Abstract
The canonical role of messenger RNA (mRNA) is to deliver protein-coding information to sites of protein synthesis. However, given that microRNAs bind to RNAs, we hypothesized that RNAs possess a biological role in cancer cells that relies upon their ability to compete for microRNA binding and is independent of their protein-coding function. As a paradigm for the protein-coding-independent role of RNAs, we describe the functional relationship between the mRNAs produced by the PTEN tumour suppressor gene and its pseudogene (PTENP1) and the critical consequences of this interaction. We find that PTENP1 is biologically active as determined by its ability to regulate cellular levels of PTEN, and that it can exert a growth-suppressive role. We also show that PTENP1 locus is selectively lost in human cancer. We extend our analysis to other cancer-related genes that possess pseudogenes, such as oncogenic KRAS. Further, we demonstrate that the transcripts of protein coding genes such as PTEN are also biologically active. Together, these findings attribute a novel biological role to expressed pseudogenes, as they can regulate coding gene expression, and reveal a non-coding function for mRNAs.
In human cancers, monoallelic mutation of PTEN without loss or mutation of the second allele is prevalent at presentation, while complete loss is observed at low frequencies with the exception of advanced cancers1. In mouse models, heterozygosity for Pten leads to multiple cancers2, and serial reduction of Pten dosage has critical consequences for the incidence and severity of epithelial cancers3,4, together suggesting that PTEN is a functionally haploinsufficient tumour suppressor gene. The identification and validation of numerous PTEN-targeting microRNAs demonstrates that post-transcriptional regulation plays a pivotal role in determining PTEN abundance in cancer cells5–11. Cells are ultrasensitive to even subtle decreases in PTEN abundance, thus highlighting the importance of microRNA-mediated PTEN regulation in cancer4. Therefore, we reasoned that the relationship between PTEN and its pseudogene PTENP1 (PTH2/ψPTEN)12 could represent a compelling test for our hypothesis (Fig. 1a).
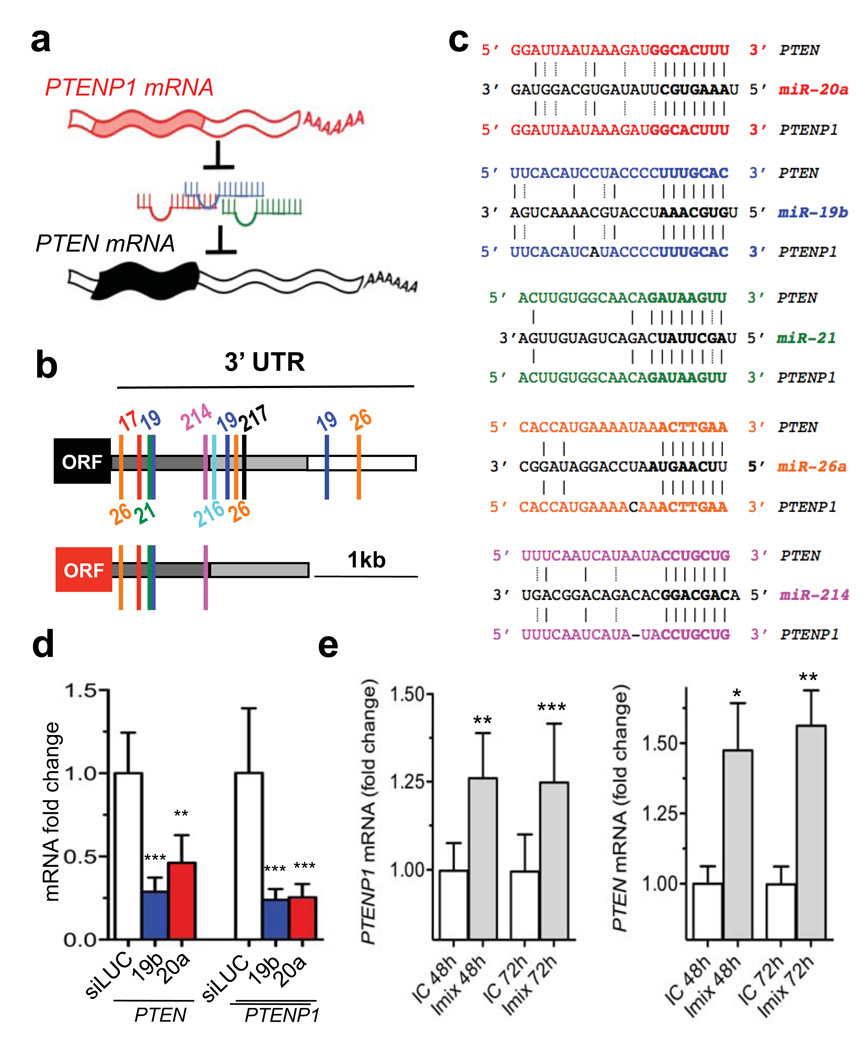
Figure 1. PTENP1 is targeted by PTEN-targeting microRNAs.
a. Working hypothesis: PTEN is protected from microRNA binding by PTENP1. microRNAs: colored squiggles; 5’and 3’UTRs: open rectangles; open reading frames: filled rectangles. b. PTEN (upper) and PTENP1 (lower) 3’UTRs contain a highly conserved (dark grey) followed by a poorly conserved (light grey) domain. PTEN-targeting microRNA seed matches within in the high homology region are conserved between PTEN and PTENP1. c. Binding of PTEN-targeting microRNAs to PTENP1. Seeds and seed matches: bold; canonical pairings: solid lines; non-canonical pairings (G:U): dotted lines. d. PTEN-targeting miR-19b and miR20a decrease PTEN and PTENP1 mRNA abundance. e. miR-17 and miR-19 family inhibitors derepress PTENP1 abundance (left). PTEN is used as positive control (right). d and e. mean ± s.d., n ≥ 3.
Pseudogenes are defined as genomic loci that resemble real genes, yet are considered biologically inconsequential because they harbour premature stop codons, deletions/insertions and frameshift mutations that abrogate their translation into functional proteins. Nevertheless, nucleotide sequences contained within pseudogenes are well preserved, suggesting that selective pressure to maintain these genetic elements exists, and that they may indeed play an important cellular role.
Pseudogenes exist as either processed or non-processed genetic elements. While non-processed pseudogenes arose from genetic duplications, processed pseudogenes were generated through retrotransposition; thus they contain no introns yet they commonly share 5’ and 3’ UTR sequences with their ancestral genes13. Pseudogenes are almost as numerous as coding genes and represent a significant proportion of the “transcriptome”14. Despite lacking canonical promoters, processed pseudogenes utilize proximal regulatory elements to mediate their transcription15. Their transcription exhibits tissue-specificity16 and is aberrantly activated in cancer17,18, suggesting that pseudogenes may contribute to carcinogenesis, although the mechanisms still remain elusive. Very few pseudogenes have been functionally characterized thus far13.
