Insertion into and release of the cytoplasmic domain of the Schizosaccharomyces pombe spindle pole body from a nuclear envelope fenestra during mitosis requires Brr6.
Abstract
The fission yeast interphase spindle pole body (SPB) is a bipartite structure in which a bulky cytoplasmic domain is separated from a nuclear component by the nuclear envelope. During mitosis, the SPB is incorporated into a fenestra that forms within the envelope during mitotic commitment. Closure of this fenestra during anaphase B/mitotic exit returns the cytoplasmic component to the cytoplasmic face of an intact interphase nuclear envelope. Here we show that Brr6 is transiently recruited to SPBs at both SPB insertion and extrusion. Brr6 is required for both SPB insertion and nuclear envelope integrity during anaphase B/mitotic exit. Genetic interactions with apq12 and defective sterol assimilation suggest that Brr6 may alter envelope composition at SPBs to promote SPB insertion and extrusion. The restriction of the Brr6 domain to eukaryotes that use a polar fenestra in an otherwise closed mitosis suggests a conserved role in fenestration to enable a single microtubule organizing center to nucleate both cytoplasmic and nuclear microtubules on opposing sides of the nuclear envelope.
Introduction
Many eukaryotes rely upon two microtubule organizing centers (MTOCs) to nucleate the antiparallel microtubule arrays of the mitotic spindle. Despite functional conservation, structure can vary dramatically (Heath, 1980). This structural variation is often accompanied by significant variation in the behavior of the nuclear envelope during mitosis (Kubai, 1976; Heath, 1980). In most higher eukaryotes, the nuclear envelope fragments to enable radial microtubule arrays to capture one set of chromosomes. In syncytial systems, complete nuclear envelope breakdown could be catastrophic, as it could facilitate chromosome exchange between neighboring spindles. A partial or complete membrane barrier is therefore retained in many of these systems. The evolutionary paths taken by fungi and protists means that conservation of nuclear envelope integrity throughout mitosis in a “closed mitosis” is a common feature of microbial cell division (Kubai, 1976; Heath, 1980).
The permanent separation of cytoplasm and nucleoplasm in closed mitoses presents significant challenges if cells possess an MTOC that executes cytoplasmic functions alongside genome segregation. Many fungi, including the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae, solve this problem by integrating planar MTOCs into the nuclear envelope so that the microtubules nucleated from the two faces can function in different compartments (McCully and Robinow, 1971; Byers and Goetsch, 1975; Heath, 1980; Ding et al., 1997). In budding yeast spindle pole body (SPB), duplication is coupled with integration of the new SPB into the membrane alongside the old. Both SPBs then reside within pores in the nuclear envelope for the remainder of the cell cycle (Byers and Goetsch, 1975; Adams and Kilmartin, 2000). In contrast, integration of the fission yeast SPB is transient and restricted to mitosis (McCully and Robinow, 1971; Ding et al., 1997).
Electron microscopy of budding yeast SPBs in G1 phase of the cell cycle reveals a triple layered structure with a “half bridge” that extends from the central plaque along the cytoplasmic face of the nuclear envelope and ends in an amorphous “satellite” structure (Byers and Goetsch, 1975; Adams and Kilmartin, 2000). The new SPB forms by elaboration of the satellite to form a duplication plaque on the cytoplasmic side of the nuclear envelope. The completely formed duplication plaque is then inserted into a pore in the nuclear envelope next to the integrated old SPB. Defects in the half bridge components Kar1, Cdc31, Mps3, and Sfi1 block half bridge formation, and hence duplication plaque formation (Adams and Kilmartin, 2000; Kilmartin, 2003; Jaspersen et al., 2006). Mps3 contains a Sad1/Unc84 (SUN) domain (Jaspersen et al., 2006). As SUN domains cooperate with Klarsicht, ANC-1, and Syne homology (KASH) domain proteins to establish connections between nuclear and cytoplasmic molecules across the nuclear envelope (Razafsky and Hodzic, 2009), Mps3 may act alongside the membrane protein Kar1 to tether half bridge components to the nuclear envelope. The membrane protein Mps2 forms a complex with Nbp1 and the Kar1-interacting protein Bbp1 that sits at the central plaque/half bridge (Schramm et al., 2000; Araki et al., 2006). This complex cooperates with the integral membrane protein Ndc1 to promote the integration of the duplication plaque into the nuclear envelope (Araki et al., 2006).
