS hyperactivity in cancer. However, the notion that subtle decreases in gene dosage or protein activity can be relevant to cancer has gained only limited acceptance in the scientific community.
One reason for this scepticism stems from the difficulty in definitively proving a haploinsufficient TSG is involved in tumourigenesis. Whereas the rare TSGs that fully conform to the two-hit hypothesis can be identified by their homozygous deletion or mutation in cancer, haploinsufficient TSGs cannot be identified in such a manner. Moreover, large regions of the genome, encompassing many genes, can be targeted by allelic loss and likewise, many genes may acquire somatic mutations during the course of tumour development. It is assumed that only a portion of these genes is responsible for the cancer, while the rest are passenger mutations. What then could be the gold standard by which to determine which gene or genes in the region are causative and which are non-involved bystanders? Unlike the analysis of traditional TSGs, no one assay or approach offers definitive proof that a haploinsufficient TSG is causally involved in tumourigenesis. Instead, an integrated approach involving human genetic analysis, in vitro and ex vivo functional studies, and murine cancer genetic modelling is critical to ascertain whether a gene may function as a haploinsufficient gene in cancer. Although there are caveats of each analysis on its own, a body of evidence from each of these analyses can strongly implicate a gene in dosage-sensitive tumour suppression.
Throughout this review, we use PTEN as a model dosage-sensitive TSG, but numerous other TSGs exhibit haploinsufficiency and dosage-sensitivity. Notably, TP53, commonly referred to as p53, exhibits haploinsufficiency. Mice with heterozygous (+/−) p53 mutation show an intermediate survival to that of p53 homozygous mutants and wild-type mice, and tumours that develop in the +/− animals do not always display loss of the remaining wild-type allele29. Similarly, tumours in patients with Li-Fraumeni syndrome, a cancer susceptibility syndrome caused by germline mutation of p53, do not always exhibit loss of the wild-type p53 allele, suggesting that haploinsufficiency of p53 may be sufficient for tumour initiation in humans as well30. A caveat with these analyses is that the effect of p53 haploinsufficiency on cancer initiation in mouse models and Li-Fraumeni patients may be due to complex non-cell autonomous effects stemming from the partial loss of p53 throughout the body. However, ex vivo analysis of murine p53 heterozygous thymocytes showed that these cells have an impaired apoptotic response to ionizing radiation or etoposide31. Similarly, a p53-deleted HCT116 isogenic cell line expressed only 25% the normal level of p53 mRNA and exhibited an impairment in induction of p53-responsive genes and apoptosis after exposure to UV radiation32. These studies demonstrate that reductions in p53 dosage and function can impact on a cells' ability to respond to oncogenic stimuli and strengthen the notion that p53 haploinsufficiency may drive tumourigenesis.
Similarly, other classic cancer susceptibility genes such as BRCA1 and BRCA2 may exhibit haploinsufficiency and/or dosage effects. Primary breast and ovarian cells from patients with heterozygous germline mutation of BRCA1 or BRCA2 have altered mRNA profiles compared to BRCA wild-type cells, suggesting that single hits in these genes can confer phenotypic differences that could in principle impact on breast and/or ovarian tumourigenesis33. Indeed, ex vivo studies of human BRCA1 mutant breast cells have found that these cells exhibit enhanced colony formation potential34 and show impaired lineage commitment in differentiation assays35. Although tumour formation in BRCA1 and BRCA2 carriers does appear to require the “second hit,” these “one-hit” premalignant changes could impact tumourigenesis by either promoting loss of the second TSG allele or of other TSGs, or by altering the cellular phenotype to predispose to certain tumour subtypes.
Another haploinsufficient TSG is the transcription factor PAX5. PAX5 is found mutated, deleted, or translocated in approximately 30% of acute lymphoblastic leukaemia cases, but the aberrant mutations/deletions are almost invariably mono-allelic and appear to function as hypomorphs, not dominant negative alleles36. Similarly, the E3 ligase FBW7 is mutated in approximately 10% of human colorectal cancers37. In 70% of these cases, the mutations are mono-allelic, suggestive of haploinsufficiency. Conditional mono-allelic deletion of Fbw7 in the murine intestine cooperated with the tumourigenic APCmin allele in intestinal tumourigenesis, albeit to a lesser degree than biallelic deletion of Fbw7, suggesting that Fbw7 is dosage-sensitive37.