MicroRNAs, a large class of small non-coding RNAs (ncRNAs), have emerged as a critical element in cellular biology and pathophysiology. microRNAs have been demonstrated to impact almost all cellular processes and cell types from plants to humans19. microRNAs function by annealing to complementary sites on coding sequences or 3’UTRs of target gene transcripts, where they promote the recruitment of protein complexes that impair translation and/or decrease the stability of mRNA leading to a decrease in target protein abundance19–22. Physiologically, aberrant expression of microRNAs has been causally linked to human diseases and cancer23.
We have tested whether pseudogene-derived RNA transcripts and mRNA transcripts possess an active biological role in cancer that is independent of their protein-coding function but would rely upon their ability to compete for microRNA binding, thereby modulating the derepression of microRNA targets (Fig. 1a).
PTENP1 is targeted by PTEN-targeting microRNAs
PTENP1 is a processed pseudogene residing at 9p13.3; it is highly homologous to PTEN, with only 18 mismatches throughout the coding sequence. A missense mutation of the initiator methionine codon prevents translation12. PTENP1 possesses a 3’-UTR that is ~1kb shorter than that of PTEN (Fig. 1b). It can be divided into 2 regions relative to its homology with the PTEN 3’UTR: a high homology (~95%) 5’ region and a low homology (<50%) 3’ region (Fig. 1b, Supplementary Fig. 1). Within the high homology region, we found perfectly conserved seed matches for the PTEN-targeting miR-17, miR-21, miR-214, miR-19 and miR-26 families (Fig. 1c, Supplementary Fig. 1). To measure the role of these microRNAs on both PTEN and PTENP1 expression, we designed specific PCR primer sets in the non-homologous 3’UTR regions (Supplementary Fig. 2a,b). In DU145 prostate cancer cells, PTEN-targeting microRNAs miR-19b and miR-20a suppress both PTEN and PTENP1 mRNA abundance (Fig. 1d, Supplementary Fig. 3a). In these cells, a pool of inhibitors of endogenously expressed PTEN-targeting microRNAs (Supplementary Fig. 3b) de-repressed both PTEN and PTENP1 transcript levels (Fig. 1e). The use of chimeric luciferase plasmids indicated the microRNA:PTENP1 interaction was direct (Supplementary Fig. 4a–c). These data indicate that PTENP1 and PTEN are subjected to the same microRNA-mediated, post-transcriptional regulation.
The 3’UTR of PTENP1 has tumour suppressive activity
We examined the ability of PTENP1 3’UTR to function as a decoy of PTEN-targeting microRNAs using a retroviral vector expressing this 3’UTR (Supplementary Fig. 5a). The 3’UTR can be transcribed, but it cannot code for protein; however it still may exert a biological role. Indeed, PTENP1 3’UTR overexpression resulted in a derepression of both PTEN transcript and protein (Fig. 2a, Supplementary Fig. 5b and 10c). Consistent with elevated PTEN, AKT phosphorylation was reduced upon stimulation of cells with EGF (Fig. 2b). These molecular observations were accompanied by growth inhibition (Fig. 2c, Supplementary Fig. 5c and 10d) and a significant reduction in the number of colonies generated in semisolid medium (Fig. 2d).
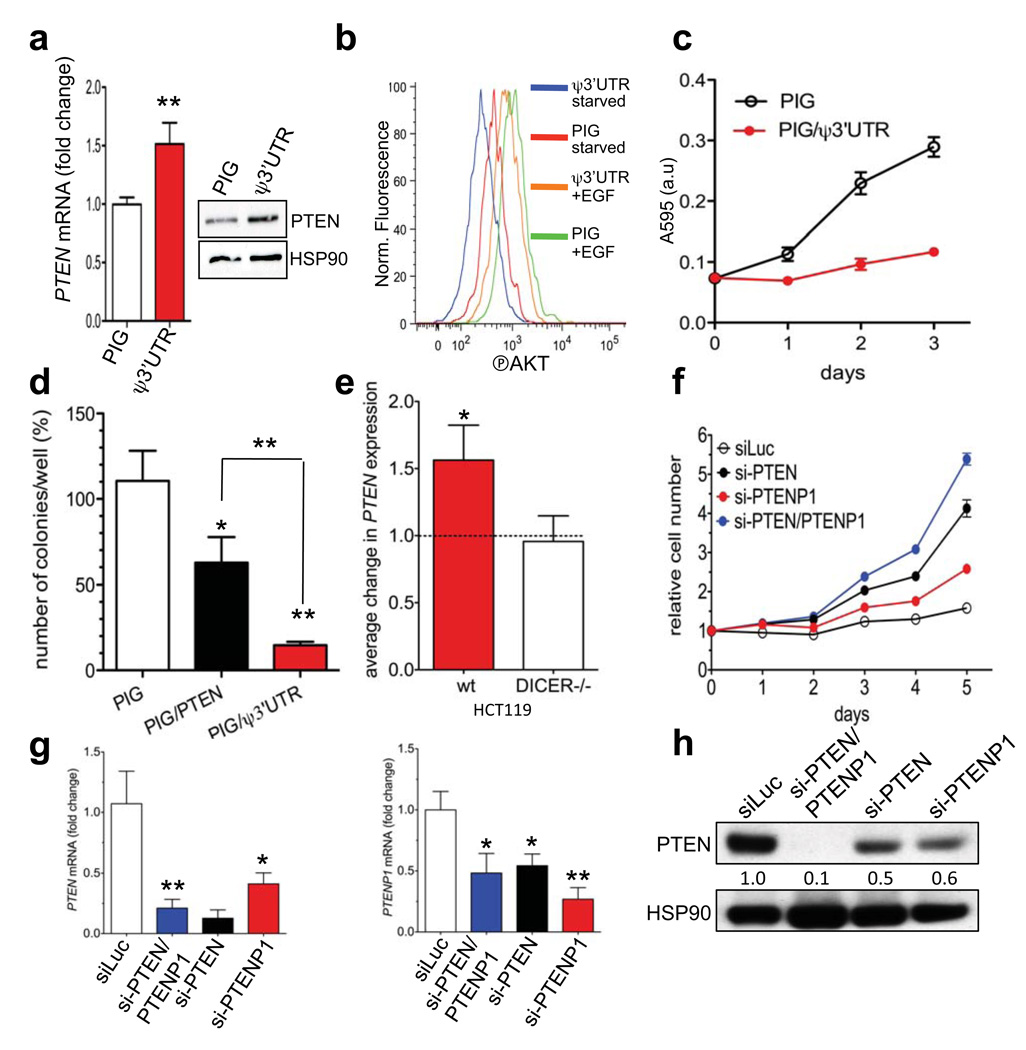
Figure 2. PTENP1 3’UTR exerts a tumour suppressive function by acting as a decoy for PTEN-targeting microRNAs.