The fission yeast interphase SPB comprises a cytoplasmic component that is separated from a nuclear component by the nuclear envelope (McCully and Robinow, 1971; Ding et al., 1997). A structure reminiscent of the S. cerevisiae half bridge/bridge extends from the cytoplasmic component over the surface of the nuclear envelope. Fine striations through the envelope connect this cytoplasmic component to a nuclear component that contains γ-tubulin and recruits centromeres to the SPB (Funabiki et al., 1993; Ding et al., 1997; Kniola et al., 2001). The SUN domain protein Sad1 and the KASH proteins Kms1 and Kms2 appear to mediate the association of centromeres with the SPB and thus the cytoplasmic microtubules (Goto et al., 2001; King et al., 2008). The ability to differentiate between the old and new SPBs with a time-sensitive fluorescent protein suggest that SPB duplication is conservative, with a new SPB forming de novo alongside the old (Grallert et al., 2004). Upon commitment to mitosis, the membrane separating the two SPB components disperses and the old and new SPBs insert into the resulting fenestra (Ding et al., 1997). Such polar fenestration is a feature shared by a subset of eukaryotes that use closed mitosis (Kubai, 1976; Heath, 1980). The local removal of the nucleoplasm/cytoplasm barrier in S. pombe is transient and rapid, as there is no detectable leakage of the nucleoplasm into the cytoplasm (Tallada et al., 2009). The two SPBs then become active, each nucleating microtubules to generate each half of the spindle (Ding et al., 1993). During anaphase B, the membrane grows back between the two components to recreate the interphase partitioning of SPB components (Ding et al., 1997).
Although less is known about SPB integration in fission yeast, Sad1 is a clear homologue of Mps3, Alm1, Mlp2, and Cdc31, and Sfi1 bridge component orthologues are also required for spindle formation (Hagan and Yanagida, 1995; Flory et al., 2002; Kilmartin, 2003; Paoletti et al., 2003; Niepel et al., 2005; Jaspersen et al., 2006). The membrane-spanning Ndc1 orthologue Cut11 is recruited to the SPB during mitosis, where it is required for the integration of the new SPB into the nuclear envelope (West et al., 1998; Tallada et al., 2009).
There are striking parallels between the integration of SPBs and nuclear pore complexes (NPCs) into the nuclear envelope such that the two systems appear to compete for assembly factors (Witkin et al., 2010). Mps2 and Mps3 associate with the nuclear periphery in addition to the SPB, Cdc31 modulates mRNA export, and the nuclear pore complex (NPC) component Mlp2 binds to SPBs and participates in SPB assembly (Muñoz-Centeno et al., 1999; Jaspersen et al., 2002; Fischer et al., 2004; Niepel et al., 2005). Most notably, Ndc1 is a component of both NPCs and SPBs (Chial et al., 1998). In NPCs, Ndc1 interacts with Pom34 and Pom152 in the membrane ring surrounding NPCs, where these molecules perform redundant functions in NPC assembly (Wozniak et al., 1994; Chial et al., 1998; Miao et al., 2006; Alber et al., 2007; Onischenko et al., 2009). Importantly, reducing translation of Pom34 mimics mutation of Ndc1 in rescuing the otherwise lethal SPB insertion arising from removal of Mps2 (Sezen et al., 2009).
We describe the recruitment of fission yeast Brr6 (Saitoh et al., 2005; Lo Presti et al., 2007) to SPBs during mitotic commitment and anaphase B/mitotic exit. We show that it is required for SPB insertion and envelope integrity during SPB extrusion. Phenotypic and genetic interactions with apq12 that mirror those reported for Saccharomyces cerevisiae (Hodge et al., 2010) suggest that it modifies the lipid composition of the envelope to facilitate SPB insertion/extrusion. We discuss the evolutionary restriction of the “Brr6 domain” to a specific subset of fungi and protists that exploit a polar fenestra to enable an MTOC to organize both cytoplasmic and spindle microtubules (Kubai, 1976; Heath, 1980).
Results
S. pombe Brr6 is required for spindle formation
Screening for mutations that formed monopolar spindles upon exposure to 4% DMSO for one generation (Broek et al., 1991; Poloni and Simanis, 2002) identified a glycine–to–aspartic acid change at position 145 of brr6+ (brr6.ds1; Fig. 1, A and B; for comparative images of wild-type cells, see Fig. S1 A and Fig. 2 A). brr6+ is an essential gene (Lo Presti et al., 2007). Inoculating spores isolated from a diploid strain in which the brr6+ ORF had been replaced with the kanR marker into medium containing geneticin to select against growth of brr6+ spores revealed monopolar rather than bipolar spindles in their first division of 88% of mitoses (unpublished data). Repressing brr6+ transcription in a haploid brr6.Δ background also blocked mitotic progression with monopolar spindles (Fig. S2, B and C).