Like complete loss of TSGs, the effect of TSG haploinsufficiency can be highly tissue-specific and context dependent (Figure 2). Thus, the cellular and molecular context in which TSG function is altered will determine the outcome of such impairment of TSG function. The differing outcomes of TSG expression level in differing situations may reflect distinct thresholds of protein expression or activity needed for certain processes or in differing cell contexts. For example, in one cell type there may be other compensatory proteins that mask the potential phenotype caused by TSG haploinsufficiency whereas in other cells these proteins are not expressed and so the haploinsufficiency of the TSG manifests as tissue-specific cancer susceptibility.
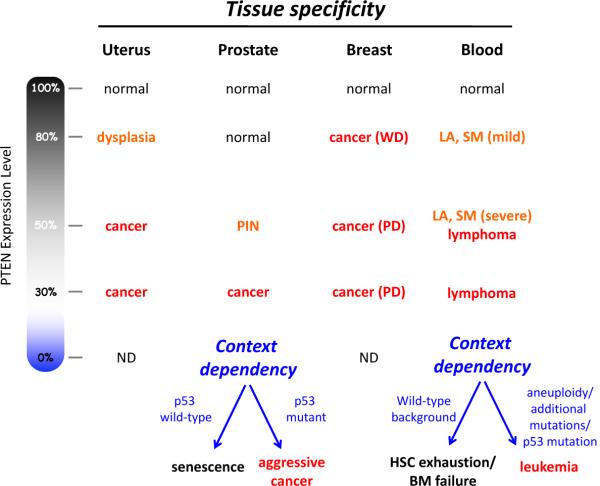
Figure 2. Tissue specificity and context dependency of tumour suppression.
The phenotypic outcome of a reduction in PTEN expression is differentially manifested depending upon tissue type and genetic background. WD, well-differentiated. PD, poorly-differentiated. LA, lymphadenopathy. SM, splenomegaly. HSC, hematopoietic stem cell. BM, bone marrow. The effect of complete loss of PTEN is highly context dependent due to the obligate haploinsufficiency caused by PTEN loss-induced cellular senescence (PICS). Data summarized here come from multiple groups and studies of genetically-engineered mice with differing PTEN alleles and expression38–40,52,65–68.
Two hits for tumour suppression
The genetic background of the individual or tumour will also impact the phenotypic outcome of TSG haploinsufficiency. In some cases, combinatorial effects could arise whereby haploinsufficiency or mutation of other genetic loci is required for a phenotype caused by TSG haploinsufficiency. A powerful example of this context dependency is provided by the interaction between Pten deficiency or haploinsufficiency and the presence or absence of mutated p5338 (Figure 2). In the context of wild-type p53, Pten haploinsufficiency is actually more tumourigenic in the prostate than complete loss of Pten, because the latter triggers a p53-dependent fail-safe senescence mechanism called Pten-loss induced cellular senescence (PICS). Haploinsufficiency of Pten enhances proliferation without activating PICS, making partial loss of Pten more detrimental in this context than complete loss of Pten (Figure 2). Similarly, complete loss of PTEN in the blood induces hematopoietic stem cell exhaustion or bone marrow failure, unless loss of PTEN is accompanied by other genetic events39. After accumulation of aneuploidy or other mutations, such as loss of p53, complete loss of PTEN in the blood induces fatal leukemias39,40 (Figure 2).
This phenomenon defines a novel paradigm of “obligate haploinsufficiency” in which selection pressure during tumour growth may favour haploinsufficiency of a TSG over complete loss in certain scenarios (Figure 1). However, in advanced cancers with p53 mutation or loss, PICS cannot be induced and complete loss of Pten enhances proliferation and tumourigenesis to a greater degree than Pten haploinsufficiency. This remarkable finding likely explains why complete loss of PTEN is typically restricted to advanced cancers and also emphasizes the importance of single-hit haploinsufficiency of TSGs in cancer initiation. Understanding the combinatorial and contextual dependencies of genetic events should not only enhance our ability to effectively treat tumours with differing combinations on genetic “hits” but will also inform chemopreventative strategies for eradicating cancer by attacking mechanisms necessary for its development.