a–c. PIG/ψ3’UTR-infected DU145 cells show (a) increased PTEN mRNA and protein levels (b) reduced phosho-AKT levels upon EGF stimulation and (c) decreased proliferation rate. d. Growth in semisolid medium of DU145 cells infected with PIG, PIG/ψ3’UTR or PIG/PTEN. e. PTEN mRNA levels 24h after the transfection of pCMV/ψ3’UTR in parental HCT116 or HCT116-DICER−/− cells. Data are normalized using pCMV empty-transfected cells. f. Growth curve of DU145 cells transfected with control siLuc, si-PTEN/PTENP1, si-PTEN or si-PTENP1. g. mRNA levels of PTEN (left) and PTENP1 (right) 24h after the transfection of siLuc (white), si-PTEN/PTENP1 (blue), si-PTEN (black), si-PTENP1 (red). i. Western blot of PTEN 48h after the transfection of the indicated siRNAs. a, c, d, e, f and g. mean ± s.d., n ≥ 3.
The derepression of PTEN abundance by PTENP1 3’UTR overexpression was blunted in HCT116-DICER−/− colon carcinoma cells (Fig. 2e). In these cells, the disruption of DICER -- the enzyme that catalyzes the last step of microRNA maturation -- leads to reduced levels of mature microRNAs compared to parental HCT116 cells24. This in turn supports the notion that the 3’UTR of PTENP1 requires mature microRNAs for its function towards PTEN.
To examine the phenotypic consequences of PTENP1 downregulation, we designed custom siRNA pools (Dharmacon) to specifically target either PTENP1 (si-PTENP1) or PTEN (si-PTEN) expression (Supplementary Fig. 6) since commercially available si-RNA pools for PTEN (si-PTEN/PTENP1) bind to common sequences in PTEN and PTENP1 (Supplementary Fig. 7a). si-PTENP1 transfection accelerated cell proliferation, suggesting that PTENP1, although expressed at lower relative levels, can exert a biological activity in DU145 cells (Fig. 2f). si-PTEN/PTENP1, which silences both PTEN and PTENP1, showed the strongest effect, indicating that PTEN and its pseudogene may have additive roles for growth suppression. PTENP1 knockdown resulted in decreased PTEN mRNA and protein abundance (Fig. 2g–h), mirroring the results obtained with overexpression of PTENP1 3’UTR (Fig. 2a).
In DU145 cells PTENP1 3’UTR is a more potent growth suppressor compared to PTEN (Fig. 2d, Supplementary Fig. 5c). This result may be explained by the fact that microRNAs for which PTENP1 functions as a decoy also bind other targets with tumour suppressive activities. For instance, the miR-17 family targets E2F1 and p2125, and miR-21 targets PDCD426. Accordingly, miR-17 and miR-21 mimics increase proliferation of PTEN-null PC3 cells (Supplementary Fig. 8a), suggesting PTEN independency. Indeed, si-PTENP1 resulted in a dose-dependent downregulation not only of PTEN, but also of p21 (Supplementary Fig. 8b). Additionally, both si-PTENP1 and si-PTEN/PTENP1 were able to suppress PTENP1 and increase proliferation in a dose-dependent manner in PTEN-null PC3 cells (Supplementary Fig. 8c–d). Conversely, stable infection of PTENP1 3’UTR in PC3 cells suppressed foci formation (Supplementary Fig. 8e), supporting the notion that PTENP1 and its 3’UTR exert a tumour suppressive role that goes beyond the regulation of PTEN abundance alone.
Expression and losses of PTENP1 in human cancer
PTEN and PTENP1 expression was explored in normal human tissues and prostate tumour samples, utilizing custom Taqman probes (see Methods, Supplementary Fig. 2c). In both the normal tissue and prostate tumour arrays, the direct correlation (r = 0.8087, p < 0.0001 and r = 0.3960, p = 0.0013, respectively) between PTEN and PTENP1 expression suggests that they may be co-regulated (Fig. 3a–b). This finding supports our molecular observations that PTENP1 can regulate PTEN expression. PTENP1 was found to be variably abundant, and in some cases expressed at higher levels than PTEN.
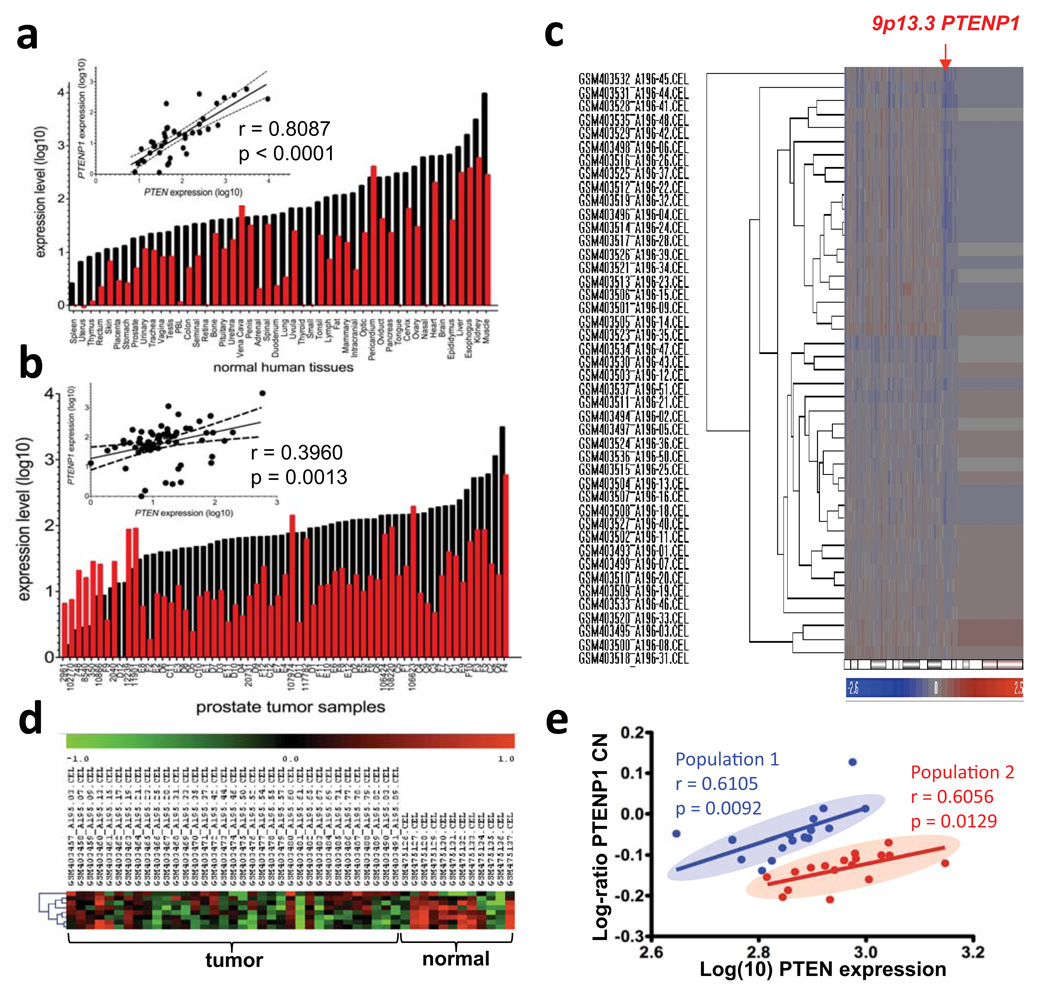
Figure 3. Loss of PTENP1 in cancer.