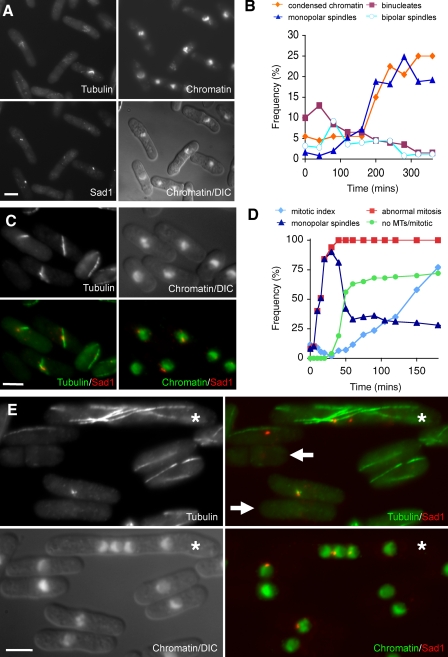
Figure 1.
Brr6 is required for bipolar spindle formation. Cells were processed for immunofluorescence with anti-tubulin, anti-Sad1, and DAPI as indicated. (A and B) brr6.ds1 cells were grown to early log phase at 32°C in rich YE5S medium before DMSO was added to a final concentration of 4%. Samples were processed for immunofluorescence at discrete time points and scored for the indicated features (B). The images in A are from the 120-min time point. Microtubules (tubulin) extend from adjacent SPBs (Sad1) as chromatin becomes highly condensed, giving it a lumpy appearance (chromatin). (C and D) brr6.ts8 cells were grown to early log phase at 25°C in rich YE5S medium before the temperature of the culture was shifted to 36°C, and samples were processed and scored for the features as indicated in D. The category “no MTs/mitotic” indicates the cells with condensed chromosomes and no microtubule staining. The images in C are from the 30-min time point. (E) brr6.ts8 cells were grown to early log phase at 25°C in rich YE5S medium before the temperature of the culture was shifted to 36°C. 2 h later, a culture of mid-log phase cdc7.A20 spg1.B8 cells (asterisks) that had been cultured at 36°C for 4 h was mixed at a ratio of 1:5 with the brr6.ts8 culture before the mixed culture was processed for immunofluorescence. Although the brr6.ds1 mutation gives rise to monopolar spindles, monopolar spindle formation is transient in the more penetrant brr6.ts8, as prolonged loss of function leads to an inability to nucleate microtubules (arrows). Bars, 5 µm.
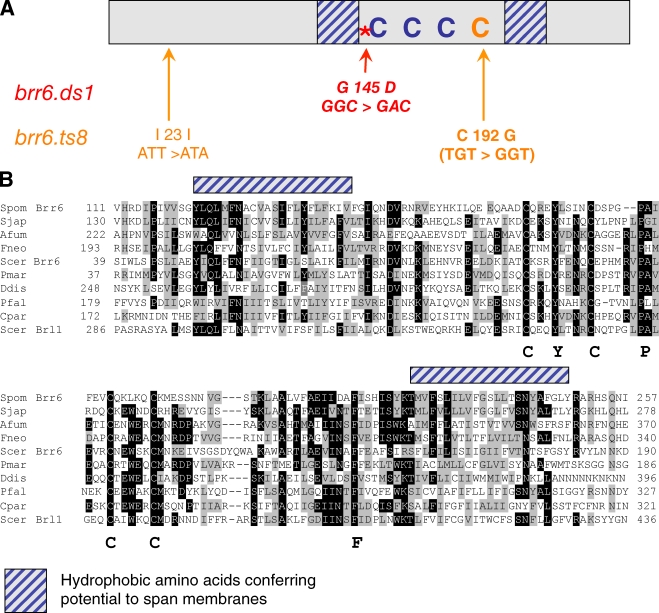
Figure 2.