Like PTEN, many other TSGs exhibit “obligate haploinsufficiency” where partial loss of the TSG is tumourigenic but complete loss is lethal to the cell. One example is the microRNA-processing enzyme DICER1. Mono-allelic deletion or mRNA downregulation of DICER1 is observed in diverse cancers41,42. In murine models, mono-allelic conditional deletion of Dicer1 enhances lung tumourigenesis driven by oncogenic Kras41, sarcoma development induced by KrasG12D and loss of p5341, and retinoblastoma formation induced by deletion of Rb1 and p10742. In the lung model, complete inactivation of Dicer1 improved survival compared to animals with mono-allelic Dicer1 deletion41. Tumours that did develop in the Dicer1flox/flox animals retained one wild-type allele and partial Dicer1 expression, strongly suggesting that partial loss of Dicer1 promotes tumourigenesis.
A recent in vivo shRNA-based screening approach identified lymphoma tumour suppressors, including one, the cell cycle regulator Rad17, which appeared to function as a TSG with obligate haploinsufficiency43. The use of multiple hairpins targeting Rad17 led to the discovery that only hairpins that partially suppressed Rad17 promoted lymphomagenesis whereas hairpins that completely repressed Rad17 expression were selected against in the lymphomas. This study highlights the utility of shRNA knockdown and screening technologies for the identification of haploinsufficient TSGs.
NPM1, found mutated and translocated in hematopoietic malignancies, also displays obligate haploinsufficiency. Whereas single-allelic deletion of NPM1 induces tumour formation in mice44, biallelic loss induces embryonic lethality and is also incompatible with cell growth after conditional deletion ex vivo45. Although more data is needed, the TSGs TAp63 and PinX1 may also display obligate haploinsufficiency; heterozygosity for TAp63 promoted tumourigenesis to a greater degree than complete loss of TAp63 in genetically engineered mice, and tumours that developed in the heterozygous mice did not display loss of the wild-type allele46. Similarly, PinX1, recently identified as a haploinsufficient tumour suppressor47, may also display obligate haploinsufficiency; heterozygous loss of PinX1 in a mouse model promoted cancer development in multiple organs, and tumours did not lose the wild-type allele or expression of PinX147.
A genome-wide screening study in yeast indicated that about 3% of the genes in the yeast genome displayed haploinsufficiency when assayed under specific growth conditions48. These haploinsufficient genes were enriched for ribosomal genes and genes that are highly expressed. Currently, mammalian tumour suppressors that exhibit haploinsufficiency do not appear to conform to one specific functional class. Besides p53 and PTEN, dozens of other TSGs exhibit haploinsufficiency for tumour suppression49,50, many of which we have discussed above. These TSGs come from many functional classes, including transcription factors (e.g. PAX5 and TAp63), cell cycle checkpoint genes (RAD17), E3 ligases (FBW7), ribosomal or associated proteins (NPM1), and other diverse genes such as Dicer1 and PinX1.
A continuum model for tumour suppression
Rather than TSG function and activity following discrete, step-wise changes, evidence is emerging that supports the dosage-dependency of TSG function. Data from an allelic series of targeted Pten alleles that results in varying levels of Pten in vivo in genetically-engineered mice51,52 demonstrate the tight correlation between Pten expression level and function. Taking advantage of a null Pten allele in combination with a Pten “hy” allele, which is expressed at a lower level than the wild-type allele, an allelic series was created in which Pten expression follows a decreasing pattern in the order Pten+/+ > Ptenhy/+ > Pten+/− > Ptenhy/−. Surprisingly, Ptenhy/+ animals, with cells expressing 80% of the normal level of Pten, did display many of the phenotypes observed in Pten+/− animals, such as lymphadenopathy, splenomegaly, and mammary gland tumours52. Moreover, survival of Ptenhy/+ animals is compromised compared to Pten+/+ controls, but not to the same degree as the survival impairment in Pten+/− animals52. Thus, the tumour incidence and survival of the animals correlate with the Pten expression level52. Critically, mammary tumours from Ptenhy/+ animals do not show loss of Pten expression or loss of the wild-type Pten allele and exhibit a Pten dose-dependent increase in activation of Akt, further demonstrating that a subtle reduction in Pten dose can result in deregulated signalling and cancer52. Similarly, in the murine prostate, a progressive lowering of Pten levels correlates with Akt activation, prostate enlargement, proliferation of prostate cells and tumourigenesis51. These in vivo studies demonstrate that even a subtle 20% reduction of protein level could constitute an important “hit” involved in the development of cancer. We termed this state: TSG “quasi-insufficiency”52 (Figure 1).