a–b. Expression level of PTEN (black) and PTENP1 (red) in a panel of normal human tissues (a) and prostate tumour samples (b). Linear regression of PTEN vs PTENP1 expression is shown in the upper left corner. c. Cluster analysis of 48 sporadic colon cancer samples interrogated by Affymetrix Human SNP Array. d. Heat map and Cluster analysis of Affymetrix Human Exon 1.0 ST Array for normalized PTEN intensity values. e. Plot of log-ratio of PTENP1 copy number (CN) against log10 PTEN expression intensity. Lines of best fit represent regression analyses of two populations. The correlation coefficient (r) measures the reliability and the p-value measures the statistical significance of the correlation between the x and y.
Next, we examined alterations of the PTENP1 genomic locus. Several array-based comparative genomic hybridization databases were mined including The Cancer Workbench (https://cgwb.nci.nih.gov/cgi-bin/heatmap) and NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/). (Supplementary Fig. 9a–b, Table S1). Remarkably, in a dataset of sporadic colon cancer (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE16125) (Fig. 3c–e), hierarchical clustering identified a clear population of samples with detectable copy number (CN) losses occurring specifically at the PTENP1 locus (Fig. 3c). Importantly, these CN losses were focal, not associated with large losses of 9p, and independent of losses at the CDKN2A locus (Supplementary Fig. 9c). T his data set formally demonstrates the existence of independent genomic CN losses at the PTENP1 locus, supporting the notion that PTENP1 exerts tumour suppressive functions and is under selective pressure to undergo CN losses in cancer.
In the same patient samples set, cluster analysis of PTEN expression showed that it was down-regulated compared to normal colon samples (p = 0.0008156; Fig. 3d). Regression analysis of PTENP1 CN variation with the expression levels of PTEN identified two discrete populations of patients in which PTENP1 CN variation and PTEN expression were directly and significantly correlated (Population 1: r = 0.6015, p = 0.0092; Population 2: r = 0.6056, p = 0.0129) (Fig. 3e). The existence of a direct relationship between PTENP1 CN and PTEN expression supports our hypothesis that PTENP1 transcript levels can regulate PTEN expression. Together, these findings constitute a proof of principle for the oncosuppressive activity of PTENP1.
A general model for endogenous mRNA-mediated biology
Based on our results, we expected that the PTEN 3’UTR would also have biological activity. Indeed, we found that PTEN 3’UTR can derepress PTENP1 abundance, as PTENP1 does on PTEN (Fig. 4a, left and Supplementary Fig. 10a-c). Importantly, PTEN 3’UTR overexpression was accompanied by growth inhibition, suggesting that PTEN exerts its tumour suppressive activity, at least in part, through its 3’UTR (Fig. 4a, right and Supplementary Fig. 10d).
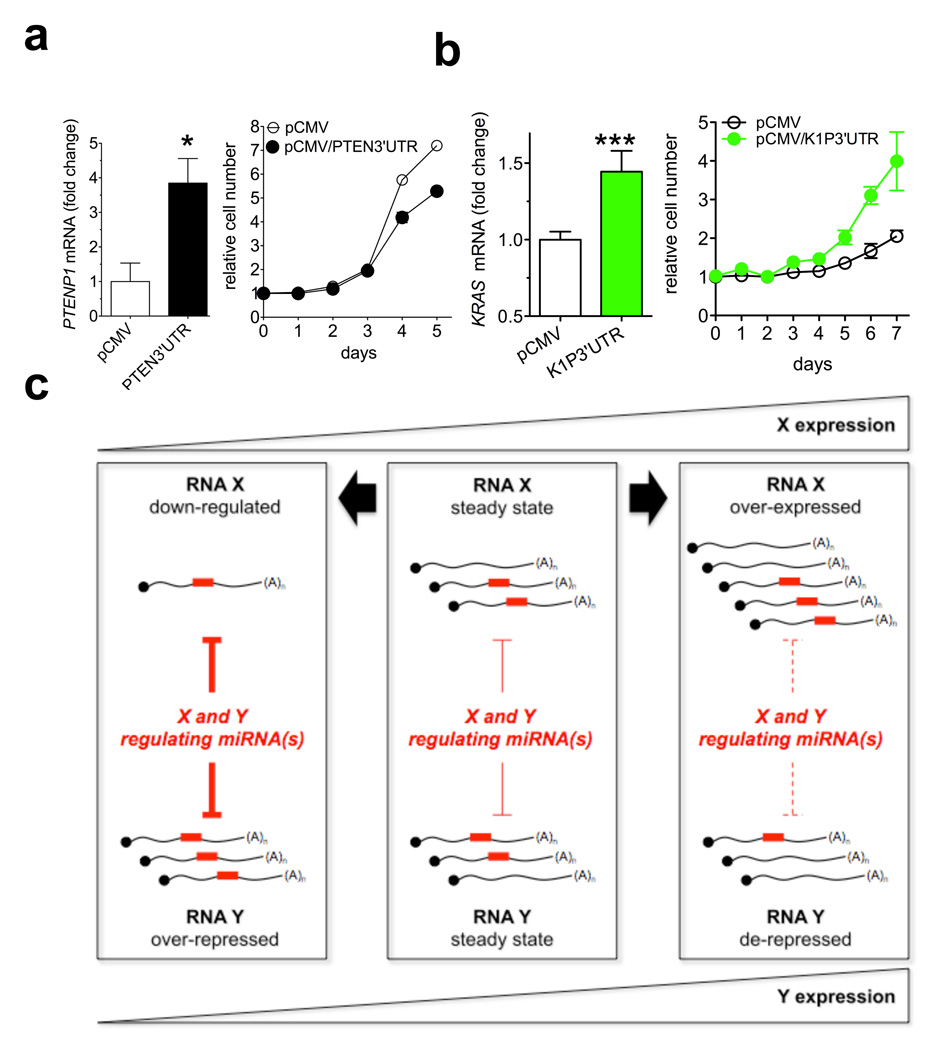
Figure 4. PTEN 3’UTR and KRAS1P 3’UTR function as decoys and a general model for endogenous microRNA decoy mechanism.
