Expression of the essential EMT inducer Snail1 is inhibited by miR-34 through a p53-dependent regulatory pathway.
Abstract
Snail1 is a zinc finger transcriptional repressor whose pathological expression has been linked to cancer cell epithelial–mesenchymal transition (EMT) programs and the induction of tissue-invasive activity, but pro-oncogenic events capable of regulating Snail1 activity remain largely uncharacterized. Herein, we demonstrate that p53 loss-of-function or mutation promotes cancer cell EMT by de-repressing Snail1 protein expression and activity. In the absence of wild-type p53 function, Snail1-dependent EMT is activated in colon, breast, and lung carcinoma cells as a consequence of a decrease in miRNA-34 levels, which suppress Snail1 activity by binding to highly conserved 3′ untranslated regions in Snail1 itself as well as those of key Snail1 regulatory molecules, including β-catenin, LEF1, and Axin2. Although p53 activity can impact cell cycle regulation, apoptosis, and DNA repair pathways, the EMT and invasion programs initiated by p53 loss of function or mutation are completely dependent on Snail1 expression. These results identify a new link between p53, miR-34, and Snail1 in the regulation of cancer cell EMT programs.
Introduction
Snail1, a zinc-finger transcriptional repressor, plays an important role in the epithelial–mesenchymal transition (EMT) and complementary morphogenetic programs that control normal development, but its inappropriate expression in cancer has been linked to disease progression (Moody et al., 2005; Peinado et al., 2007; Debies et al., 2008; Kudo-Saito et al., 2009; Thiery et al., 2009; Vincent et al., 2009; Wu et al., 2009; Mak et al., 2010). By co-opting developmental processes, neoplastic cells mobilize Snail1 to promote EMT-like programs and the associated tissue-invasive phenotype characteristic of aggressive cancers (Ota et al., 2009; Polyak and Weinberg, 2009). Associations between increased Snail1 expression and cancer progression raise the possibility that oncogenic events promote Snail-dependent EMT programs by as yet uncharacterized mechanisms (Peinado et al., 2007; Polyak and Weinberg, 2009). In this regard, the tumor suppressor, p53, is mutated in >50% of human cancers, wherein loss of function is associated with a more aggressive disease phenotype, raising the possibility that functional links may exist between p53 and Snail1 activity (Gunther et al., 2003; Chen et al., 2007; Gadea et al., 2007; He et al., 2007b; Debies et al., 2008; Godar et al., 2008; Morton et al., 2008; Riley et al., 2008).
Herein, we demonstrate that p53 loss of function or mutation promotes EMT-like programs in human cancer cells by de-repressing Snail1 protein expression and activity. In the absence of wild-type (wt) p53, Snail1 activity is up-regulated as a consequence of a decrease in miRNA-34 (miR-34) levels, which normally serve to antagonize Snail1 expression by binding to highly conserved 3′ untranslated regions (UTRs) within the Snail1 transcript itself as well as a series of accessory targets that directly control Snail1 protein half-life and activity. Together, these data identify a new link between p53, EMT, and the activation of Snail1-dependent invasion programs in neoplastic states.
Results
p53 controls Snail1 expression
Wt HCT116-p53+/+ carcinoma cells form confluent monolayers and display prominent E-cadherin staining (Fig. 1 A). In contrast, an isogenic HCT116 cell line in which both alleles of p53 are inactivated by homologous recombination (HCT116-p53−/−; Bunz et al., 1998) expresses only low levels of E-cadherin at cell–cell borders (Fig. 1 A). Western blot analyses confirm that E-cadherin protein expression is largely ablated in HCT116-p53−/− cells, whereas transduction of null cells with a p53 expression vector rescues both E-cadherin staining and protein expression (Fig. 1, A and B). Consistent with the possible induction of an EMT program coincident with the loss of p53 expression, HCT116-p53−/− cells also express lower mRNA levels of the epithelial markers claudin-3 and occludin, coupled with increased expression of the mesenchymal markers, fibronectin and vimentin (Fig. 1 C). As multiple EMT-inducing transcription factors can repress E-cadherin expression through the tandem E-box domains located in its proximal promoter region (Peinado et al., 2007), HCT116 p53+/+ and p53−/− cells were transfected with E-cadherin promoter constructs, which contain either the wt or mutated domains (Yook et al., 2005). Under these conditions, wt E-cadherin promoter activity is repressed in HCT116-p53−/− cells via a process that is reversed completely after wt p53 transduction (Fig. 1 D). Although Snail1, Snail2, Zeb1, and Zeb2 can each mediate E-box–dependent repression of E-cadherin expression (Peinado et al., 2007), only Snail1 mRNA and protein levels are increased in HCT116-p53−/− cells by Western blot analysis or nuclear staining (Fig. 1, B and E; and Fig. S1). The dominant role of Snail1 in regulating E-cadherin is established by the fact that siRNA-mediated Snail1 silencing in HCT116-p53−/− cells rescues E-cadherin promoter activity (Fig. 1 D). In contrast, knockdown of Slug/Snail2, Zeb1, Zeb2, or Twist1 does not affect E-cadherin promoter activity in a p53-dependent fashion (Fig. S1).
Figure 1.
p53-dependent reciprocal regulation of E-cadherin and Snail1 expression. (A) Laser confocal images of E-cadherin expression in wt HCT116 (top left) and isogenic p53−/− cells. E-cadherin is reexpressed in p53-null HCT116 cells 7 d after transduction with a wt p53-expressing adenovirus. Bar, 10 µm. (B) Immunoblot analyses of endogenous Snail1 and E-cadherin protein levels in wt versus p53-null HCT116 cells (left). After adenoviral transduction with a control or wt p53 expression vector, endogenous Snail1 and E-cadherin expression levels were determined in p53-null HCT116 cells after a 7-d culture period (right). Tubulin is used as the loading control. (C) Claudin-3, occludin, fibronectin, and vimentin expression levels were determined by RT-PCR analysis in wt and p53-null HCT-116 cells before or after Snail1 silencing. GAPDH levels were determined as internal control. (D) E-cadherin promoter activities of wt (blue) or E-box mutant (red) firefly luciferase constructs were determined in HCT116-p53+/+ and HCT116-p53−/− cells. Decreases in E-cadherin promoter activity of HCT116-p53−/− cells are reversed 7 d after adenoviral transduction with wt p53 (Ad-p53) or 48 h after electroporation with either of two independent siRNAs directed against Snail1 (siSnail1 and unpublished data). Activity of the E-cadherin promoter constructs were normalized to the activity of a cotransfected SV-40 promoter renilla luciferase construct. Results are expressed as the mean ± 1 SD of three or more experiments (error bars; *, P ≤ 0.01). (E) Confocal images of endogenous Snail1 expression in HCT116-p53+/+ and p53−/− cells. Endogenous Snail1 expression was detected with a monoclonal antibody directed against Snail1 in HCT116-p53+/+ and HCT116-p53−/− cells after adenoviral transduction of the p53-null cells with a control (HCT116-p53−/−/mock) or p53 (HCT116-p53−/−/Ad-p53) expression vector. Bar, 10 µm.
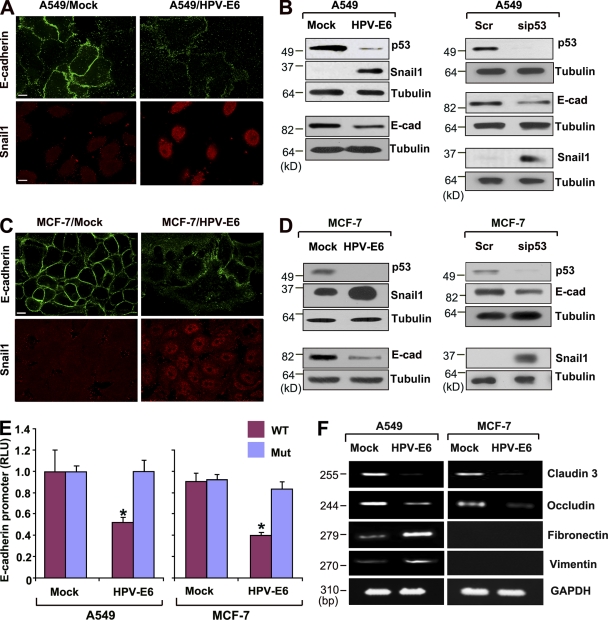
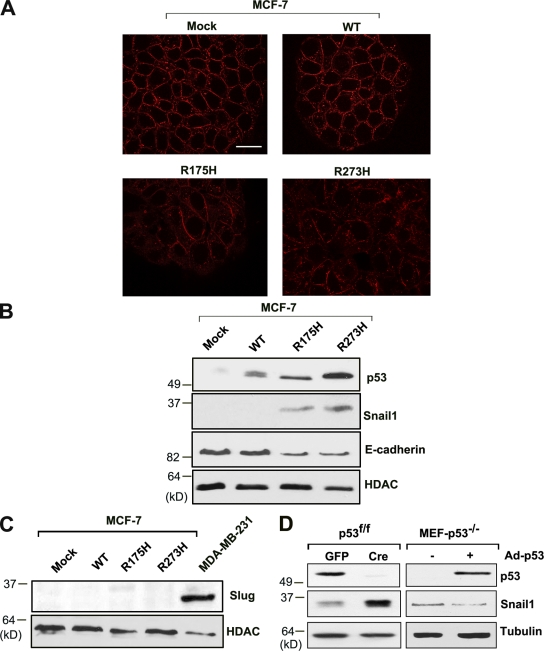
The ability of p53 inactivation to induce Snail1-dependent E-cadherin repression is not limited to HCT116 cells, as degradation of wt p53 by the oncogenic HPV-E6 protein (Scheffner et al., 1990) or siRNA-mediated knockdown of p53 similarly trigger EMT programs in A549 non-small lung carcinoma cells as well as MCF-7 breast carcinoma cells (Fig. 2, A–D). After p53 suppression in A549 or MCF-7 cells, E-cadherin protein expression and reporter activity decrease in tandem with (a) the down-regulation of claudin-3 and occludin expression, (b) increases in fibronectin and vimentin mRNA levels, and (c) the induction of Snail1 protein expression (Fig. 2, A–F; and Fig. S1). Alternatively, similar results are observed when MCF-7 cells harboring wt p53 are transfected with either a p53 conformational mutant (e.g., R175H) or DNA-contact mutant (e.g., R273H; Brosh and Rotter, 2009), whereby Snail1 protein expression is induced along with decreases in E-cadherin staining (Fig. 3, A and B). Though p53 mutants have been reported to increase Slug/Snail2 protein levels (Wang et al., 2009), no changes in its expression are observed under these conditions (Fig. 3 C). Of note, the ability of p53 to regulate Snail1 is not restricted to cells of epithelial origin alone, as p53 wt fibroblasts harboring floxed p53 alleles up-regulate Snail1 after Cre-mediated excision, whereas p53−/− fibroblasts down-regulate Snail1 protein expression when wt p53 expression is restored (Fig. 3 D).