Thus, protein function can be a continuum that is related to the level of expression or activity of the TSG rather than to discrete step-by-step changes in gene copy number. We therefore propose a shift from the classical discrete model of tumour suppression to a continuum model (Figure 3). This model takes into account the fact that subtle regulation of TSGs can have profound consequences on cancer susceptibility and/or progression, and in turn has important implications for cancer diagnosis and therapy. Note that while the function of PTEN appears to be strongly coupled to its expression level, for other TSGs it will be the activity of the protein and not the expression level that may be critical. This situation is readily observed in tumours with p53 mutation where the mutant allele is highly expressed but functionally inactive or acting as a dominant negative.
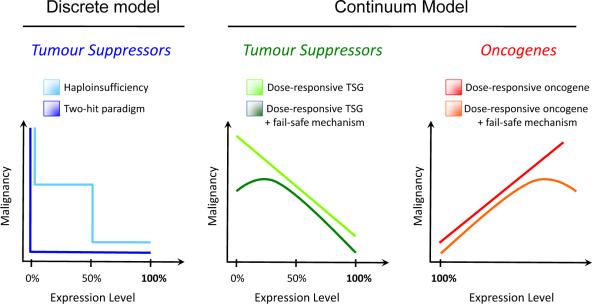
Figure 3. The continuum model of tumour suppression.
The classical discrete, stepwise model of tumour suppression (left) is contrasted with a continuum model of tumour suppression and oncogenesis (centre and right, respectively). In the discrete model, tumourigenesis is induced by either complete loss of a TSG (two-hit paradigm, dark blue) or after single-copy loss of a TSG (haploinsufficiency, light blue). In contrast, we propose a continuum model (centre and right), in which tumour suppression is related to a continuum of TSG expression, rather than to discrete changes in DNA copy number. A continuum of increasing TSG expression will generally be negatively correlated with malignancy (centre, light green) whereas increasing oncogene expression will generally be positively correlated with malignancy (right, red). A linear relationship is depicted for schematic purposes, but the dose-response relationship need not be linear. In some cases, fail-safe mechanisms are induced by complete loss of TSG expression or by massive oncogene overexpression. In these cases, complete loss of TSG expression (centre, dark green) or massive overexpression of an oncogene (right, orange) will be negatively correlated with malignancy, as shown.
Like the effect on TSGs, dosage can be extremely important for regulating the cellular effects of oncogene expression. Dramatically different consequences for the cell and for cancer development can occur depending on the level of oncogene expression. For example, high expression of oncogenic, mutated RAS genes drives senescence, not proliferation53. Similarly, robust expression of MYC induces apoptosis, not proliferation54. These examples mirror the paradigm of PICS and “obligate haploinsufficiency” and suggest that tumours may select for optimal expression of oncogenes. Data to support this notion exist already in the rampant amplification of mutated oncogenes, such as the amplification of the mutated EGFR gene in human lung cancer55,56. Such data suggests that it is not only mutation of an oncogene that is important for cancer, but also its expression level. In this respect, the continuum model could also apply to oncogenes (Figure 3).
It appears now that some TSGs are exquisitely sensitive to dose (e.g. PTEN), some are intermediately sensitive (e.g. p53) and others are more resistant to expression level and/or activity changes (e.g. RB1). Those that are most resistant to changes in expression or activity, such as RB1, will be most likely to conform to the two-hit hypothesis whereas those that are exquisitely sensitive to dose may exhibit frequent monoallelic deletion/mutation or be misregulated in cancer via other means, such as miRNA overexpression. Moreover, tumour suppressors that exhibit “obligate haploinsufficiency” due to the lethality of homozygous inactivation of the TSG will never exhibit complete inactivation in cancer unless there are other context- or genotype-dependent events that favour their complete inactivation (as in the case of Pten/p53 discussed above).
The dogma that every TSG must behave according to the two-hit paradigm could significantly slow the pace of cancer research by dismissing potentially critical genes as unimportant. The pace of cancer drug development will in turn be delayed by a lack of appreciation of the involvement of these potentially druggable pathways in tumour development and maintenance. A very real challenge for cancer biologists in the next decade will be first to define the TSGs affected by haploinsufficiency and then to model these subtle dosage effects in a way that can yield quantitative models of cancer risk or outcome. Since human genetics analysis alone is often not enough to unequivocally identify critical haploinsufficient genes, mouse models will prove essential to define which genes exhibit haploinsufficiency for TSG function and in which specific cellular and genetic contexts.