Brr6 structure, mutations, and homology domains. (A) A cartoon of the Brr6 protein. The brr6.ds1 and brr6.ts8 mutations are indicated in red and orange, respectively. The domains predicted to span membranes are shown with blue hatching and the highly conserved cysteines are indicated by “C.” (B) Multiple sequence alignment of the conserved domains of Brr6-related proteins from the following organisms: Spom, Schizosaccharomyces pombe; Sjap, Schizosaccharomyces japonicus; Afum, Aspergillus fumigatus; Scer, Saccharomyces cerevisiae; Fneo, Filobasidiella neoformans var. neoformans; Pmar, Perkinsus marinus; Ddis, Dictyostelium discoideum; Pfal, Plasmodium falciparum; and Cpar, Cryptosporidium parvum. Amino acid sequences were aligned with the ClustalW program. Identical residues are indicated by black shading, conserved residues by gray. The single, large letters below the alignment highlight some key conserved amino acids, whereas the blocks of stripped blue shading above the alignments correspond to the hydrophobic stretches indicated in A.
brr6.ds1 was not a penetrant allele (Fig. 1 B). We therefore generated tighter temperature-sensitive mutations, the tightest of which, brr6.ts8, gave no binucleate cells after a shift to the restrictive temperature from 25°C to 36°C (unpublished data). brr6.ts8 behaved as a recessive mutation in a heterozygous diploid (Fig. S2 D). Monitoring spindle formation in an asynchronous brr6.ts8 culture shifted from 25°C to 36°C by immunofluorescence established that, after a wave of monopolar spindles, cells with condensed chromosomes devoid of any microtubule staining appeared 40 min after shift to the restrictive temperature (Fig. 1, C–E). To ensure that the lack of tubulin staining within these cells was not an artifact of defective processing, we mixed brr6.ts8 cells that had been held at 36°C for 2 h with spg1.B8 cdc7.A20 cells that had been incubated at 36°C for 4 h, and processed the mixed culture for immunofluorescence microscopy. Microtubules were present in the long multinucleated spg1.B8 cdc7.A20 cells (Fig. 1 E, asterisks) but not in the shorter mononucleated brr6.ts8 cells within the same sample (Fig. 1 E, arrows), which indicates that brr6.ts8 cells were unable to maintain a mitotic spindle after prolonged incubation at the restrictive temperature.
Compromising Brr6 blocks SPB insertion into the nuclear envelope
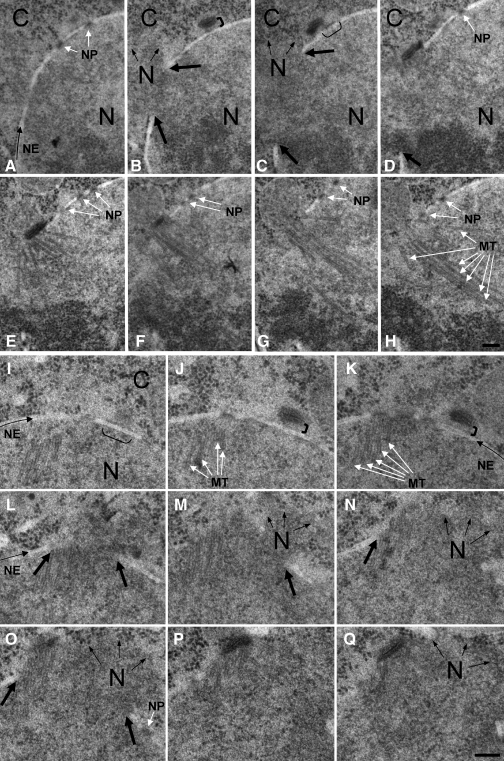
To determine the basis for the brr6− monopolar spindle phenotype, brr6.ts8 cells were processed for electron microscopy. Size-selected G2 cells were used for the shift to 36°C to maximize the frequency of mitotic cells when sampling took place between 110 and 150 min later. Serial sections indicated that SPB duplication was normal; however, the cytoplasmic component of one SPB failed to insert into the nuclear envelope and appeared to drift away from it (Fig. 3 and Fig. S3, for comparative images of an inserted pro-metaphase wild-type SPB, see Fig. S1 B). There was a striking concordance in the distance between the nonfunctional SPB and the nuclear envelope in all 12 cells examined (thick brackets in Fig. 3, B and J; and Fig. S3, D and E). Furthermore, some residual osmophilic material persisted on the outer face of the nuclear envelope (outer NE) below this SPB in the same, or subsequent section (thin brackets Fig. 3, C and I; and Fig. S3, C and O). Thus, this nonfunctional SPB may retain some connection with the nuclear envelope. In all cases, the second SPB inserted into the nuclear envelope and nucleated an extensive microtubule array; however, the insertion of this active SPB was defective, as it was associated with a large hole in the nuclear envelope in 11 out of 12 complete sets of serial sections of brr6.ts8 cells in which both SPBs could clearly be identified (the exception is shown in Fig. S3, M–Q). In contrast, the nuclear envelope was intact in all 15 mitotic wild-type control cells (Fig. S1 B and not depicted).
Figure 3.