Figure 2.
E-cadherin and Snail1 expression after HPV-E6–dependent p53 degradation. (A) A549 cells were stably transfected with control (Mock) or HPV-E6 expression vectors. Cells were stained with anti–E-cadherin and anti-Snail1 antibodies for confocal imaging. Bar, 10 µm. (B) Endogenous p53, E-cadherin, and Snail1 levels were determined by immunoblot analysis in A549 cells transfected with control (Mock) versus HPV-E6 expression vectors, cells transfected with control versus p53-specific siRNA. Tubulin is used as the loading control. (C) MCF-7 cells were stably transfected with control (Mock) or HPV-E6 expression vectors. Cells were stained with anti–E-cadherin or anti-Snail1 antibodies for confocal imaging. Bar, 10 µm. (D) Endogenous p53, E-cadherin, and Snail1 levels were determined in MCF-7 cells as described above in B. (E) E-cadherin promoter activity of wt (blue) or E-box mutant (red) firefly luciferase constructs was determined in mock- or HPV-E6–transfected cells. Activity of the E-cadherin promoter construct was normalized to the activity of a cotransfected SV-40 promoter renilla luciferase complex. Results are expressed as the mean ± 1 SD (error bars; n = 3, *, P ≤ 0.01). (F) Claudin-3, occludin, fibronectin, and vimentin expression levels were determined by RT-PCR analysis in mock- or HPV-E6–transfected A549 as well as MCF-7 cells.
Figure 3.
Induction of Snail1 expression by mutant p53. (A) MCF-7 cells were transfected with lentiviral vectors expressing wt p53, p53 mutant R175H, or p53 mutant R273H. Confocal images of E-cadherin levels in control MCF-7 cells and each of the transfected cell populations are shown. Bar, 20 µm. (B) p53, Snail1, and E-cadherin protein levels were determined in each transfected cell population by Western blot analysis. (C) Changes in Slug/Snail2 expression in MCF-7 cells after transfection with mock, wt p53, R175H, or R273H mutant p53 lentiviral expression vectors as assessed by Western blot analysis. MDA-MB-231 cells were used as a positive control for Slug/Snail2 expression. (D) Snail1 expression in p53fl/fl mouse fibroblast cells transduced with Ad-GFP or Ad-Cre as well as p53−/− MEFs after transduction with a control or p53 adenoviral expression vector. The Snail1 doublet represents the phosphorylated and nonphosphorylated forms, respectively.
p53 regulates Snail1 by modulating miR-34 expression
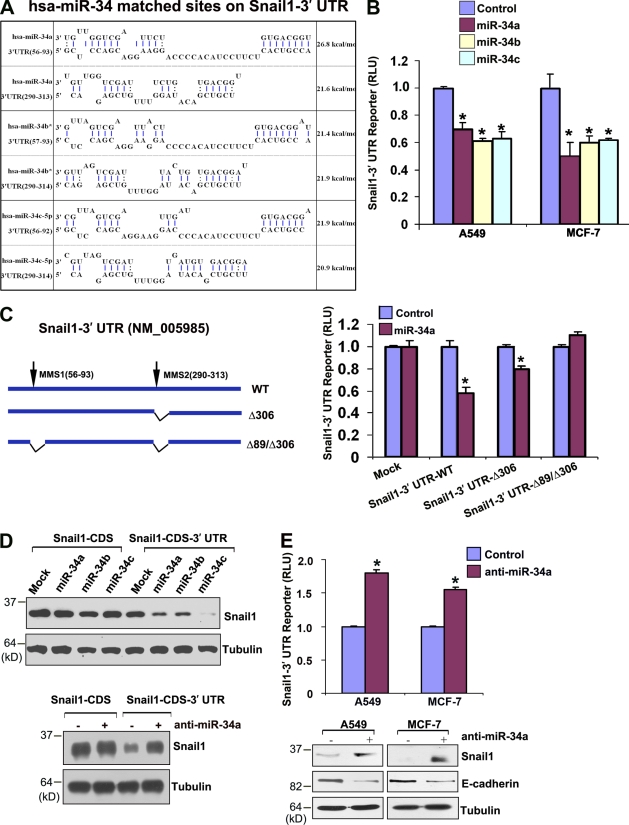
Though p53 does not directly regulate Snail1 mRNA expression (Fig. S1), bioinformatic analysis of the 3′ Snail1 UTR (Lee et al., 2009) identified two potential targeting sites for the p53-regulated microRNAs miR-34a, miR-34b, and miR-34c (Fig. 4 A; Bommer et al., 2007; Chang et al., 2007; Corney et al., 2007; He et al., 2007a; Raver-Shapira et al., 2007; Tarasov et al., 2007). Importantly, both the Snail1-binding domains in miR-34 family members as well as the miR-34–binding domains in the 3′ UTR of Snail1 orthologues are highly conserved (Table S1). To determine whether miR-34 family members are able to directly inhibit Snail1 expression, the 3′ UTR of Snail1 was cloned downstream of a luciferase reporter construct and its expression monitored in the absence or presence of miR-34a, miR-34b, or miR-34c. Reporter expression is suppressed significantly by each of the miR-34 species in either A549 or MCF-7 cell lines, whereas reporter activity increases when p53 levels are suppressed (Fig. 4 B and Fig. S2). Furthermore, miR-34–dependent repression of the Snail1 3′ UTR reporter is ablated after mutation of the predicted microRNA targeting sequences (Fig. 4 C). Each of the miRNAs also inhibited Snail1 protein expression when cells were transfected with a Snail1 expression vector harboring the 3′ UTR of the endogenous transcript (Fig. 4 D). Likewise, knockdown of endogenous miR-34a levels only increases Snail1 protein levels when the expression vector includes the 3′ UTR (Fig. 4 D). Consistent with the demonstrated ability of p53 to regulate miR-34 (Bommer et al., 2007; Chang et al., 2007; Corney et al., 2007; He et al., 2007a; Raver-Shapira et al., 2007; Tarasov et al., 2007), miR-34a and miR-34c expression increase as a function of wt p53 levels in A549 or MCF-7 cells as well as fibroblasts (Fig. S2; miR-34b is not detected under these conditions). In turn, when miR-34a expression (the dominant miR-34 family member detected in A549 or MCF-7 cell lines; unpublished data) is silenced, repression of the Snail1 3′ UTR reporter is relieved and the accumulation of endogenous Snail1 protein is enhanced in association with the suppression of E-cadherin protein levels, claudin-3 and occludin mRNA levels, as well as the induction of fibronectin and vimentin mRNA levels (Fig. 4 E and Fig. S3). Conversely, although low levels of Snail1 can be detected in A549 or MCF-7 cells (Fig. 5 A), further decreases in endogenous Snail1 protein content and 3′ UTR reporter activity are observed when p53 and miR-34a levels are increased in response to the MDM-2 inhibitor, nutlin-3a (Fig. 5, A–C; Kumamoto et al., 2008).
Figure 4.
Snail1 is a direct target of the miR-34 family. (A) Predicted interactions between miR-34a, -34b, and -34c, and Snail1 3′ UTR sequences. Extended seed matches between the 5′ end of miR-34 family (miR-34a, -34b*, and -34c-5p) and the 3′ UTR binding sites of Snail1 (available from GenBank/EMBL/DDBJ under accession no. NM_005985) are shown. Binding energy was calculated by RNAhybrid. (B) Snail1 3′ UTR reporter activity repression by miR-34 family expression vectors. The Snail1 3′ UTR reporter construct cloned downstream of the firefly luciferase was transfected into A549 or MCF-7 cells with control or miR-34 family expression vectors. Activity of the 3′ UTR reporter construct was normalized to the activity of a cotransfected SV-40 promoter renilla luciferase construct after a 2-d culture period. Results are expressed as the mean ± 1 SD for three or more experiments (error bars; *, P ≤ 0.01). (C) Schematic diagram depicting firefly luciferase reporter constructs containing the wt Snail1 3′ UTR or deletion mutants of putative miR-34 family interaction sites (top). Each of the 3′ UTR reporter constructs was cotransfected with a synthetic control or miR-34a into MCF-7 cells, and normalized activity was assessed as described in A after a 2-d culture period. (*, P ≤ 0.01). (D) Immunoblot blot analysis of 293 cells transfected with FLAG-tagged human Snail1 expression vector without the Snail1 3′ UTR (Snail-CDS) or an expression vector harboring the Snail1 3′ UTR (nt +1 to +675; Snail-CDS 3′ UTR). The Snail1 expression vectors (300 ng each) were cotransfected with miR-34 family expression vectors (1 µg each) or anti–miR-34a oligonucleotides (40 pmol each). Whole-cell lysates were analyzed by immunoblotting with anti-FLAG monoclonal antibody after a 2-d culture period. Tubulin is shown as the loading control. (E) Transfection of A549 or MCF-7 cells with anti–miR-34a increased Snail1 3′ UTR reporter activity after a 4-d culture period as described in A, with results presented as the mean ± 1 SD (n = 3; *, P ≤ 0.01). Immunoblot analyses of Snail1 and E-cadherin protein expression levels were performed in A549 or MCF-7 cells 4 d after transfection with control or anti–miR-34a (bottom). Tubulin is shown as the loading control.