Regulation of TSG activity
The continuum model of tumour suppression holds that precise regulation of TSG expression and activity is critical and thus, mechanisms of regulation of expression and function may powerfully contribute to tumour suppression. One of the most widespread networks of expression regulation is the post-transcriptional regulatory network of microRNAs (miRNAs) and competing endogenous RNAs (ceRNAs). In addition to these networks, TSG dosage/activity is regulated by transcriptional, translational, and post-translational mechanisms (Figure 4). These networks open up the realm of genes that could be contextually important for tumourigenesis, especially when considering mechanisms that tune TSG activity and not only those that function as on/off switches.
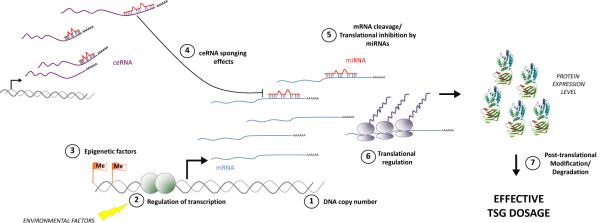
Figure 4. Mechanisms of regulation of TSG dosage.
The interaction between coding and non-coding factors determines final TSG dosage. Classic mechanisms such as DNA copy number (1), transcriptional regulation (2), and epigenetic silencing can impact expression of TSG mRNA. TSG mRNA level or translation into protein is then regulated by miRNAs (4). The availability of miRNAs for TSG downregulation is further regulated by ceRNA-mediated sponging effects (3). Finally, additional translational regulation (6) or post-translational modifications contribute to the final protein expression, function and effective dosage. The protein structure shown is PTEN (PDB ID: 1D5R).
As an example, a powerful PTEN-targeting miRNA network was recently identified. MiRNAs are small non-coding RNA molecules that regulate protein expression through inhibition of protein translation or induction of direct cleavage of their mRNA targets via RISC57. Many of the PTEN-targeting miRNAs are amplified or overexpressed in cancer such as miR-26 in glioma58 and miR-22 in prostate cancer59. The regulation of coding RNAs by miRNAs is made additionally complex through the interaction of miRNAs with ceRNAs60,61. We coined the term “competing endogenous RNA” (ceRNA), to refer to an RNA – coding or noncoding – that shares miRNA binding sites with another RNA with which it competes for miRNA binding60,61. Any two or more mRNAs with shared miRNA recognition elements (MRE) can act as ceRNAs towards each other by competing for miRNA binding and thereby co-regulating each other's expression. As the expression of the ceRNA increases, the miRNA binding sites on the ceRNA compete with the miRNA binding sites on the other transcript, and less miRNA is available for binding to the TSG transcript (Figure 4). CeRNA expression (e.g. expression of the PTEN pseudogene60), hence results in sustained TSG expression by preventing miRNA binding to the TSG and subsequent mRNA or protein downregulation. Conversely, loss of ceRNA expression could result in low TSG expression through diversion of additional miRNA molecules to the TSG transcript60,61. Even non-coding ceRNAs that are not expressed at the protein level could function as TSGs by regulating expression of protein-coding TSGs, and mutation or deletion of ceRNA genes could be important in cancer60,61.
Implications for cancer susceptibility and therapy
An appreciation of the continuous nature of tumour suppressive function must in turn lead to a re-evaluation of the role of TSG dosage in cancer susceptibility, diagnosis, and treatment. Subtle variation in TSG expression or activity could underlie genetic variation in cancer susceptibility in the population. Inherent, constitutive differences in gene expression could be caused by polymorphic variation in TSG promoter regions or in miRNA binding sites or other regulatory regions. Importantly, synonymous SNPs that do not change the protein sequence of an expressed gene could still have a powerful impact by altering the level of TSG expression through alteration of miRNA binding. Moreover, SNPs in non-coding RNAs could impact tumour development through ceRNA effects. Alternatively, on the basis of a continuum model, susceptibility to cancer could be induced by environmental factors through subtle transcriptional up- or down-regulation of relevant genes. Development of drugs that prevent and counter these fluctuations could be used as chemopreventative agents to reduce the impact of environmental carcinogens or to correct the effects of inherited predisposing factors.