a. PTENP1 mRNA level 24h after the transfection of the empty pCMV or pCMV/PTEN3’UTR plasmid in DU145 cells (left) and growth curve (right). b. KRAS mRNA level 24h after the transfection of the empty pCMV or pCMV/K1P3’UTR plasmid in DU145 cells (left) and growth curve (right). c. Model. X and Y are different transcripts targeted by the same microRNA(s). In the steady state (middle), equilibrium exists between the microRNA molecules and their targets X and Y. Downregulation of X (left) leads to increased availability of microRNA molecules to bind to Y, thus decreasing its abundance. By contrast, overexpression of X (right) leads to less microRNA molecules free to bind to Y, and thus Y abundance increases. Red rectangles: microRNA molecules. X and Y can be a pseudogene and its cognate protein-coding gene. a and b. mean ± s.d., n ≥ 3.
To extend our studies beyond PTEN and its pseudogene, we examined other cancer-related pseudogenes and genes (Table S2 and S3, Supplementary Fig. 11–17). Alignments of gene and pseudogenes sequences show that microRNA-binding sites are well conserved; for example, the miR-145 binding site on OCT4 and its pseudogenes OCT4-pg1, 3, 4 and 5 (Supplementary Fig. 11a); miR-1 family binding sites on CONNEXIN 43 (CX43) and its pseudogene (Supplementary Fig. 11b). Notably, OCT4-pg1 and -pg5 are exclusively expressed in cancer tissues and not in normal tissues17. Furthermore OCT4-pg5 is truncated at the 5’ end and expresses only a partial open reading frame region followed by the 3’UTR27. Further examples of such conservation include: miR-34 family binding site on CDK4PS (Supplementary Fig. 12); miR-182 binding site on FOXO3B (Supplementary Fig. 13); miR-17 family binding site on E2F3P1 (Supplementary Fig. 14); miR-143 and let-7 family binding sites on KRAS1P (Supplementary Fig. 15).
Since the 3’UTR of PTENP1 was growth suppressive like its parental gene PTEN, we hypothesized a similar relationship between KRAS and its pseudogene KRAS1P. Indeed, KRAS1P 3’UTR overexpression in DU145 cells resulted in increased KRAS mRNA abundance (Fig. 4b, left and Supplementary Fig. 18a,b) and accelerated cell growth (Fig. 4b, right). We also found that the KRAS and KRAS1P transcript levels are positively correlated in prostate cancer (Supplementary Fig. 18c). Notably, the KRAS1P locus at 6p11–12, is amplified in different human tumours, including neuroblastoma, retinoblastoma and hepatocellular carcinoma28–30. Together these findings point to a putative proto-oncogenic role for KRAS1P, and support the notion that pseudogene functions mirror the functions of their cognate genes as explained by a microRNA decoy mechanism.
Discussion
The findings presented in this study have allowed us to reach a number of important conclusions. First, the discovery of a microRNA-decoy function for pseudogenes identifies these transcripts as biologically active units. We show that PTENP1 and KRAS1P affect the levels of their cognate genes and are possibly involved in disease pathogenesis. Thus, the analysis of pseudogene expression level and genomic status in tumourigenesis needs to be undertaken systematically to further our understanding of disease progression.
Processed pseudogenes in mouse oocytes have been previously reported to generate endogenous small interfering RNAs (endo-siRNAs) that downregulate the expression of cognate genes through conventional RNA interference31. However, endo-siRNA production has yet to be identified in somatic human cells32. Notably, while only few pseudogenes undergo antisense transcription, all transcribed pseudogenes can in principle compete with cognate genes for microRNA binding. Similarly, the microRNA-decoy capacity of pseudogenes is likely to be more widespread than their cleavage into piRNAs, which have been recently discovered only in germ cells of many organisms, including mouse33.
We also demonstrate that pseudogenes such as PTENP1 can derepress their cognate genes, even when expressed at lower levels (Supplementary Fig. 3a and Fig. 2f–h). We propose that pseudogenes are “perfect decoys” for their ancestral genes, because they retain many of the microRNA binding sites and can compete for the binding of many microRNAs at once. It has been hypothesized that suboptimal “pseudotargets” may compete with authentic targets for microRNA binding34. By contrast, we propose that pseudogenes have an intrinsic biological activity in microRNA networks because they are legitimate microRNA targets and compete with other legitimate targets for microRNA binding (Fig. 2e). This notion is corroborated by the “target mimicry” process that in plants is achieved by the expression of non-protein coding genes that sequester microRNAs35.
Exogenously administered microRNA sponges have recently emerged as effective and specific inhibitors of microRNAs36,37. Pseudogenes act like “endogenous sponges”, able to affect the distribution of microRNA molecules on all their targets. They may be particularly effective precisely because they are non-coding, thus active translation does not interfere with microRNA binding38.
The ability of pseudogenes to regulate the biology of a cell goes beyond their ability to modulate the levels of their cognate gene (Supplementary Fig. 8). This phenomenon is consistent with the fact that each microRNA has multiple targets and can lead to widespread homeostatic effects. Also, given that a single gene often has numerous differentially regulated pseudogenes (e.g. OCT4, NPM1 (Supplementary Figs. 11a, 17) and ribosomal protein pseudogenes39), such networks can become intricately dynamic.
Cellular microRNA abundance is dictated by total genomic CN and by their biogenesis process40. Less is known about the regulation of mature microRNA activity. microRNAs can increase their spectrum of targetable mRNAs by undergoing deamination41, while shortening of 3’UTRs42 and polymorphisms can prevent microRNA binding to mRNAs43. Pseudogene-mediated microRNA decoys offer a new dimension regulating the cross-talk between microRNAs and their targets. Indeed the greater the number of pseudogenes that a protein-coding gene has, the more it is protected from microRNAs.
Our discovery of a functional role for PTENP1 is relevant to PTEN biology since minute changes in PTEN can have tumourigenic consequences3. In our analysis we found that PTENP1 positively regulates PTEN levels. Furthermore, we found that the PTENP1 locus undergoes CN losses in human cancer and this correlates with decrease in PTEN; thus we propose that PTENP1 is a bona fide tumour suppressor gene. In light of this, better tools must be developed to detect pseudogene abundance in cancer. For instance, pseudogenes including PTENP1 have been overlooked to such an extent that pseudogene-specific probes are absent in some microarray platforms (Supplementary Figure 7b).