Figure 5.
MDM2 inhibition represses Snail1 expression. (A) Immunoblot analysis of p53 and Snail1 levels after treatment of A549 or MCF-7 cells with 10 µM nutlin-3a for 48 h. Tubulin is shown as the loading control. (B) Quantitative RT-PCR analysis of miR-34a transcript levels after treatment with nutlin-3a. Results are expressed as the mean ± 1 SD of three experiments. (error bars; *, P ≤ 0.01). (C) 3′ UTR activity of the Snail1 gene is repressed by treatment with nutlin-3a for 48 h (red) compared with the DMSO control (blue) in A549 or MCF-7 cells. Results are expressed as the mean ± 1 SD of three experiments (error bars; *, P ≤ 0.01).
A novel p53/miR-34 axis dictates Snail1 protein stability and activity
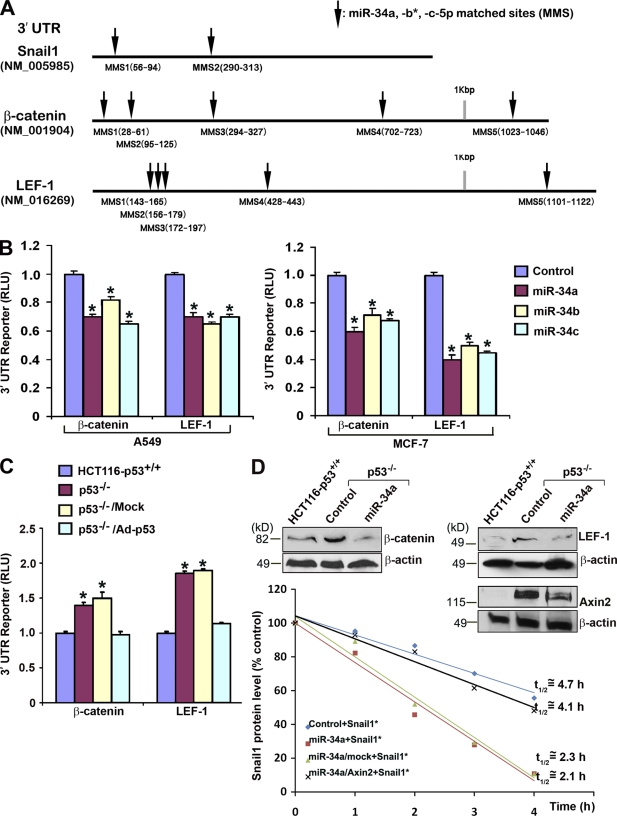
Independent of microRNA-dependent Snail1 translational control, Snail1 function is also regulated posttranslationally by the Wnt canonical pathway (Yook et al., 2005, 2006). Under baseline conditions, Snail1 undergoes rapid GSK3β-dependent phosphorylation, which initiates its subsequent ubiquitination and proteasomal destruction (Zhou et al., 2004; Yook et al., 2005; Yook et al., 2006; Wu et al., 2009; MacPherson et al., 2010; Mak et al., 2010). Snail1 protein levels are, however, stabilized by activating the canonical Wnt signaling cascade wherein induction of the β-catenin–LEF-1 transcriptional complex leads to the inhibition of GSK3β-dependent Snail1 phosphorylation (Yook et al., 2005, 2006). In this regard, the 3′ UTRs of β-catenin and LEF-1 contain miR-34 binding sites that are also sensitive to miR-34a, miR-34b, and miR-34c–dependent regulation (Fig. 6, A and B; and unpublished observation). As HCT116 cancer cells harbor an endogenous β-catenin mutation that results in the constitutive activation of the β-catenin–LEF-1 pathway (Morin et al., 1997), the impact of the p53/miR-34a axis on Snail1 protein stability was examined. As predicted, 3′ UTR reporter activity of β-catenin or LEF-1 is significantly increased in HCT116-p53−/− relative to the p53+/+ control cells (Fig. 6 C). Further, p53-null cells also express higher protein levels of β-catenin and LEF-1 (Fig. 6 D). After ectopic expression of miR-34a in HCT116-p53−/− cells, however, β-catenin and LEF-1 protein levels decrease (Fig. 6 D). In coincident fashion, when p53−/− HCT116 cells are transfected with epitope-tagged Snail1 (wherein the 3′ UTR is deleted to monitor effects on Snail1 protein stability alone) in tandem with miR-34a, Snail1 protein half-life is reduced by more than twofold (Fig. 6 D). Consistent with recent findings that the β-catenin–LEF-1 axis regulates Snail1 stability via an Axin2-dependent process (Yook et al., 2006), the 3′ UTR of Axin2 also contains miR-34–binding sites that directly control Axin2 levels (Fig. 6 D; Lee et al., 2009). Indeed, after expression of 3′ UTR–deleted Axin2 in HCT116-p53+/+ cells, the half-life of Snail1 protein is stabilized to levels observed in HCT116-p53−/− cells (Fig. 6 D). Hence, p53 inactivation and the resultant decrease in miR-34 expression affect Snail1 protein levels, not only by allowing for increased Snail1 translation, but also by increasing Snail1 protein stability via the regulation of canonical Wnt pathway components.
Figure 6.
p53 and miR-34 regulate Snail1 protein stability. (A) Predicted target sites in Snail1 (NM_005985), β-catenin (NM_001904), and LEF-1 (NM_016269) by miR-34a, miR-34b*, and miR-34c-5p are indicated by the arrows. (B) miR-34 family members specifically repress β-catenin 3′ UTR or LEF-1 3′ UTR firefly luciferase reporter activity (normalized to the activity of a cotransfected SV-40 promoter renilla luciferase construct) after a 2-d culture period in A549 (left) or MCF-7 cells (right). Results are expressed as the mean ± 1 SD (error bars; n = 3; *, P ≤ 0.01). (C) 3′ UTR activity of β-catenin and LEF-1 reporter constructs are regulated by p53 status in HCT116 cells. β-catenin 3′-UTR or LEF-1 3′-UTR reporter constructs were transfected into HCT116-p53+/+ or p53−/− cells alone or into HCT116-p53−/− cells in combination with a control adenoviral vector (p53−/−/Mock) or a p53-expressing adenoviral vector (p53−/−/Ad-p53). After a 7-d culture period, reporter activities were normalized as described in A and presented as the mean ± 1 SD (error bars; n = 3; *, P ≤ 0.01). (D) Endogenous β-catenin, LEF-1, and Axin2 protein levels were determined after a 4 d culture period by immunoblot analysis in HCT116-p53+/+ cells and HCT116-p53−/− cells transfected with control or miR-34a oligonucleotides. β-actin is used as the loading control (insert). Snail1 protein half-life was determined in HCT116-p53−/− cells transfected with control or miR-34a oligonucleotides, or p53−/− cells transfected with miR-34a together with a mock or Axin2 expression vector after a 2-d culture period by quantifying residual levels of FLAG-tagged Snail1 (Snail1*) in cycloheximide-treated cells.
p53/miR-34–dependent regulation of the Snail1 EMT program
In neoplastic states, the activation of cancer cell EMT programs by Snail1 triggers motile responses as well as tissue-invasive machinery (Yook et al., 2005, 2006; Peinado et al., 2007; Ota et al., 2009; Thiery et al., 2009). As such, the ability of the p53–miR-34a axis to modulate Snail1-dependent cancer cell migration in A549 or MCF-7 cells, before or after transfection with an HPV-E6 expression vector, was assessed. In the presence of HPV-E6, Snail1 3′ UTR reporter activity is up-regulated in association with increased levels of endogenous Snail1 protein, E-cadherin promoter activity is suppressed, and motility is induced in a Snail1 siRNA-sensitive fashion (Fig. 7, A–C). Conversely, when miR-34a is ectopically introduced into HPV-E6–expressing cells, Snail1 3′ UTR reporter activity is inhibited, Snail1 protein levels decreases to baseline, and E-cadherin promoter activity returns to control levels as increases in Snail1-dependent motility are reversed (Fig. 7, A–C). In concert with the observed increases in E-cadherin promoter activity induced by miR-34a, a similar up-regulation of the epithelial markers claudin-3 and occludin is accompanied by decreases in fibronectin and vimentin expression (unpublished data). Although miR-34a can exert complex effects on cell cycle kinetics or apoptosis that might indirectly impact cancer cell migration (Bommer et al., 2007; Chang et al., 2007; Corney et al., 2007; He et al., 2007a; Raver-Shapira et al., 2007; Tarasov et al., 2007), the motile response of miR-34a–expressing cells is rescued fully after Snail1 re-expression (Fig. 7 C). As expected, the increased migratory activity displayed by anti–miR-34a–transduced A549 cells is lost after Snail1 silencing (Fig. 7 C).