SPB insertion defect of brr6.ts8. brr6.ts8 cells that had been grown to mid-log phase at 25°C were synchronized with respect to cell cycle progression by centrifugal elutriation and shifted to 36°C. Cells were fixed by high-pressure freezing 110–150 min later and processed for electron microscopy. A–H and I–Q are consecutive, serial sections through two mitotic cells, respectively. In both cells, a nonfunctional SPB has detached from the nuclear envelope (B and C, and J and K). Although the active SPB is anchored to the nuclear envelope on just one side in the upper cell (right side in D and E), it is not possible to discern an association of the active SPB with the envelope in the lower cell in O–Q. In both cells, the active SPB is associated with a hole in the envelope (to its left in D and E and around it in L–Q). The following features are indicated: The dense brackets in B and J indicate the separation between the nonfunctional SPB and the nuclear envelope. The thin brackets underneath the nuclear envelope in C and I indicate a region occupied by osmophilic material below the SPB on the outer, cytoplasmic face of the nuclear envelope. The pale linear structure curving through the panels indicated by the arrow is the nuclear envelope (NE). The thick black arrows in B–D and L–O indicate the edges of the hole in the nuclear envelope through which the nucleoplasm (N) spills into the more granular cytoplasm (C). The arrows in B, C, M–O, and Q indicate the boundary of the nucleoplasm extruded into the cytoplasm. NP, nuclear pore; MT, microtubules. Bars, 200 nm.
The new SPB fails to insert into the envelope of brr6.ts8 cells at 36°C
As the old and new SPBs differ in their potential to recruit elements of the septum initiation network (SIN) and nucleate microtubules in cut12.1 and cut11.1 mutations (Bridge et al., 1998; Sohrmann et al., 1998; West et al., 1998; Grallert et al., 2004; Tallada et al., 2009), we asked whether SPB age accounted for the heterogeneity in SPB functions during insertion in brr6 mutants. pcp1.RFP atb2.GFP brr6.ts8 and pcp1.RFP atb2.GFP brr6+ cells were isolated by filtration and resuspended in medium lacking nitrogen at 25°C to arrest cell cycle progression in G1 for 16 h. This arrest gave sufficient time for the slow folding RFP to fold into an actively fluorescent configuration. Addition of a nitrogen source as the temperature was increased to 36°C induced cell cycle reentry whereupon a new SPB was synthesized. The RFP within the new SPB did not have time to fold into a fluorescent configuration before division so that the old SPB, but not the new one, emitted a red signal (Fig. S1 C, i). The red signal, in all 17 brr6.ts8 cells in which a single spot of Pcp1.RFP fluorescence could be seen, presided at the base of a microtubule array (Fig. S1 C, ii–iv), which indicates that it was the old SPB that was active.
Brr6 is not required for spindle formation after SPB insertion
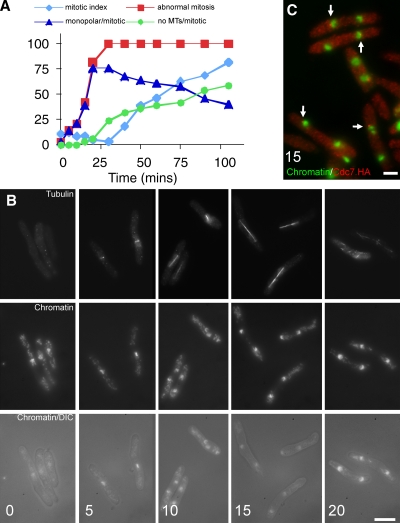
To ask whether Brr6 was required for spindle function after the insertion of the SPB into the nuclear envelope, we used the cold-sensitive nda3.KM311 β-tubulin mutation. Incubation of nda3.KM311 cells at 20°C arrests mitotic progression in prophase after the SPBs have inserted into the nuclear envelope (Kanbe et al., 1990), after which a temperature shift between 30°C and 37°C restores tubulin function, generating a synchronized mitosis (Uemura et al., 1987). For this approach to really test Brr6 function, we had to be confident that Brr6.ts8 protein was inactivated immediately upon temperature shift of brr6.ts8. The stress response invoked when a brr6.ts8 culture is shifted from 25°C to 36°C transiently blocks mitotic commitment at precisely the point at which we wish to study Brr6 requirement in the nda3.KM311 experiment (Fig. 1 D). We therefore monitored spindle integrity as wee1.50 brr6.ts8 cells were shifted from 25°C to 36°C because Wee1 inactivation overrides this stress response, driving all cells into mitosis (Nurse, 1975). All spindles formed over the 20 min after the heat shift were monopolar (Fig. 4 A), which indicates that Brr6.ts8 function was indeed immediately inactivated by the temperature shift. We therefore shifted brr6.ts8 nda3.KM311 cells from 20°C to 36°C and observed normal segregation of chromosomes on bipolar spindles (Fig. 4 B). Furthermore, cell cycle progression through anaphase B was unaffected, as the recruitment of SIN kinase Cdc7 to the new SPB in anaphase B (Grallert et al., 2004; Sohrmann et al., 1998) was not perturbed (Fig. 4 C). We conclude that Brr6 is not required for chromosome segregation after the SPB has inserted into the nuclear envelope.