An important implication of our findings is that the decoy mechanism may not be limited to pseudogenes, but may include other long ncRNA transcripts including ribosomal RNAs, lincRNAs and coding gene mRNAs39,44 (Fig. 4a and Supplementary Fig. 10). Beyond their function as cis regulatory elements that impact the stability of their own transcripts, UTRs are also trans modulators of gene expression through microRNA binding. Furthermore, since binding sites for microRNAs are also located in open reading frame sequences21, the entire transcript of coding genes, and not only the 3’UTR may possess decoy function (see working model: Fig. 4c).
As our model suggests, mRNA introduced into a cell can potentially perturb the interaction between microRNAs and their multiple targets and thus, have a biological activity independent of the translation of the protein they encode45. Importantly, the same applies to the transcriptional induction or repression of endogenous mRNA levels, which can lead to changes in the number of mRNA molecules present within a cell in the scale of several orders of magnitude46.
Our findings indicate that, when studying specific nonsense or frameshift mutations and genomic alterations leading to “readthrough” or fusion transcripts, one must consider this new RNA-regulated biological dimension. For example, chromosomal fusion events such as the t(15;17) translocation of APL which generates PML-RARα and RARα-PML fusion transcripts or recurrent “readthrough” transcripts in melanoma such as CDK2-RAB5B could exert oncogenic activities through an aberrant microRNA “sponging” activity47,48. This phenomenon could also occur as a consequence of somatic genomic rearrangements, which are emerging as grossly unappreciated events in many cancers49. Moreover, the shortening of 3’UTRs as observed in human cancer cells42 would not solely affect microRNA-dependent mRNA regulation, but on the flipside, also alter the “sponging” capacity of a given RNA transcript. Finally, in the case of PTEN-loss associated cancers, there is little known of the molecular consequences of PTEN mutations where the PTEN transcript is retained, compared to complete genetic loss of PTEN where no transcript remains1. While these events were previously thought to alter protein abundance, protein signaling and protein networks, they will also have a significant impact on cellular RNA and microRNA homeostasis. In this study, we have therefore identified a novel dimension by which cellular and tumour biology can be regulated.
Methods Summary
Cell lines were cultured under standard conditions. microRNA overexpression was measured by transient transfection (si-miRNAs). PTENP1/PTEN/KRAS1P 3’UTR overexpression was achieved by transient transfection (pCMV expression vectors) or stable infection with MSCV-PIG retroviral constructs. miRNA/target interaction was measured by a luciferase reporter assay. PTENP1, PTEN, KRAS, KRAS1P and miRNA expression level was detected by real time PCR. Proliferation, foci and soft agar assay were performed according standard protocols.
METHODS
Reagents
Anti-HSP90 antibody #61041 (Becton Dickinson); anti-PTEN antibody #9559, antip21 antibody #2947, anti-Tubulin antibody #2125; siGENOME non-targeting siRNA #2 (siLuc), si-PTEN, si-PTENP1, si-PTEN/PTENP1, siGLO RISC-free control siRNA, si-miR-17, si-miR-19b, si-miR20a, si-miR-21, si-miR-26b, si-miR-214, microRNA inhibitor negative control #1 (IC), miR-19b inhibitor, miR-93 inhibitor, miR-106b inhibitor, Dharmafect 1 (Dharmacon); lipofectamine 2000, Trizol Reagent, DNAseI amplification grade, SuperScript II reverse transcriptase, Dulbecco’s Modified Eagle Medium (D-MEM), RPMI-1640, foetal bovine serum (FBS) (Invitrogen); Tissue Scan Normal Tissue qPCR Arrays: Human Major Tissue (HMRT103); Tissue Scan Disease Tissue qPCR Arrays: Prostate Cancer II (HPRT102) (Origen); pGL3-Control, pRL-TK, Dual-Luciferase reporter assay (Promega); polybrene, puromycin (Sigma); QuantiTect Sybr Green PCR kit, Effectene (Qiagen); EGF (R&D); QuikChange II XL Site-Directed Mutagenesis Kit, Herculase Taq polymerase (Stratagene).
Plasmids
The 3’UTR of PTENP1 (NM_023917) was amplified by PCR from the genomic DNA of PC3 cells and cloned into the BamHI and XhoI sites of pCMV-MCS expression plasmid. In this way, pCMV/ψ3’UTR plasmid was obtained. The primers used for PCR amplification were: F 5’-GAGGAGCCGTCAAATCCAGAG-3’ and R 5’-TCGTCAATGTGTGAGGTTCC-3’. The 3’UTR of PTENP1 was then subcloned into the BglII and XhoI sites of MSCV-PIG retroviral vector50 to obtain PIG/ψ3’UTR plasmid. The 3’UTR of PTEN (NM_000314.4) was amplified by PCR from the genomic DNA of HeLa cells and cloned into the BamHI and XhoI sites of pCMV-MCS expression plasmid. In this way, pCMV/PTEN3’UTR plasmid was obtained. The primers used for PCR amplification were: F 5 ’-TAGAGGAGCCGTCAAATCCA-3 ’ and R 5’-TGGACATCTGATTGGGATGA-3’. The 3’UTR of KRAS1P (NC_000006.11) was amplified by PCR from the genomic DNA of HeLa cells and cloned into the BamHI and XhoI sites of pCMV-MCS expression plasmid. In this way, pCMV/K1P3’UTR plasmid was obtained. The primers used for PCR amplification were : F 5 ’-AACCAGCAAAGACAGGGTGT-3’ and R 5’-GTTCAATTGCTCAACGCAGA-3’. The homology between wt KRAS and its pseudogene is very high (> 90%) across the whole mRNA sequence. The primers used for the amplification of KRAS1P 3’UTR contain many mismatches which made them specific for the pseudogene. In order to construct pGLU/ψ3’UTR chimeric luciferase plasmid, the multicloning site of pGL3-Control plasmid was removed from its original position and inserted into the XbaI site located downstream of Luciferase STOP codon (pGLU). PTENP1 3’UTR was then subcloned from pCMV/ψ3’UTR plasmids using the SmaI and XhoI sites of pGLU. The QuikChange II XL Site-Directed Mutagenesis Kit was used to generate the mutated version of this plasmid (pGLU/ψ3’UTRmut).
Cells and culture conditions
Phoenix A, 293T and PC3 cells were grown in DMEM +10% FBS. RWPE-1, PWR-1E and VCaP were grown in keratinocyte medium + EGF + BPE. 22Rv1, DU14551 and LnCaP were grown in RPMI 1640 + 10% FBS. All cell lines were obtained from ATCC and grown in penicillin/streptomycin and glutamine containing medium, at 37°C in a humidified atmosphere with 6% CO2.