Figure 7.
miR-34 blocks cell migration and EMT-associated invasion by silencing Snail1. (A) Immunoblot analysis and Snail1 3′ UTR reporter activities 48 h after transduction of an miR-34a expression vector into HPV-E6–expressing A549 or MCF-7 cells. Snail1 3′ UTR reporter activity (normalized to a cotransfected XV-40 promoter renilla luciferase construct) was determined after HPV-E6 expression alone in A549 or MCF-7 cells, or alternatively, after cotransfection with HPV-E6 in tandem with a control (control) or miR-34a (miR-34a) synthetic RNA oligonucleotide. Results are expressed as the mean ± 1 SD of three or more experiments (*, P ≤ 0.01). (B) Reporter activities of a wt E-cadherin promoter construct (blue) or its E-box mutant (red) were determined in HPV-E6–expressing A549 or MCF-7 cells 48 h after transfection with a control or miR-34a expression vector. E-cadherin promoter activity was determined as described in Fig. 2 E (mean ± 1 SD; n = 3; *, P ≤ 0.01). (C) The migratory activity of HPV-E6–expressing A549 cells relative to cells transfected with a control vector (Mock) alone or cells transfected with HPV-E6 and either a Snail1 siRNA alone, miR-34a oligonucleotide alone, or miR-34a oligonucleotide and a Snail1 expression vector in tandem was determined after a 2-d culture period. Inhibition of endogenous miR-34a in wt A549 cells with a miR-34a antisense oligonucleotide (anti–miR-34a) increased cell migration after a 4-d culture period via a process reversed by Snail1 siRNA (anti–miR-34a/siSnail1) or a Snail1 shRNA (not depicted). Results are expressed as the mean ± 1 SD (n = 5; *, P ≤ 0.01). (D) HCT116-p53−/− cells were transfected with a control RNA oligonucleotide (control) alone, miR-34a oligonucleotide alone, or miR-34a in combination with mock (miR-34a/Mock) or Snail1 expression vectors (miR-34a/Snail1). After a 24-h culture period, the cells were labeled with fluorescent nanobeads and cultured atop the chorioallantoic membrane of live chick embryos for 3 d. Tissues were then removed, H&E-stained cross sections were prepared for light microscopy, or unstained frozen sections were examined by fluorescence microscopy. The boxed areas in the H&E-stained sections (the first and third rows in the panel) were magnified and examined by fluorescence microscopy to visualize directly the invasive activity of the green-colored cancer cells. The upper face of the chorioallantoic membrane is indicated by the broken lines in the fluorescent images. Bar, 50 µm. (E) HCT116-p53−/− cells were transfected with control or miR-34a oligonucleotides, and Snail1 or E-cadherin protein expression levels were monitored by immunoblot analysis after a 24-h culture period. (F) After transfection with an siRNA control (Scr) or one of two Snail1-specific siRNAs (si1Snail1 and si2Snail1), HCT116-p53−/− cells were cultured for 24 h in vitro before being inoculated on the chick chorioallantoic membrane. Relative invasion depth was monitored after a 3-d assay period as described in D. Alternatively, HCT116-p53−/− cells were transfected with control or miR-34a oligonucleotide alone, or with a miR-34a oligonucleotide in tandem with either a control expression vector (miR-34a/Mock) or a Snail1 expression vector (mi-R34a/Snail1), and cultured for 24 h in vitro before being placed atop the chorioallantoic membrane. Results are expressed as the mean ± 1 SD (error bars; n = 3; *, P ≤ 0.01).
Loss of p53 function has been associated with the acquisition of an aggressive, tissue-invasive phenotype (Gunther et al., 2003; Lewis et al., 2005; Chen et al., 2007; Debies et al., 2008; Godar et al., 2008; Morton et al., 2008). During cancer cell EMT or EMT-like programs, neoplastic epithelial cells must first perforate the underlying basement membrane to access underlying stromal tissues, a process that cannot be visualized readily in mouse models (Rowe and Weiss, 2008). Recently, however, a human-chick xenograft model has been developed where cancer cells can be cultured atop the chorioallantoic basement membrane of live animals to quantify invasive processes (Ota et al., 2009). Under these conditions, fluorescently labeled p53-null HCT116 cells traverse basement membrane barriers (indicated by the broken lines in Fig. 7 D) and infiltrate the underlying stromal tissues as assessed in H&E-stained tissue sections as well as by immunofluorescence (Fig. 7 D). After transfection with a miR-34a expression vector, invasion by p53-null HCT116 cells is inhibited by >80% as Snail1 protein levels fall and E-cadherin expression increases (Fig. 7, E and F). The tissue-invasive potential of p53−/− HCT116 cells is similarly blocked when Snail1, but not Slug/Snail2, expression is silenced directly (Fig. S3), whereas the inhibitory effect exerted by miR-34a on invasion is reversed completely after transfection with a Snail1 expression vector (Fig. 7, D and F). These findings are not restricted to p53-null colon cancer cells alone, as the invasive phenotype displayed by MDA-MB-231 cells, a breast carcinoma cell line that expresses mutant p53 (Bykov et al., 2002), is similarly inhibited either by silencing Snail1 or after ectopic expression of miR-34a (Fig. 8 A). Likewise, Snail1 silencing or miR-34a expression each trigger the reexpression of epithelial cell markers in tandem with the down-regulation of mesenchymal cell markers (Fig. 8 B). In contrast, decreasing endogenous miR-34a levels in wt MCF-7 by expressing mutant p53 not only increases Snail1 protein levels (Fig. 3), but induces a tissue-invasive response as well (Figs. S1 and S2). Together, these data support the proposition that decreases in miR-34 levels activate EMT programs wherein Snail1 serves as a dominant effector of the invasive phenotype.
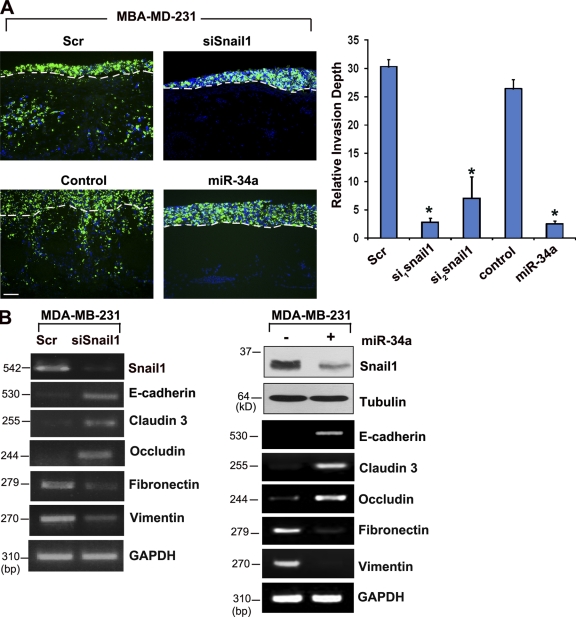
Figure 8.
Snail1- and miR-34- dependent invasion program of MDA-MB-231 cells. (A) MDA-MB-231 cells were transfected with control siRNA (Scr), Snail1-specific siRNA (siSnail1), a control RNA oligonucleotide (control), or a miR-34a RNA oligonucleotide, and after a 24-h culture period in vitro, labeled with fluorescent nanobeads, and cultured atop the live chick chorioallantoic membrane for 3 d. Frozen sections were examined by fluorescence microscopy with labeled MDA-MB-231 cells colored green. The upper face of the chorioallantoic membrane is indicated by broken lines. Relative invasion depth was monitored after a 3-d assay period (n = 3; *, P ≤ 0.01). Error bars indicate mean ± 1 SD. Bar, 50 µm. (B) Snail1, E-cadherin, claudin 3, occludin, fibronectin, and vimentin mRNA transcript levels in MDA-MB-231 cells after transfection with control or Snail1 siRNAs, or control versus an miR-34a RNA oligonucleotide were determined by RT-PCR. Endogenous Snail1 protein levels were also assessed by immunoblot analysis after transfection with control (−) or miR-34a oligonucleotides (+) in MDA-MB-231 cells. Tubulin serves as the loading control. E-cadherin, claudin-3, occludin, fibronectin, and vimentin mRNA levels were determined in control or miR-34a–treated MDA-MB-231 cells by RT-PCR.