Just as transcriptional or post-transcriptional regulation may determine cancer susceptibility, it could also be harnessed for cancer prevention and therapy (Table 1). As an example, statins, which are already in clinical use, were recently found to upregulate PTEN through PPARγ signaling62. Statins or PTEN-enhancing drugs could therefore be utilized not only to treat dyslipidemic cases but also to elevate PTEN expression in patients with tumours with lowered levels of PTEN (Figure 5). Importantly, such an approach would require a quantitative analysis of the expression levels of TSGs and oncogenes and not solely a genetic assessment of their presence or absence.
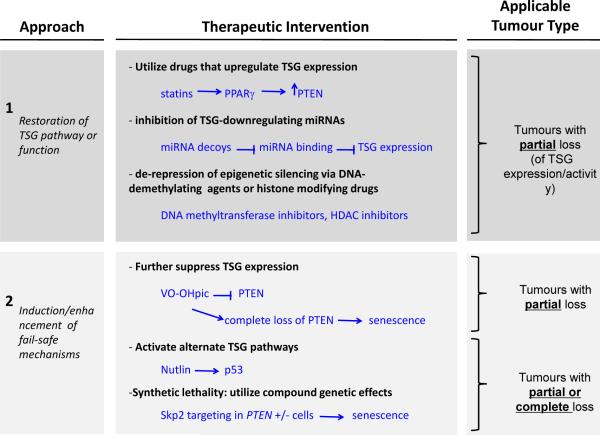
Figure 5. Opportunities for therapeutic intervention after partial or complete loss of TSG expression.
Tumour genotype ultimately determines which therapies can be considered for intervention. Broadly, two general approaches can be taken: restoration of the TSG pathway or function (upper panel) or induction of fail-safe mechanisms by complete downregulation of the TSG (lower panel). In tumours with downregulation of TSG expression in the absence of complete mutation or deletion, expression of the TSG can be attempted using drugs that induce the TSG transcription, inhibit miRNA-induced TSG downregulation, or relieve epigenetic silencing. Alternatively, fail-safe mechanisms may be induced by further downregulation of the TSG, such as inhibition of PTEN by the small molecule VO-OHpic69. In tumours with mutation or complete loss of the TSG, activation of other TSG pathways could be pursued. Alternatively, if a synthetic lethality relationship with the TSG is known, then the synthetic lethal gene can be targeted to induce cell death in the cells with complete loss of the TSG.
TSG-targeting microRNAs add another dimension to genotype/phenotype considerations in targeted cancer prevention and therapy. Overexpression of one or more of TSG-targeting miRNAs in cancer could confer cancer susceptibility, and tumours not showing genetic alterations in the TSG but with aberrant TSG levels due to miRNA misregulation could behave similarly to tumours with genetic deletion or mutation of the gene. Mapping the interactions between miRNAs and TSGs could be useful for defining and predicting tumour response to therapy.
Provocatively, tumours with partial loss of TSGs like PTEN that display “obligate haploinsufficiency” might be treated with agents to fully suppress the expression of the TSG, thereby suppressing tumour growth via senescence or other fail-safe mechanisms (Figure 5). Alternatively, when a TSG is completely lost, activation of alternative tumour suppressive pathways or synthetic lethality approaches could be considered63 (Figure 5). The success of such strategies would obviously depend on the integrity of the fail-safe response and the expression level of the TSG, once again requiring a careful annotation of the molecular characteristics of the cancer lesion subjected to treatment. The need for the same careful annotation is not restricted to targeted therapies but applies to any cancer therapy since variations of oncogene or TSG dosage may well be at the core of failure of more conventional radio- or chemo-therapies.
On this basis, the scientific challenge going forward will be to develop quantitative assays that can assess and score subtle differences in TSG gene expression to allow accurate diagnostics and prediction of risk/prognosis. Such diagnostics will also be important for analysis and prediction of response to therapeutics, since subtle variations could also determine response to therapy. A tumour with a partial downregulation of a TSG could respond differently from a tumour that has either no alteration of the TSG or complete mutation/loss of the gene (Figure 5). Critically, in cases where a tumour shows downregulation of a TSG via transcriptional repression, epigenetic downregulation, or aberrant miRNA and ceRNA regulation, there is an opportunity to explore therapeutic restoration of TSG expression.
The recent explosion of knowledge in the field of non-coding RNA biology has opened a new frontier in understanding gene dosage regulation and its involvement in tumourigenesis. In the months and years to come, careful experimentation is required to map out the networks of gene regulation of TSGs and oncogenes to allow drug development and creation of diagnostics from this information. Forty years after the twohit hypothesis, we are refining and extending our understanding of TSG inactivation in cancer. Milestones made in these past forty years have contributed to the decline in cancer deaths observed in the US since 199064. The next decades promise further relief from the burden of cancer as we continue to tease out its molecular genetic basis.