Transient transfection
For the transfection of si-miRNAs / microRNA inhibitors, DU145 (1.5×105) or PC3 (1×105) were seeded in 12-well dishes. The following day they were transfected with 100nM siRNAs/si-microRNAs or 400nM microRNA inhibitors using Dharmafect 1 according to the manufacturer’s recommendations. With this protocol more than 90% of cells were positive to the fluorescent siGLO RISC-free control siRNA (data not shown). The day after the transfection cells were trypsinized and reseeded in 12 well plates for subsequent collection and analysis.
For plasmid transfection, 293T and DU145 were seeded in 6cm dishes (2.5 and 3.5 × 105, respectively) and the day after they were transfected with Effectene. 6h later, they were trypsinized and seeded for the various assays.
Dual luciferase reporter assay
DU145 cells were seeded at a density of 6×104 cells per 24-well dish. 24 hours later, 720 ng of pGLU/ψ3’UTR or pGLU/ψ3’UTRmut were cotransfected with 80 ng of pRL-TK. Lipofectamine 2000 was used as transfectant. 24h after transfection, luciferase activity was measured and normalized as in Ref. 7.
Retroviral infection
Phoenix A cells were plated in 10cm poly-D-Lysine coated dishes (3 × 106/dish) and, 16 hours later, were transfected with PIG retroviral plasmids using Lipofectamine 2000. 48 hours later, the virus-containing medium (10 ml) was filtered, mixed with 5 ml of freshly prepared medium, supplemented with 4 µg/ml polybrene and added to 5 × 105 DU145 or PC3 cells plated in a 10 cm dish the day before. Puromycin (2 µg/ml) was administered 48 hours after infection. The cells were selected for 2 days and then utilized for the various assays. Selection medium was changed everyday.
PCR analysis
Total RNA was extracted using Trizol reagent according to the manufacturer’s instructions. It was then subjected to DNase treatment and retrotranscription (1 µg RNA/vial).
Regular PCR was performed using Herculase Taq Polymerase.
Real time PCR of wt PTEN, PTENP1, KRAS and KRAS1P was carried out using Sybr Green fluorescence. 2 µl of RT were used in a 20 µl reaction. ACTIN was used as an internal standard. Relative quantification of gene expression was performed with the comparative CT method52. PTEN primers: F 5’-GTTTACCGGCAGCATCAAAT-3’; R 5’-CCCCCACTTTAGTGCACAGT-3’. PTENP1 primers: F 5’-TCAGAACATGGCATACACCAA-3’; R 5’-TGATGACGTCCGATTTTTCA-3’. KRAS primers: F 5’-ATTGTGAATGTTGGTGT-3 ’ ; R 5 ’-GAAGGTCTCAACTGAAATT-3’. KRAS1P primers: F 5’-AAGGTTTCTTCCAGTTCT-3 ’ ; R 5 ’-ATTTGGGAATTTTGTGAG-3’. ACTIN primers: F 5’-CATGTACGTTGCTATCCAGGC-3’; R 5’-CTCCTTAATGTCACGCACGAT-3’.
The real time PCR of mature microRNAs was performed according to Ref. 53 with some modifications. Briefly, an independent retrotranscription reaction was set up for each microRNA using 0.05µM of the microRNA-specific RT primer and 0.05µM of SNORD44 RT primer (5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACagtcag-3 ’). Real-time PCR of both the microRNA and SNORD44, which was used as an internal standard, were then carried out using Sybr Green fluorescence (2 µl of RT in a 20 µl reaction). For the microRNA, a specific forward primer and the universal R primer (5’- GTGCAGGGTCCGAGGT-3’) were used. For SNORD44, 5’-CGGCGGtggcgatgaggaggtacc-3’ forward primer and the universal reverse primer were used. The microRNA-specific RT and forward PCR primers are listed in Supplementary Fig. 19. The Real-time PCR reaction comprised 40 cycles of 95°C for 15sec followed by 60°C for 1min. Relative quantification of gene expression was performed with the comparative CT method as described above.
TaqMan RT PCR was performed at the HMS Biopolymers Facility utilizing an Applied Biosystems 7900 HT Fast instrument.
Western blot
Cells were collected and lysed (50mM Tris pH8.0, 1mM EDTA, 1mM MgCl2, 150mM NaCl, 1% NP-40, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM NaF, protease inhibitors). Proteins (30 µg/lane) were separated on 10% SDS-polyacrylamide gel and transferred to nitrocellulose membrane. Immunoblotting of the membranes was performed using the following primary antibodies: anti-PTEN (1:1000), anti-p21 (1:1000), anti-HSP90 (1:1000), anti-Tubulin (1:2000). Signals were revealed after incubation with recommended secondary antibody coupled to peroxidase by using enhanced chemiluminescence. Scanned images were quantified using ImageJ software.
FACS analysis
After 10 min treatment with 50 ng/ml EGF, cells were scraped from 10 cm dishes, immediately fixed in 4%PFA and permeabilized with ice cold Methanol. After rehydrating with 0.1%BSA in PBS, cells were stained with Phospho-Akt (Thr308) Rabbit mAb -Alexa Fluor® 647 Conjugate (Cell Signaling). Cells were analyzed on an LSRII flow cytometer (BD).
Cell proliferation
At the end of the selection period (infection) or 6h post-transfection, 2 × 105 DU145 cells were trypsinized, resuspended in 50ml and seeded in 8 sets of 3 wells of a 12-well plate. Starting from the following day (d0), 1 set of wells per day was washed once with PBS, fixed in 10% formalin solution for 10min at room temperature and then kept in PBS at 4°C. At day 7, all the wells were stained with crystal violet. After lysis with acetic acid 10%, O.D. was read at 590 nm.
Foci assay
At the end of the selection period (infection) or 6h post-transfection, DU145 or PC3 cells were trypsinized. 5 × 103 cells were plated on 10cm dishes. 14–21 days later, the plates were stained with crystal violet and the foci were counted.
Growth in semisolid medium
The bottom layer was obtained by covering 6-well dishes with 3 ml of 0,6% agar in DMEM. The day after, 5×104 infected cells were seeded on top in triplicate in 2 ml of 0,3% agar in DMEM + 10%FBS. Colonies were counted after 3–4 weeks at 40× magnification.
Analysis of PTEN and PTENP1 genomic status and expression
Breast Cancer: Affymetrix GeneChip® Human Mapping 500K Array datasets GSE7545 and GSE16619 were downloaded from NCBI GEO and analyzed with the Partek Genomic Suite (Partek Inc) for detection of genomic regions with alterations and data visualization. Copy number aberrations were scored with the Partek segmentation algorithm with default paramaters: p-value cutoff at 0.001 for neighboring regions with significantly different means, 10 minimum number of probe sets required for any candidate region, 0.3 signal to noise difference as minimum magnitude of change, and p-value threshold 0.01 for one-sided t-test of probes in each region to be considered as significantly deviated from the expected normal. All aberrations were calculated with respect to a set of 270 HapMap-normal persons. 118 breast cancer samples and 44 normal samples were included in the study.