Snail1 target gene expression signatures stratify miR-34a–deleted tumors
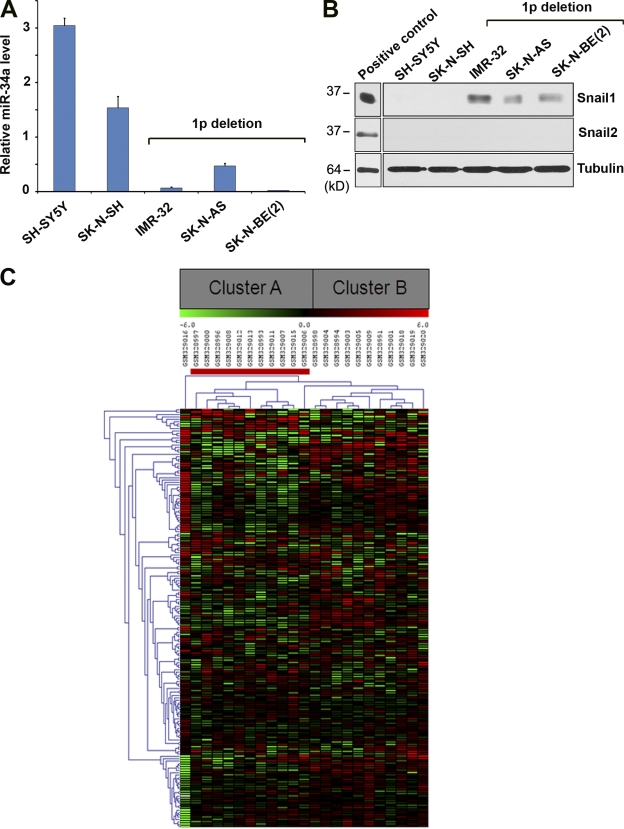
Based on the ability of the p53/miR-34 axis to induce a Snail1-dependent EMT program in vitro, changes in miR-34 expression that occur as a consequence of p53 loss of function in vivo would be predicted to engage a Snail1-induced gene signature. However, efforts to link p53 status to Snail1 expression in vivo are complicated by at least two issues. First, p53 loss of function or mutation alone does not necessarily associate with changes in miR-34 expression, as the microRNA is regulated by both p53-dependent and independent pathways (Bommer et al., 2007; Chang et al., 2007; Gallardo et al., 2009; Corney et al., 2010; Hermeking, 2010). Second, the complex posttranslational regulation of Snail1 protein levels precludes attempts to predict Snail1 activity as a function of its mRNA expression levels alone, whereas more recent studies have demonstrated that Snail1 protein phosphorylation, whose status cannot be determined by immunohistochemistry, further regulates its transcriptional activity in a positive or negative fashion (MacPherson et al., 2010). As such, a publicly available patient cohort was interrogated containing neuroblastoma tumor samples harvested from tissues characterized as having retained, or deleted, chromosome 1p36, a region encoding miR-34a (Łastowska et al., 2007; Cole et al., 2008). Although neural crest-derived in origin, neuroblastoma tumors also undergo EMT-like changes when adopting an invasive or metastatic phenotype whose aggressive characteristics correlate with decreased miR-34a expression (Cole et al., 2008; Vitali et al., 2008). Indeed, neuroblastoma tumor cell lines harboring 1p36 deletions express only low levels of miR-34a while up-regulating Snail1, but not Slug, protein levels (Fig. 9, A and B). Using unsupervised hierarchical cluster analysis, the ability of a 179-member Snail1 gene expression signature (derived from genes differentially expressed in wt vs. Snail1-deleted fibroblasts; Rowe et al., 2009) to stratify miR-34a–positive or –negative patient samples was determined. As shown in Fig. 9 C, the Snail1 signature correctly segregates patients based on the chromosomal loss of miR-34a with a high level of significance (P = 9.61483 × 10−6), which is consistent with a model wherein the p53-dependent as well as p53-independent regulation of miR-34 controls Snail1-mediated EMT programs.
Figure 9.
Cluster analysis of neuroblastoma clinical cohort. (A) miR-34a expression levels in 1p36-positive and 1p36-deleted human neuroblastoma cells. (B) Snail1 and Slug/Snail2 protein levels in 1p36-positive and 1p36-deleted neuroblastoma cells as determined by Western blot analysis. Cell lines expressing recombinant Snail1 or recombinant Slug/Snail2 were used as positive controls. (C) Heat map shows average-linkage hierarchical clustering, using a Euclidean distance metric, performed on 23 neuroblastoma patient samples. Clinical samples harboring a deletion of chromosome 1p are indicated with a red bar. Two distinct clusters, A and B, are highlighted in the gray boxes.
Discussion
Snail1 expression has been linked to events ranging from the induction of carcinoma EMT-like programs and cancer recurrence to the generation of cancer stem cells (Moody et al., 2005; Olmeda et al., 2007; Mani et al., 2008; Kudo-Saito et al., 2009; Massoumi et al., 2009; Wu et al., 2009). Nevertheless, potential intersections between oncogenic processes, microRNA expression, and the regulation of Snail1 activity have not been defined previously. Herein, we demonstrate that p53, a potent yet frequently mutated tumor suppressor, controls Snail1 activity via its ability to transactivate miR-34 family members. As such, modulation of Snail1 function is not only linked to alterations in p53 activity, but also to p53-independent changes in miR-34 expression via gene deletion or epigenetic silencing (Hermeking, 2010). Although loss of p53 or miR-34 function has been reported to impact multiple targets that may lie outside of the Snail1 target range (e.g., Rho/Rock, c-Met, Notch, caldesmon, WISP-2, C-myc, and Id-2; Xia and Land, 2007; Gadea et al., 2007; Dhar et al., 2008; Lujambio et al., 2008; Yan et al., 2008; Li et al., 2009; Mukhopadhyay et al., 2009), none of these accessory pathways are able to bypass an absolute requirement for Snail1 during the induction of tissue-invasive EMT activity in either p53- or miR-34–silenced cancer cells. Interestingly, although mutant p53 has recently been proposed to act as a pro-invasive factor by virtue of its ability to suppress p63 function in a cell context–dependent fashion (Adorno et al., 2009; Muller et al., 2009), Snail1 directly represses the expression of ΔNp63α, the dominant p63 isoform responsible for maintaining the epithelial phenotype (Higashikawa et al., 2007; Koster et al., 2007). Indeed, in our hands, cancer cells that do not express mutant p53 (i.e., HCT116-p53−/− colon carcinoma cells as well as HPV-E6–treated MCF-7 or A549 cells) undergo Snail1-dependent EMT and display tissue-invasive activity after either the loss of wt p53 function or the silencing of endogenous miR-34. These findings do not, however, preclude the ability of p53 mutants to confer expressing cells with additional gain-of-function activities that complement Snail1-dependent activities (Adorno et al., 2009; Muller et al., 2009). Nevertheless, given the ability of Snail1 to directly regulate EMT programs in multiple neoplastic cell populations (Peinado et al., 2007; Debies et al., 2008; Thiery et al., 2009; Vincent et al., 2009; Wu et al., 2009), the identification of this transcriptional repressor as a downstream effector of a dysfunctional p53–miR-34 network unveils a potentially important axis in tumor progression. Caution should, however, be exercised in assuming that Snail1 activity is linked solely to EMT-associated, pro-invasive events. Snail1 can also impact cell cycle activity and apoptotic pathways, key targets for both p53 and miR-34 (Barrallo-Gimeno and Nieto, 2005; Hermeking, 2010). In our hands, however, Snail1 directly induces EMT as well as invasive activity independently of any effects on tumor cell proliferation or apoptosis (Ota et al., 2009). Instead, Snail1 operates as a critical regulator of motile activity in combination with the activation of membrane-type matrix metalloproteinases that play necessary roles in conferring cancer cells with tissue-invasive activity during EMT (Ota et al., 2009; Rowe and Weiss, 2009). Given the fact that Snail1 has been detected within the invasive front of human breast, colon, and prostate carcinomas (Francí et al., 2009; Vincent et al., 2009; Wu et al., 2009; Mak et al., 2010), our data lend further support to models of invasion wherein the reversible and local activation of EMT or EMT-like programs promote cancer cell invasion (Polyak and Weinberg, 2009; Thiery et al., 2009).
Independent of the ability of the p53/miR-34 axis to regulate Snail1 activity and function, p53 has also been reported to regulate Slug/Snail2 (and, more recently, Snail1 protein levels) via a novel process involving the MDM2-dependent degradation of these transcriptional repressors (Wang et al., 2009; Lim et al., 2010). In our studies, however, neither loss of p53 expression nor expression of mutant p53 induced Slug/Snail2 expression. Further, silencing Slug/Snail2 expression in HCT116 cells failed to affect invasive activity. In addition, the MDM2 inhibitor, nutlin-3a, which has been reported to stabilize Snail1/2 levels by preventing their ubiquitination and subsequent proteasomal destruction (Wang et al., 2009; Lim et al., 2010), instead decreases Snail1 protein levels by increasing miR-34a levels as a consequence of stabilizing p53. Though p53 may nevertheless regulate Slug/Snail2 by divergent, miR-34–independent mechanisms under select circumstances, our studies reinforce accumulating evidence that Snail1 plays a dominant role in regulating EMT-associated cancer invasion programs under conditions in which the transcription factors are coexpressed (Olmeda et al., 2008). Interestingly, although our work was under review, Chang et al. (2011) likewise reported that p53 can regulate EMT, but via a distinct mechanism linked to miR-200c and Zeb1 expression. These studies were largely confined to analyses of normal epithelial cell lines, and the impact of Zeb1 silencing on p53-induced EMT was not assessed (Chang et al., 2011). However, as Snail1 can also regulate miR-200c (Burk et al., 2008; Gill et al., 2011), it seems likely that p53 controls EMT through multiple miRNA-dependent routes. Nevertheless, we were unable to detect changes in Zeb1 expression in p53 wt versus p53-null cancer cells (Fig. S1), and our data demonstrate clearly that silencing Snail1 or expressing miR-34 in p53-null cells is sufficient to reverse EMT programs in multiple cell populations.
Although alterations in p53 or miR-34 status directly impact on Snail1 activity and function, we find that neither of these regulatory axes affects Snail1 transcription directly. Hence, the induction of Snail1-mediated EMT programs falls under the control of binary events wherein p53 or miR-34 function is altered in combination with microenvironmental changes known to support Snail1 mRNA expression, including increases in TGFβ, TNF, HGF, EGF, FGF, or hypoxia-inducible factor signaling (Peinado et al., 2007; Sahlgren et al., 2008; Yang et al., 2008; Vincent et al., 2009; Wu et al., 2009; Mak et al., 2010). Likewise, p53 loss-of-function or mutation occur frequently as a later event in cancer progression (Vogelstein et al., 2000), which suggests that the acquisition of invasive activity occurs in conjunction with additional oncogenic events, particularly with activating mutations in Ras and/or the PI3 kinase axis (Lewis et al., 2005; Xia and Land, 2007; Morton et al., 2008). Our experimental conditions did not require the addition of exogenous growth factors or cytokines to activate Snail1-dependent EMT programs after loss of wt p53 function or miR-34 silencing, but we have reported previously that serum alone is a potent stimulus of Snail1 transcription (Rowe et al., 2009). Given the fact that conditions permissive for Snail1 transcription and activation operate within a range of neoplastic states (Rowe and Weiss, 2009), we sought to extend our findings into the clinical setting where correlations between p53 status, miR-34 levels, Snail1 expression, and disease progression might be uncovered. However, several caveats complicated these efforts. First, in human tumor tissues, p53 mutations do not correlate strictly with miR-34 levels (Bommer et al., 2007; Chang et al., 2007; Gallardo et al., 2009; Corney et al., 2010). For example, miR-34 silencing via promoter methylation is more frequently increased in p53 wt, as opposed to p53 mutant, tumor samples (Corney et al., 2010; Hermeking, 2010). Second, although the complex posttranslational regulation of Snail1 protein levels precludes attempts to predict Snail1 activity as a function of its mRNA expression levels alone, few, if any, commercial antibodies have undergone the rigorous testing needed to validate their use in the clinical arena (Becker et al., 2007). This issue notwithstanding, more recent studies indicate that the Snail1 protein can reside within the nuclear compartment in either active or inactive states dependent on its phosphorylation status, a functional change that cannot be resolved by immunohistochemistry alone (MacPherson et al., 2010).