Figure 4.
Brr6 is not required for spindle formation after SPB insertion into the nuclear envelope. (A) wee1.50 brr6.ts8 cells were grown to early log phase in rich YE5S medium at 25°C before the temperature of the culture was shifted to 36°C. Cells were processed for immunofluorescence with anti-tubulin, anti-Sad1, and DAPI, and scored as indicated. (B and C) brr6.ts8 nda3.KM311 (B) or brr6.ts8 nda3.KM311 cdc7.HA cells (C) were grown to early log phase at 30°C before the temperature of the culture was dropped to 20°C. 8 h later, the temperature of each culture was shifted to 36°C and samples were processed at the indicated times (min) for immunofluorescence with anti-tubulin (B) or 12CA5 antibodies (C), then stained with DAPI. (B) From top to bottom, microtubules, chromatin, and combined chromatin and differential interference contrast (DIC). (C) Combined staining of Cdc7.HA (red) and chromatin (green). Arrows indicate Cdc7.HA at new SPBs. Bars: (B) 10 µm; (C) 5 µm.
Extensive genetic interactions between brr6 and genes encoding SPB components
brr6.ds1 and brr6.ts8 mutants exhibited significant genetic interactions with mutations in SPB components (Fig. S4 A). The inclusion of brr6.ts8 blocked growth of sad1.2, cut12.1, and kms2.Δ mutants at temperatures at which either single mutant was viable. In contrast, perturbation of Brr6 function with brr6.ts8 (or brr6.ds1; unpublished data) reduced the requirement for the Ndc1 homologue Cut11. Conversely, the inclusion of a temperature-sensitive allele of cdc31+, cdc31.1, or deletion of kms1+ alleviated the cold sensitivity of the brr6.ds1 mutation. Monitoring spindle integrity in cut11.1 brr6.ts8 mutants 3 h after a shift to the semipermissive temperatures of 28°C and 30°C confirmed the results of the spot test analysis, which showed that compromised Brr6 function alleviates the impact of defective Cut11 function (Fig. S4 B). The recruitment of Cut12, Sad1, or Cut11 to SPBs was unaffected by the brr6.ts8 mutation (Fig. S4 C; Hagan and Yanagida, 1995; Bridge et al., 1998; West et al., 1998).
Brr6 is recruited to SPBs during SPB insertion and extrusion
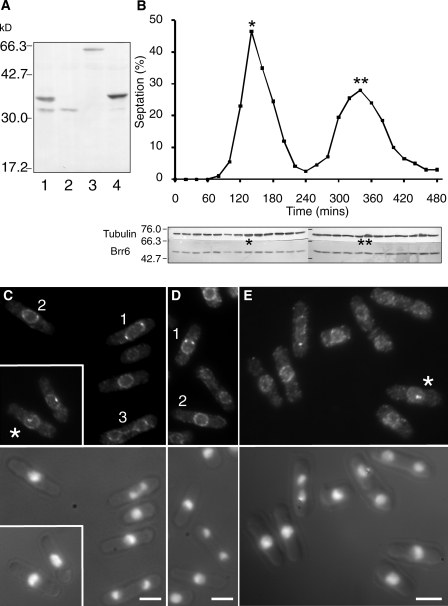
Brr6 is a member of a protein family that contains the Pfam motif PF10104 (Finn et al., 2008; http://pfam.sanger.ac.uk//family/PF10104#tabview=tab6; Brr6_like_C_C), which has members in fungi and protists, but not higher eukaryotes (Lo Presti et al., 2007). Brr6 family members contain two regions predicted to span membranes that are separated by a conserved region that contains, among other conserved residues, four cysteine residues (Fig. 2; brr6.ts8 is a mutation of the conserved cysteine at 192 to glycine [Fig. 2 A]). Immunofluorescence with antibodies to the nonconserved domains of Brr6 (Fig. 5 A) revealed a signal around the nuclear periphery throughout the cell cycle and an association with the mitotic SPB (Fig. 5, C–E; not depicted). The SPB fluorescence was initially a single dot, indicating that Brr6 was recruited at the earliest stages of insertion of the adjacent two SPBs (Ding et al., 1997). This dot staining broadened to give two foci that were barely resolved from one another and that often resided within the circle defined by the staining at the nuclear periphery, after which the SPB staining was at the nuclear periphery (Fig. 5, C and E). The signal persisted at both SPBs until midway through anaphase, whereupon it is diminished asymmetrically. Labeling the new SPB with Cdc7.HA (Sohrmann et al., 1998; Grallert et al., 2004) established that the loss of Brr6 from the SPB was stochastic and did not correlate with SPB age (Fig. 6 A). There was a striking variation in the intensity of SPB staining. Early mitotic and mid-anaphase SPBs had stronger signals than metaphase or late mitotic cells (Fig. 5, C and D; cells are numbered to indicate descending intensity levels).