Preface
2011 marks the forty-year anniversary of the statistical analysis of retinoblastoma that provided the first evidence that tumourigenesis could be initiated by as few as two mutations. This work provided the foundation for the “two-hit” hypothesis that explained the role of recessive tumour suppressor genes (TSGs) in dominantly inherited cancer susceptibility syndromes. However, four decades later, it is now known that even partial inactivation of tumour suppressors can critically contribute to tumourigenesis. Here we analyse this evidence and propose a continuum model of TSG function to explain the full range of TSG mutations found in cancer.
Acknowledgements
We thank L. Salmena, L. Poliseno, and all Pandolfi lab members for advice and critical discussions. Our work is supported by NIH grants awarded to A.G.K. and P.P.P.
References
- 1.Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- 2.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–3. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 3.Heisterkamp N, et al. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983;306:239–42. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 4.Rowley JD, Golomb HM, Dougherty C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet. 1977;1:549–50. doi: 10.1016/s0140-6736(77)91415-5. [DOI] [PubMed] [Google Scholar]
- 5.Pandolfi PP, et al. Structure and origin of the acute promyelocytic leukemia myl/RAR alpha cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6:1285–92. [PubMed] [Google Scholar]
- 6.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–3. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 7.Oppermann H, Levinson AD, Varmus HE, Levintow L, Bishop JM. Uninfected vertebrate cells contain a protein that is closely related to the product of the avian sarcoma virus transforming gene (src) Proc Natl Acad Sci U S A. 1979;76:1804–8. doi: 10.1073/pnas.76.4.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris H. The analysis of malignancy by cell fusion: the position in 1988. Cancer Res. 1988;48:3302–6. [PubMed] [Google Scholar]
- 9.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]; The original publication that led to the “two-hit hypothesis.” Based on the Poisson distribution of the multiplicity of tumours found in hereditary retinoblastoma cases, it could be estimated that the number of mutations required for tumour formation is two.
- 10.Lee WH, et al. Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science. 1987;235:1394–9. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- 11.Fung YK, et al. Structural evidence for the authenticity of the human retinoblastoma gene. Science. 1987;236:1657–61. doi: 10.1126/science.2885916. [DOI] [PubMed] [Google Scholar]
- 12.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–6. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]; Building on several years of work by many labs to localize the gene responsible for hereditary retinoblastoma, this study is the first to identify and clone the responsible gene, RB1, and to show that it is altered in retinoblastoma tumours.
- 13.Sparkes RS, et al. Gene for hereditary retinoblastoma assigned to human chromosome 13 by linkage to esterase D. Science. 1983;219:971–3. doi: 10.1126/science.6823558. [DOI] [PubMed] [Google Scholar]
- 14.Benedict WF, et al. Patient with 13 chromosome deletion: evidence that the retinoblastoma gene is a recessive cancer gene. Science. 1983;219:973–5. doi: 10.1126/science.6336308. [DOI] [PubMed] [Google Scholar]
- 15.Dryja TP, et al. Homozygosity of chromosome 13 in retinoblastoma. N Engl J Med. 1984;310:550–3. doi: 10.1056/NEJM198403013100902. [DOI] [PubMed] [Google Scholar]
- 16.Cavenee WK, et al. Genetic origin of mutations predisposing to retinoblastoma. Science. 1985;228:501–3. doi: 10.1126/science.3983638. [DOI] [PubMed] [Google Scholar]
- 17.Cavenee WK, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–84. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 18.Baker SJ, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–21. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 19.Levy DB, et al. Inactivation of both APC alleles in human and mouse tumors. Cancer Res. 1994;54:5953–8. [PubMed] [Google Scholar]
- 20.Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet. 1992;2:128–31. doi: 10.1038/ng1092-128. [DOI] [PubMed] [Google Scholar]
- 21.Gudmundsson J, et al. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12-q13. Cancer Res. 1995;55:4830–2. [PubMed] [Google Scholar]
- 22.Volinia S, et al. Genome wide identification of recessive cancer genes by combinatorial mutation analysis. PLoS One. 2008;3:e3380. doi: 10.1371/journal.pone.0003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–8. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 25.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–41. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 26.Hagstrom SA, Dryja TP. Mitotic recombination map of 13cen-13q14 derived from an investigation of loss of heterozygosity in retinoblastomas. Proc Natl Acad Sci U S A. 1999;96:2952–7. doi: 10.1073/pnas.96.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher E, Scambler P. Human haploinsufficiency--one for sorrow, two for joy. Nat Genet. 1994;7:5–7. doi: 10.1038/ng0594-5. [DOI] [PubMed] [Google Scholar]