As an alternative approach, we reasoned that an unequivocal decrease in miR-34a expression, an event precipitated by the deletion of chromosome region 1p36 in multiple cancer types (Yamakuchi et al., 2008), could result in the induction of a Snail1 gene signature. A search of public databases indicated that a large cohort of neuroblastoma tissues, a pediatric tumor type wherein miR-34a has been identified as a candidate tumor suppressor gene on the basis of frequent 1p36 deletions (Welch et al., 2007; Cole et al., 2008), had previously undergone expression profiling as a function of their chromosome 1p status (i.e., either 1p-positive or negative; Łastowska et al., 2007). Using a Snail1 gene expression signature derived from earlier profiling studies performed in our laboratory comparing Snail1-floxed to Snail1-deleted cells (Rowe et al., 2009), all 11 of the chromosome 1p–negative/miR-34a–deleted neuroblastoma patients were identified correctly, with only one false positive recorded at a high level of statistical significance. Though we cannot rule out the possibility that additional tumor regulatory molecules lie encoded within the chromosome 1p36 region, no other conventional tumor suppressor genes have been identified in this region (Cole et al., 2008; Yamakuchi et al., 2008). As such, analysis of this patient cohort provides additional support for our contention that p53-dependent or independent regulation of miR-34 expression can impact Snail1-initiated transcriptional programs in vivo (Fig. 10), as well as a range of upstream Wnt targets (Kim et al., 2011). Although further studies are required to define in comprehensive fashion those p53 mutant phenotypes that are mediated by changes in miR-34 expression, the studies outlined herein clearly identify Snail1 as a key, and as of yet unsuspected, downstream effector of the dysregulated p53 axis in cancer.
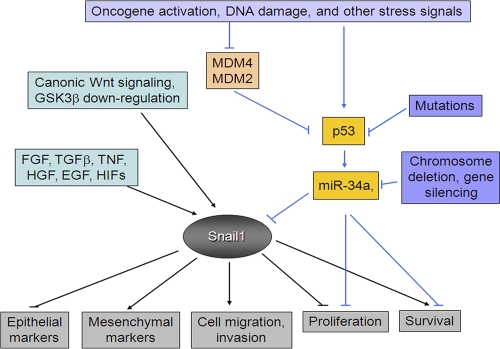
Figure 10.
Schematic cartoon outlining the regulation of Snail1 activity by the p53/miR-34 axis.
Materials and methods
Cells and cell assays
HCT116 p53+/+ and isogenic HCT116 p53−/− cell lines (provided by B. Vogelstein, Johns Hopkins University, Baltimore, MD); p53−/− mouse embryonic fibroblasts (MEFs); MCF-7, A549, MDA-MB-231, and 293 cells; and neuroblastoma cell lines (all from the American Type Culture Collection) were cultured as described previously (Yook et al., 2005, 2006). The predicted miR-34 family target sites in the 3′ UTR of Snail1, β-catenin, and LEF-1 were identified as described previously (Lee et al., 2009). siRNA duplexes or small hairpin RNA (shRNA) expression vectors for Snail1 or p53 silencing as well as scrambled controls have been described previously (Yook et al., 2005, 2006; Bommer et al., 2007). siRNA duplexes directed against human p53 as well as scrambled controls were obtained from Santa Cruz Biotechnology, Inc. Adenoviral expression vectors for GFP (Ad-GFP, #1060) and GFP-p53 (Ad-GFP-p53, #1260) were obtained from Vector Biolabs. For migration assays, A549 or HPV-E6–transfected A549 cells were transfected with control siRNA, Snail1 siRNA, miR-34a, or anti-miR-34a with Lipofectamine. After 24 h, cells were trypsinized, 105 cells were seeded into Transwell inserts (3.0 µm pore; BD), and the number of cells migrating to the basal side of the membrane insert was determined at 18 h. Cancer cell invasion into the live chorioallantoic membrane of 11-d-old chick embryos was performed as described previously (Ota et al., 2009). Invasion was quantified after a 3-d culture period in cross sections of the fixed chorioallantoic membrane by fluorescent microscopy (DM-LB; Leica) using either a 10× or 20× objective lens, 0.50 numerical aperture (Leica). Phase contrast and chick chorioallantoic membrane images were acquired and analyzed with a SPOT camera and software (Diagnostic Instruments, Inc.; Ota et al., 2009).
Confocal microscopy and immunoblot analysis
Confocal imaging and Western blotting were performed as described previously (Yook et al., 2005) with antibodies directed against E-cadherin (Invitrogen), β-catenin (BD), LEF-1 (Abcam), Snail1 (Cell Signaling Technology), Snail2/Slug (Cell Signaling Technology), Axin2 (Cell Signaling Technology), tubulin (Labfrontier Co., Ltd.), p53 (Santa Cruz Biotechnology, Inc.), or FLAG-tagged proteins (Sigma-Aldrich). Confocal images of cells were acquired on a confocal microscope (FV500) using a 60× or 100× water immersion lens, 1.20 numerical aperture, using FluoView software (all from Olympus). Control and test images for each experimental set were acquired with equal photomultiplier tube intensity and gain settings.
Expression and reporter constructs
An HPV-16 E6 open reading frame (provided by K.H. Kim, Yonsei University, South Korea; Kim et al., 1995) and retroviral expression vector of HPV-E6 were constructed by cloning HPV-E6 into pQCXIP retroviral expression vector (Takara Bio, Inc.) using Kozak consensus sequences. FLAG-tagged human Snail1 expression vector has been described previously (Kim et al., 1995; Lee et al., 2009). Wt and E-box mutated E-cadherin reporter gene constructs (nt-108 to +125), Ecad(-108)-Luc and Ecad(-108)/E.BoxA.MUT/E.BoxB.MUT/E.BoxC.MUT-Luc, were used as described previously (Yook et al., 2005, 2006). pMSCV-miR-34a, -34b, and -34c expression vectors were constructed as described previously (Bommer et al., 2007). The 3′ UTR reporter constructs for Snail1 (nt +1 to +675), β-catenin (nt +1 to +950), and LEF-1 (nt +6 to +712) were cloned by previously described methods (Lee et al., 2009). In brief, the luciferase coding sequences were amplified from a pGL3 vector (Promega) and subcloned between HindIII and BamHI sites of a pcDNA3.1-Hyg mammalian expression vector (Invitrogen) to generate firefly luciferase expression constructs with multiple cloning sites. The 3′ UTRs of Snail1, β-catenin, and LEF-1 were amplified from genomic DNA of MCF-7 cells, and subcloned into the BamHI and NotI sites downstream of luciferase to generate the respective 3′ UTR reporter constructs. Snail1 expression vectors in which the 3′ UTR was deleted or retained were constructed by substituting FLAG-tagged Snail1 cDNA for firefly luciferase in a control luciferase vector (Snail-CDS) or a Snail1 3′ UTR luciferase reporter construct (Snail-CDS-UTR). The potential complementary sites for a 6-mer match by the 3′ end of miR-34 on the Snail1 3′-UTR (nt +86 to +91 and +308 to +313) were deleted by PCR-based methods to generate the mutant 3′ UTR reporter constructs Snail1-UTR-D306 and Snail1-UTR-D89/306. All expression and reporter constructs were verified by sequencing.
Generation of retrovirus and HPV-E6–expressing cells
A pQCXIP-HPV-E6 expression vector was used to generate retroviral stocks in 293 cells for infecting MCF-7 and A549 cells. Stable HPV-E6 transfectants were obtained after selection with puromycin (1.0 µg/ml).
RT-PCR and real-time quantitative PCR (qPCR)
For monitoring occludin, claudin-3, vimentin, and fibronectin expression, primer sequences for RT-PCR were: occludin forward, 5′-ATGGTTCACAAATATATGCCCTCT-3′; occludin reverse, 5′-TCATCATAAATGTGTTCCTTGTCC-3′; claudin-3 forward, 5′-AACACCATTATCCGGGACTTCTAC-3′; claudin-3 reverse, 5′-GTCTGTCCCTTAGACGTAGTCCTT-3′; vimentin forward, 5′-CGTACGTCAGCAATATGAAAGTGT-3′; vimentin reverse, 5′-GTGTCTTGGTAGTTAGCAGCTTCA-3′; fibronectin forward, 5′-AAGAGACAGCTGTAACCCAGACTT-3′; and fibronectin reverse, 5′-AAGTGCAATCAGTGTAATTGTGGT-3′.
The PCR reactions were performed at 25 cycles for occludin, claudin-3, vimentin, fibronectin, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Amplified products were visualized with a UV illuminator, followed by cloning into a TA vector to verify PCR product sequences. Real-time qPCR analysis was performed using an ABI-7300 instrument (Applied Biosystems) under standard conditions.