Figure 5.
Stage-specific differential association of Brr6 with mitotic SPBs. (A) Antibodies raised and purified against a equimolar mixture of the first 79 and last 44 amino acids of the Brr6 recognized a single band at 34 kD on blots of cell extracts prepared from wild-type cells (lane 2; brr6+). This band was replaced by a band at 37 kD and 65 kD in strains in which three Pk epitopes or a single EGFP had been inserted at the start of the brr6+ ORF (lanes 3 [brr6.NEGFP] and 4 [brr6.N3Pk]) and was supplemented by a band at 37 kD when a Brr6.3Pk fusion protein was expressed in wild-type cells (lane 1; leu1::ura4+nmt41brr6N3Pk). (B) Size selected mid-log phase cells were inoculated at a density of 106 cells/ml, and samples were processed for calcofluor staining to score septa (graph) or anti-Brr6 Western blot to monitor Brr6 protein. The sets of one and two asterisks above the blot in B show the peaks of maximal septation in the first and second cell cycles, respectively, as indicated on the graph. (C–E) Anti-Brr6 immunofluorescence microscopy of mid-log phase cells reveals punctate staining around the nuclear periphery and association with the SPBs of mitotic cells. The intensity of the signal varies at different stages of mitosis. Cells are numbered in descending order of brightness in a particular field. The intensity of the two metaphase SPBs in cell 2 (D) is markedly lower than that in the earlier mitotic stage in cell 1 in the same field, which indicates that the amount of Brr6 associating with SPBs diminishes at metaphase. Similarly, the reduced level of staining in the early and late anaphase cells 2 and 3 (C) shows that Brr6 association with the SPB peaks in mid-anaphase (cell 1) at a point before the nuclear envelope can define two distinct yet connected spheres (cell 3). At the earliest stages of mitosis, Brr6-stained SPBs appear to reside within the area defined by the nuclear peripheral staining (asterisks in C and E). Bars, 5 µm.
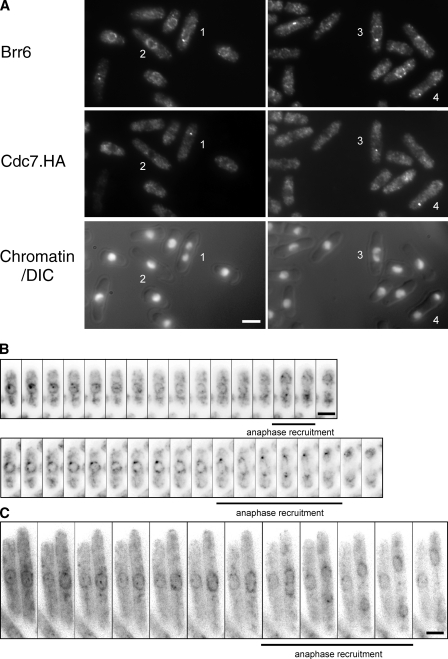
Figure 6.
Stage-specific differential association of Brr6.NEGFP with mitotic SPBs. (A) Mid-log phase cdc7.HA cells were processed for immunofluorescence microscopy and stained with antibodies to Brr6, 12CA5 antibodies to the HA epitope on Cdc7.HA, and DAPI. Although the Cdc7.HA staining of the new SPB recognizes the pole opposite the one recognized by the Brr6 antibodies in cell 1, the two signals coincide on the same SPB in cells 3 and 4. Note that although the HA staining of cell 2 indicates that this cell is at metaphase, there is no Brr6 staining detected at this SPB. This highlights the departure of Brr6 from SPBs after insertion into the nuclear envelope. (B and C) Live cell imaging of brr6.NEGFP (B) and brr6.NEGFP cdc25.22 (C) cells. Although there is variation in absolute timing of events in individual cells, in each case, the initially strong association of Brr6.NEGFP with the SPB diminishes as cells get to metaphase to rise once more midway through anaphase B (indicated by the thick lines under the panels). The cells in B were arrested at the G2/M boundary for 4.25 h by incubation at 36°C before the temperature was returned to 25°C to induce mitosis. The increased cell volume arising from the prolonged arrest results in an extension in the duration of anaphase B and so extends the duration between the first and second phases of Brr6.NEGFP recruitment. Bars, 5 µm.