- 28.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–11. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 29.Venkatachalam S, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–67. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varley JM, Evans DG, Birch JM. Li-Fraumeni syndrome--a molecular and clinical review. Br J Cancer. 1997;76:1–14. doi: 10.1038/bjc.1997.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke AR, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–52. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 32.Lynch CJ, Milner J. Loss of one p53 allele results in four-fold reduction of p53 mRNA and protein: a basis for p53 haplo-insufficiency. Oncogene. 2006;25:3463–70. doi: 10.1038/sj.onc.1209387. [DOI] [PubMed] [Google Scholar]
- 33.Bellacosa A, et al. Altered gene expression in morphologically normal epithelial cells from heterozygous carriers of BRCA1 or BRCA2 mutations. Cancer Prev Res (Phila) 2010;3:48–61. doi: 10.1158/1940-6207.CAPR-09-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burga LN, et al. Altered proliferation and differentiation properties of primary mammary epithelial cells from BRCA1 mutation carriers. Cancer Res. 2009;69:1273–8. doi: 10.1158/0008-5472.CAN-08-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proia TA, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–63. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 37.Sancho R, et al. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–41. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper defined the paradigm of “obligate haploinsufficiency” of the PTEN gene by demonstrating that homozygous loss of PTEN is less tumourigenenic than heterozygous loss of PTEN due to the induction of a p53-dependent senescence response.
- 39.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–82. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 40.Lee JY, et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work presents the analysis of the contextual dependencies of leukaemia induced by loss of PTEN. This paper extends the “obligate haploinsufficiency” paradigm of PTEN to the hematopoietic system where it is found that, as in the prostate, loss of PTEN cooperates with p53 mutation.
- 41.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambertz I, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 17:633–41. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bric A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–35. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sportoletti P, et al. Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood. 2008;111:3859–62. doi: 10.1182/blood-2007-06-098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grisendi S, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–53. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 46.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–90. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou XZ, et al. The telomerase inhibitor PinX1 is a major haploinsufficient tumor suppressor essential for chromosome stability in mice. J Clin Invest. 2011 doi: 10.1172/JCI43452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deutschbauer AM, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–25. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fodde R, Smits R. Cancer biology. A matter of dosage. Science. 2002;298:761–3. doi: 10.1126/science.1077707. [DOI] [PubMed] [Google Scholar]
- 50.Payne SR, Kemp CJ. Tumor suppressor genetics. Carcinogenesis. 2005;26:2031–45. doi: 10.1093/carcin/bgi223. [DOI] [PubMed] [Google Scholar]
- 51.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 42:454–8. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]; A subtle reduction in Pten expression is shown to induce cancer in mice in a tissue-specific manner, demonstrating that very small changes in expression can promote cancer, and thereby defining the phenomenon of “quasi-insufficiency.”
- 53.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]; An example of the complex dosage effects of oncogenes, this paper demonstrates that aberrantly high expression of an oncogene can promote senescence, rather than proliferation.
- 54.Evan GI, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–28. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 55.Takano T, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–37. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 56.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huse JT, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poliseno L, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of a coding-independent functions of mRNAs whereby they act as ceRNAs that “sponge” miRNAs to regulate distinct mRNAs in trans.
- 61.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. The ceRNA hypothesis: the new Rosetta stone of a hidden RNA language. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teresi RE, Planchon SM, Waite KA, Eng C. Regulation of the PTEN promoter by statins and SREBP. Hum Mol Genet. 2008;17:919–28. doi: 10.1093/hmg/ddm364. [DOI] [PubMed] [Google Scholar]
- 63.Lin HK, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 464:374–9. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 65.Di Cristofano A, et al. Impaired Fas response and autoimmunity in Pten+/− mice. Science. 1999;285:2122–5. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki A, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–78. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 67.Podsypanina K, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–8. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–55. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 69.Alimonti A, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 120:681–93. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]