3′ UTR reporter assays
Cells were transiently transfected with 3′ UTR reporter constructs (1–5 ng) and miR-34 family expression vectors (500 ng) as indicated. Alternatively, cells were transfected with 40 nM of a control RNA oligonucleotide (AM17110; Invitrogen) or mature miR-34a oligonucleotide (product ID PM11030; Ambion) in combination with 3′ UTR reporter constructs and a SV40-renilla luciferase construct using Lipofectamine 2000 (Invitrogen). For targeting of endogenous miR-34a, cells were transfected with 40 nM of control miRNA or anti–miR-34a (product ID AM11030; Ambion), and after a 48 h culture period, retransfected. After a final 48-h culture period, the cells were used for assay. The activity of 3′ UTR reporter constructs was normalized to the activity of the cotransfected SV40-renilla luciferase construct (2 ng; Promega). Cells were lysed at 48 h after transfection, and the relative ratio of renilla luciferase to firefly luciferase activity was measured in a dual luciferase assay (Promega). Snail1 half-life was determined by electroporating HCT116-p53−/− cells with a FLAG-tagged Snail1 expression vector in tandem with a control RNA oligonucleotide or miR-34a oligonucleotide. After a 48-h culture period, new protein synthesis was blocked with 50 µg/ml cycloheximide and residual Snail1 protein levels were determined by immunoblot analysis and densitometry.
Snail1 signature and microarray data analysis
The Snail1 gene signature, comprised of 179 genes having a twofold relative change in Snail1 knockout fibroblasts versus Snail1 wt fibroblasts, was obtained from previously published results (Rowe et al., 2009). A Perl script was used to parse an Affymetrix annotation file (http://www.affymetrix.com/Auth/analysis/downloads/na30/ivt/Mouse430_2.na30.ortholog.csv.zip) to identify the human orthologue for each mouse probes within the Snail1 signature. Publicly available primary neuroblastoma cancer profiling data (GSE13141; Łastowska et al., 2007), using the HG-U133A Plus 2.0 array platform, were downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). To ensure that profiling was limited to the effects exerted by miR-34a, seven samples harboring potential losses in miR-34b/c expression because of chromosome 11q deletions were removed from the sample set. Unsupervised hierarchical cluster analysis was conducted using TIGR Multi Experiment Viewer (Saeed et al., 2006). Genes were log transformed and mean centered before performing average-linkage hierarchical clustering, with a Euclidean distance metric, on both samples and genes. Using the R statistical package, a two-tailed Fisher’s exact test was performed to assess the statistical significance between the hierarchical clusters and 1p deletion.
Online supplemental material
Fig. S1 shows p53-dependent regulation of Snail1, Snail2, Zeb1, and Twist1 expression; and EMT. Fig. S2 shows p53-dependent control of miR-34 expression. Fig. S3 shows Snail1- and Snail2-dependent regulation of EMT. Table S1 shows miR-34 and Snail1 3′ UTR sequence conservation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103097/DC1.
Acknowledgments
J.I. Yook and H.S. Kim were supported by a grant from the National R&D Program for Cancer Control (0720270), a grant from the Korea Health Technology (A080916), Ministry of Health & Welfare, a grant from the National Research Foundation of Korea (NRF) funded by the South Korean government (MEST; 2011-0000358, 2009-0067248, 2011-0002620, 2010-00297, R15-2004-024-00000-0), and support from the Korea Research Institute of Chemical Technology (SI-1105). S.J. Weiss is supported by a grant from the National Institutes of Health (5R01CA116516-5) and the Breast Cancer Research Foundation.
Footnotes
Abbreviations used in this paper:
- EMT
- epithelial–mesenchymal transition
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- H&E
- hematoxylin and eosin
- MEF
- mouse embryonic fibroblast
- miR-34
- miRNA-34
- qPCR
- quantitative PCR
- shRNA
- small hairpin RNA
- UTR
- untranslated region
- wt
- wild type
References
- Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M.B., Guzzardo V., et al. 2009. A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 137:87–98 10.1016/j.cell.2009.01.039 [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Nieto M.A. 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 132:3151–3161 10.1242/dev.01907 [DOI] [PubMed] [Google Scholar]
- Becker K.F., Rosivatz E., Blechschmidt K., Kremmer E., Sarbia M., Höfler H. 2007. Analysis of the E-cadherin repressor Snail in primary human cancers. Cells Tissues Organs (Print). 185:204–212 10.1159/000101321 [DOI] [PubMed] [Google Scholar]
- Bommer G.T., Gerin I., Feng Y., Kaczorowski A.J., Kuick R., Love R.E., Zhai Y., Giordano T.J., Qin Z.S., Moore B.B., et al. 2007. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17:1298–1307 10.1016/j.cub.2007.06.068 [DOI] [PubMed] [Google Scholar]
- Brosh R., Rotter V. 2009. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 9:701–713 [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 282:1497–1501 10.1126/science.282.5393.1497 [DOI] [PubMed] [Google Scholar]
- Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9:582–589 10.1038/embor.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov V.J., Issaeva N., Shilov A., Hultcrantz M., Pugacheva E., Chumakov P., Bergman J., Wiman K.G., Selivanova G. 2002. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8:282–288 10.1038/nm0302-282 [DOI] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 26:745–752 10.1016/j.molcel.2007.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.J., Chao C.H., Xia W., Yang J.Y., Xiong Y., Li C.W., Yu W.H., Rehman S.K., Hsu J.L., Lee H.H., et al. 2011. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 13:317–323 10.1038/ncb2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Klimstra D.S., Mongeau M.E., Tatem J.L., Boyartchuk V., Lewis B.C. 2007. Loss of p53 and Ink4a/Arf cooperate in a cell autonomous fashion to induce metastasis of hepatocellular carcinoma cells. Cancer Res. 67:7589–7596 10.1158/0008-5472.CAN-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole K.A., Attiyeh E.F., Mosse Y.P., Laquaglia M.J., Diskin S.J., Brodeur G.M., Maris J.M. 2008. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 6:735–742 10.1158/1541-7786.MCR-07-2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney D.C., Flesken-Nikitin A., Godwin A.K., Wang W., Nikitin A.Y. 2007. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 67:8433–8438 10.1158/0008-5472.CAN-07-1585 [DOI] [PubMed] [Google Scholar]
- Corney D.C., Hwang C.I., Matoso A., Vogt M., Flesken-Nikitin A., Godwin A.K., Kamat A.A., Sood A.K., Ellenson L.H., Hermeking H., Nikitin A.Y. 2010. Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 16:1119–1128 10.1158/1078-0432.CCR-09-2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debies M.T., Gestl S.A., Mathers J.L., Mikse O.R., Leonard T.L., Moody S.E., Chodosh L.A., Cardiff R.D., Gunther E.J. 2008. Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16 Ink4a loss. J. Clin. Invest. 118:51–63 10.1172/JCI33320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar G., Banerjee S., Dhar K., Tawfik O., Mayo M.S., Vanveldhuizen P.J., Banerjee S.K. 2008. Gain of oncogenic function of p53 mutants induces invasive phenotypes in human breast cancer cells by silencing CCN5/WISP-2. Cancer Res. 68:4580–4587 10.1158/0008-5472.CAN-08-0316 [DOI] [PubMed] [Google Scholar]
- Francí C., Gallén M., Alameda F., Baró T., Iglesias M., Virtanen I., García de Herreros A. 2009. Snail1 protein in the stroma as a new putative prognosis marker for colon tumours. PLoS ONE. 4:e5595 10.1371/journal.pone.0005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea G., de Toledo M., Anguille C., Roux P. 2007. Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. J. Cell Biol. 178:23–30 10.1083/jcb.200701120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo E., Navarro A., Viñolas N., Marrades R.M., Diaz T., Gel B., Quera A., Bandres E., Garcia-Foncillas J., Ramirez J., Monzo M. 2009. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 30:1903–1909 10.1093/carcin/bgp219 [DOI] [PubMed] [Google Scholar]
- Gill J.G., Langer E.M., Lindsley R.C., Cai M., Murphy T.L., Kyba M., Murphy K.M. 2011. Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godar S., Ince T.A., Bell G.W., Feldser D., Donaher J.L., Bergh J., Liu A., Miu K., Watnick R.S., Reinhardt F., et al. 2008. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 134:62–73 10.1016/j.cell.2008.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther E.J., Moody S.E., Belka G.K., Hahn K.T., Innocent N., Dugan K.D., Cardiff R.D., Chodosh L.A. 2003. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 17:488–501 10.1101/gad.1051603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., et al. 2007a. A microRNA component of the p53 tumour suppressor network. Nature. 447:1130–1134 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He X., Lowe S.W., Hannon G.J. 2007b. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat. Rev. Cancer. 7:819–822 10.1038/nrc2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. 2010. The miR-34 family in cancer and apoptosis. Cell Death Differ. 17:193–199 10.1038/cdd.2009.56 [DOI] [PubMed] [Google Scholar]
- Higashikawa K., Yoneda S., Tobiume K., Taki M., Shigeishi H., Kamata N. 2007. Snail-induced down-regulation of DeltaNp63alpha acquires invasive phenotype of human squamous cell carcinoma. Cancer Res. 67:9207–9213 10.1158/0008-5472.CAN-07-0932 [DOI] [PubMed] [Google Scholar]
- Kim K.H., Park T.K., Yoon D.J., Kim Y.S. 1995. The effects of wild type p53 tumor suppressor gene expression on the normal human cervical epithelial cells or human epidermal keratinocytes transformed with human papillomavirus type 16 DNA. Yonsei Med. J. 36:287–298 [DOI] [PubMed] [Google Scholar]