Colon Cancer: GSE16125 sporadic colon cancer raw datasets were downloaded from NCBI GEO (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE16125). There are two chip platforms employed in this dataset: 48 sporadic colon cancer samples interrogated by Affymetrix GeneChip(r) Human Mapping 250K Nsp SNP Array and 36 of them analyzed by Affymetrix Human Exon 1.0 ST Array. The SNP array raw datasets were analyzed with the Partek Genomic Suite (Partek Inc) for detection of genomic regions with alterations and data visualization (Partek smoothing algorithm was based on 46 probes). Fortyeight normal samples from the HapMap project supplied by Affymetrix were used as an un-paired reference set [http://www.affymetrix.com/support/technical/sample_data/500k_data.affx]. Raw exon array intensity CEL files for colon cancer and normal colon were analyzed by Affymetrix Power Tools (APT, v. 1.12.0). Normalized intensity value for PTEN was calculated as average of 8 probes corresponding to two PTEN specific exon probe set "3256703" and "3256704". These values were extracted out by APT software. Affymetrix Human Exon 1.0 ST Array dataset for normal colon epithelial cells was downloaded from NCBI GEO GSE1916 dataset (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19163). The correlation plot with r and p values between log10 PTEN expression intensity and log-ratio of PTENP1 copy number was generated in GraphPad Prism (GraphPad Software, Inc.). The log-ratio of PTENP1 was based on average of 14 SNP probes flanking PTENP1 gene.
Statistical analysis
In vitro data were analyzed using unpaired t-test (GraphPad Prism, GraphPad Software, Inc.) Values of p < 0.05 were considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001. The mean ± s.d. of three or more independent experiments is reported. Regression analyses and correlation coefficients were generated using GraphPad Prism, GraphPad Software, Inc.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pandolfi lab members for critical discussions, in particular A. Carracedo for critical input; S. Feng for technical assistance. We thank Bert VogeIstein for DICER−/− cells; T. Yuan for assistance with FACS analysis; A. Tuccoli for assistance with microRNA RT-PCR; I. Osman for support and insightful suggestions. L.P. was supported by fellowships from the Istituto Toscano Tumori and the American Italian Cancer Foundation. L.S. was supported by fellowships from the Human Frontier Science Program and the Canadian Institutes of Health Research. This work was supported by NIH grant R01 CA-82328-09 to P.P.P.
Footnotes
AUTHOR CONTRIBUTIONS
P.P.P. spearheaded and supervised the project; L.P., L.S. and P.P.P. designed experiments; L.P., L.S. and W.H. performed experiments; B.C. provided prostate cancer patient sample cDNAs. J.Z. performed all bioinformatic analyses. L.P., L.S., and P.P.P. analyzed the data and wrote the paper. All authors critically discussed the results and the manuscript.
References
- 1.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Di Cristofano A, et al. Impaired Fas response and autoimmunity in Pten+/− mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- 3.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010 doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takakura S, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–1154. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 8.Meng F, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 9.Huse JT, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009 doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 12.Fujii GH, Morimoto AM, Berson AE, Bolen JB. Transcriptional analysis of the PTEN/MMAC1 pseudogene, psiPTEN. Oncogene. 1999;18:1765–1769. doi: 10.1038/sj.onc.1202492. [DOI] [PubMed] [Google Scholar]
- 13.D'Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief Funct Genomic Proteomic. 2004;3:157–167. doi: 10.1093/bfgp/3.2.157. [DOI] [PubMed] [Google Scholar]
- 14.Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005;33:2374–2383. doi: 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bristow J, Gitelman SE, Tee MK, Staels B, Miller WL. Abundant adrenal-specific transcription of the human P450c21A "pseudogene". J Biol Chem. 1993;268:12919–12924. [PubMed] [Google Scholar]
- 17.Suo G, et al. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun. 2005;337:1047–1051. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 22.Lal A, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrocca F, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Lu Z, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pain D, Chirn GW, Strassel C, Kemp DM. Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. J Biol Chem. 2005;280:6265–6268. doi: 10.1074/jbc.C400587200. [DOI] [PubMed] [Google Scholar]
- 28.van der Wal JE, et al. Comparative genomic hybridisation divides retinoblastomas into a high and a low level chromosomal instability group. J Clin Pathol. 2003;56:26–30. doi: 10.1136/jcp.56.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimonjic DB, Keck CL, Thorgeirsson SS, Popescu NC. Novel recurrent genetic imbalances in human hepatocellular carcinoma cell lines identified by comparative genomic hybridization. Hepatology. 1999;29:1208–1214. doi: 10.1002/hep.510290410. [DOI] [PubMed] [Google Scholar]
- 30.Plantaz D, et al. Gain of chromosome 17 is the most frequent abnormality detected in neuroblastoma by comparative genomic hybridization. Am J Pathol. 1997;150:81–89. [PMC free article] [PubMed] [Google Scholar]
- 31.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamura K, Chung WJ, Lai EC. The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs. Cell Cycle. 2008;7:2840–2845. doi: 10.4161/cc.7.18.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robine N, et al. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 35.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 36.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee DY, et al. A 3'-untranslated region (3'UTR) induces organ adhesion by regulating miR-199a* functions. PLoS One. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balasubramanian S, et al. Comparative analysis of processed ribosomal protein pseudogenes in four mammalian genomes. Genome Biol. 2009;10:R2. doi: 10.1186/gb-2009-10-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 41.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, Bartel DP. Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nat Biotechnol. 2009;27:472–477. doi: 10.1038/nbt.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frith MC, et al. Pseudo-messenger RNA: phantoms of the transcriptome. PLoS Genet. 2006;2:e23. doi: 10.1371/journal.pgen.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang SL, Lozanski G, Samols D, Kushner I. Induction of human serum amyloid A in Hep 3B cells by IL-6 and IL-1 beta involves both transcriptional and post-transcriptional mechanisms. J Immunol. 1995;154:825–831. [PubMed] [Google Scholar]
- 47.Scaglioni PP, Pandolfi PP. The theory of APL revisited. Curr Top Microbiol Immunol. 2007;313:85–100. doi: 10.1007/978-3-540-34594-7_6. [DOI] [PubMed] [Google Scholar]
- 48.Berger MF, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 50.Maeda T, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433(7023):278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 51.Myers MP, et al. P-TEN, the tumour suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci U S A. 1997;94(17):9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drabkin HA, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16(2):186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 53.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.