- Kim N.H., Kim H.S., Kim N.-G., Lee I., Choi H.S., Li X.-Y., Kang S.E., Cha S.Y., Ryu J.K., Na J.N., Park C., Kim K., Lee S., Gumbiner B.M., Yook J.I., Weiss S.J. 2011. P53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster M.I., Dai D., Marinari B., Sano Y., Costanzo A., Karin M., Roop D.R. 2007. p63 induces key target genes required for epidermal morphogenesis. Proc. Natl. Acad. Sci. USA. 104:3255–3260 10.1073/pnas.0611376104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo-Saito C., Shirako H., Takeuchi T., Kawakami Y. 2009. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 15:195–206 10.1016/j.ccr.2009.01.023 [DOI] [PubMed] [Google Scholar]
- Kumamoto K., Spillare E.A., Fujita K., Horikawa I., Yamashita T., Appella E., Nagashima M., Takenoshita S., Yokota J., Harris C.C. 2008. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 68:3193–3203 10.1158/0008-5472.CAN-07-2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łastowska M., Viprey V., Santibanez-Koref M., Wappler I., Peters H., Cullinane C., Roberts P., Hall A.G., Tweddle D.A., Pearson A.D., et al. 2007. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene. 26:7432–7444 10.1038/sj.onc.1210552 [DOI] [PubMed] [Google Scholar]
- Lee I., Ajay S.S., Yook J.I., Kim H.S., Hong S.H., Kim N.H., Dhanasekaran S.M., Chinnaiyan A.M., Athey B.D. 2009. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 19:1175–1183 10.1101/gr.089367.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.C., Klimstra D.S., Socci N.D., Xu S., Koutcher J.A., Varmus H.E. 2005. The absence of p53 promotes metastasis in a novel somatic mouse model for hepatocellular carcinoma. Mol. Cell. Biol. 25:1228–1237 10.1128/MCB.25.4.1228-1237.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Guessous F., Zhang Y., Dipierro C., Kefas B., Johnson E., Marcinkiewicz L., Jiang J., Yang Y., Schmittgen T.D., et al. 2009. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69:7569–7576 10.1158/0008-5472.CAN-09-0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.O., Kim H., Jung G. 2010. p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS Lett. 584:2231–2236 10.1016/j.febslet.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Lujambio A., Calin G.A., Villanueva A., Ropero S., Sánchez-Céspedes M., Blanco D., Montuenga L.M., Rossi S., Nicoloso M.S., Faller W.J., et al. 2008. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA. 105:13556–13561 10.1073/pnas.0803055105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson M.R., Molina P., Souchelnytskyi S., Wernstedt C., Martin-Pérez J., Portillo F., Cano A. 2010. Phosphorylation of serine 11 and serine 92 as new positive regulators of human Snail1 function: potential involvement of casein kinase-2 and the cAMP-activated kinase protein kinase A. Mol. Biol. Cell. 21:244–253 10.1091/mbc.E09-06-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P., Leav I., Pursell B., Bae D., Yang X., Taglienti C.A., Gouvin L.M., Sharma V.M., Mercurio A.M. 2010. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell. 17:319–332 10.1016/j.ccr.2010.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R., Kuphal S., Hellerbrand C., Haas B., Wild P., Spruss T., Pfeifer A., Fässler R., Bosserhoff A.K. 2009. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J. Exp. Med. 206:221–232 10.1084/jem.20082044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody S.E., Perez D., Pan T.C., Sarkisian C.J., Portocarrero C.P., Sterner C.J., Notorfrancesco K.L., Cardiff R.D., Chodosh L.A. 2005. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 8:197–209 10.1016/j.ccr.2005.07.009 [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 275:1787–1790 10.1126/science.275.5307.1787 [DOI] [PubMed] [Google Scholar]
- Morton J.P., Klimstra D.S., Mongeau M.E., Lewis B.C. 2008. Trp53 deletion stimulates the formation of metastatic pancreatic tumors. Am. J. Pathol. 172:1081–1087 10.2353/ajpath.2008.070778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay U.K., Eves R., Jia L., Mooney P., Mak A.S. 2009. p53 suppresses Src-induced podosome and rosette formation and cellular invasiveness through the upregulation of caldesmon. Mol. Cell. Biol. 29:3088–3098 10.1128/MCB.01816-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., Caswell P.T., Doyle B., Iwanicki M.P., Tan E.H., Karim S., Lukashchuk N., Gillespie D.A., Ludwig R.L., Gosselin P., et al. 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell. 139:1327–1341 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Olmeda D., Moreno-Bueno G., Flores J.M., Fabra A., Portillo F., Cano A. 2007. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res. 67:11721–11731 10.1158/0008-5472.CAN-07-2318 [DOI] [PubMed] [Google Scholar]
- Olmeda D., Montes A., Moreno-Bueno G., Flores J.M., Portillo F., Cano A. 2008. Snai1 and Snai2 collaborate on tumor growth and metastasis properties of mouse skin carcinoma cell lines. Oncogene. 27:4690–4701 10.1038/onc.2008.118 [DOI] [PubMed] [Google Scholar]
- Ota I., Li X.Y., Hu Y., Weiss S.J. 2009. Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc. Natl. Acad. Sci. USA. 106:20318–20323 10.1073/pnas.0910962106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Olmeda D., Cano A. 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 7:415–428 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- Polyak K., Weinberg R.A. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 9:265–273 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 26:731–743 10.1016/j.molcel.2007.05.017 [DOI] [PubMed] [Google Scholar]
- Riley T., Sontag E., Chen P., Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9:402–412 10.1038/nrm2395 [DOI] [PubMed] [Google Scholar]
- Rowe R.G., Weiss S.J. 2008. Breaching the basement membrane: who, when and how? Trends Cell Biol. 18:560–574 10.1016/j.tcb.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Rowe R.G., Weiss S.J. 2009. Navigating ECM barriers at the invasive front: the cancer cell-stroma interface. Annu. Rev. Cell Dev. Biol. 25:567–595 10.1146/annurev.cellbio.24.110707.175315 [DOI] [PubMed] [Google Scholar]
- Rowe R.G., Li X.Y., Hu Y., Saunders T.L., Virtanen I., Garcia de Herreros A., Becker K.F., Ingvarsen S., Engelholm L.H., Bommer G.T., et al. 2009. Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J. Cell Biol. 184:399–408 10.1083/jcb.200810113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., Quackenbush J. 2006. TM4 microarray software suite. Methods Enzymol. 411:134–193 10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
- Sahlgren C., Gustafsson M.V., Jin S., Poellinger L., Lendahl U. 2008. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA. 105:6392–6397 10.1073/pnas.0802047105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Werness B.A., Huibregtse J.M., Levine A.J., Howley P.M. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 63:1129–1136 10.1016/0092-8674(90)90409-8 [DOI] [PubMed] [Google Scholar]
- Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A., Meister G., Hermeking H. 2007. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 6:1586–1593 10.4161/cc.6.13.4436 [DOI] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Vincent T., Neve E.P., Johnson J.R., Kukalev A., Rojo F., Albanell J., Pietras K., Virtanen I., Philipson L., Leopold P.L., et al. 2009. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat. Cell Biol. 11:943–950 10.1038/ncb1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali R., Mancini C., Cesi V., Tanno B., Mancuso M., Bossi G., Zhang Y., Martinez R.V., Calabretta B., Dominici C., Raschellà G. 2008. Slug (SNAI2) down-regulation by RNA interference facilitates apoptosis and inhibits invasive growth in neuroblastoma preclinical models. Clin. Cancer Res. 14:4622–4630 10.1158/1078-0432.CCR-07-5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A.J. 2000. Surfing the p53 network. Nature. 408:307–310 10.1038/35042675 [DOI] [PubMed] [Google Scholar]
- Wang S.P., Wang W.L., Chang Y.L., Wu C.T., Chao Y.C., Kao S.H., Yuan A., Lin C.W., Yang S.C., Chan W.K., et al. 2009. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat. Cell Biol. 11:694–704 10.1038/ncb1875 [DOI] [PubMed] [Google Scholar]
- Welch C., Chen Y., Stallings R.L. 2007. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 26:5017–5022 10.1038/sj.onc.1210293 [DOI] [PubMed] [Google Scholar]
- Wu Y., Deng J., Rychahou P.G., Qiu S., Evers B.M., Zhou B.P. 2009. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 15:416–428 10.1016/j.ccr.2009.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Land H. 2007. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat. Struct. Mol. Biol. 14:215–223 10.1038/nsmb1208 [DOI] [PubMed] [Google Scholar]
- Yamakuchi M., Ferlito M., Lowenstein C.J. 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA. 105:13421–13426 10.1073/pnas.0801613105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Liu G., Scoumanne A., Chen X. 2008. Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer Res. 68:6789–6796 10.1158/0008-5472.CAN-08-0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M.H., Wu M.Z., Chiou S.H., Chen P.M., Chang S.Y., Liu C.J., Teng S.C., Wu K.J. 2008. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 10:295–305 10.1038/ncb1691 [DOI] [PubMed] [Google Scholar]
- Yook J.I., Li X.Y., Ota I., Fearon E.R., Weiss S.J. 2005. Wnt-dependent regulation of the E-cadherin repressor snail. J. Biol. Chem. 280:11740–11748 10.1074/jbc.M413878200 [DOI] [PubMed] [Google Scholar]
- Yook J.I., Li X.Y., Ota I., Hu C., Kim H.S., Kim N.H., Cha S.Y., Ryu J.K., Choi Y.J., Kim J., et al. 2006. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 8:1398–1406 10.1038/ncb1508 [DOI] [PubMed] [Google Scholar]
- Zhou B.P., Deng J., Xia W., Xu J., Li Y.M., Gunduz M., Hung M.C. 2004. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 6:931–940 10.1038/ncb1173 [DOI] [PubMed] [Google Scholar]