Although we were unable to tag the carboxyl terminus of Brr6 with GFP (unpublished data), fusion of the EGFP sequences to the amino terminus generated a functional fusion protein that showed very weak association with cytoplasmic membranes, strong enrichment around the nuclear periphery, and very strong associations with mitotic SPBs. Fluctuations in the intensity of this SPB association of Brr6.NEGFP confirmed that SPB recruitment of Brr6 was transient, first during mitotic commitment and then during anaphase B/mitotic exit (Fig. 6 B). The lack of recruitment between these two phases was graphically illustrated in the greatly extended anaphase B that follows transient arrest at the G2/M boundary of cdc25.22 cells (Fig. 6 C; Hagan et al., 1990). Thus, at times when the SPB enters or leaves the nuclear envelope, Brr6 association with the SPB increased. Western blot analysis of extracts from cells synchronized with respect to cell cycle progression by centrifugal elutriation with anti-Brr6 antibodies established that the changes in Brr6 localization were not accompanied by marked alteration of Brr6 levels or migration in SDS-PAGE (Fig. 5 B).
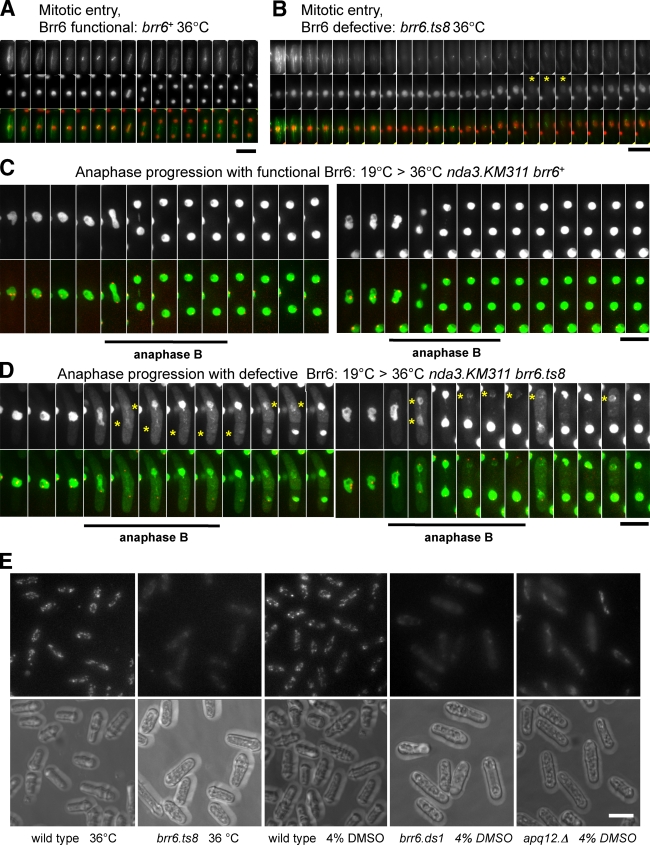
The re-recruitment of Brr6 to SPBs in anaphase B suggested that it may play a role in coordinating membrane dynamics with SPB extrusion. We therefore addressed the impact of brr6.ts8 upon nuclear integrity with a GFP–β-Gal NLS fusion protein (Yoshida and Sazer, 2004). As this NLS-tagged fusion protein is constantly imported into the nucleus, cytoplasmic fluorescence is negligible throughout the closed mitosis of S. pombe (Fig. 7, A and C; and Videos 1 and 3). Any breach of nuclear integrity at any point in the cell cycle is therefore easily detected as the fluorescent signal disperses throughout the cell (Yoshida and Sazer 2004; Gonzalez et al., 2009; Tallada et al., 2009). As expected from the electron microscopic analysis of the brr6.ts8 phenotype (Figs. 3 and S3), the GFP signal flooded out of brr6.ts8 nuclei, as nuclear integrity was compromised during mitotic commitment (Fig. 7 B, asterisks; and Video 2).
Figure 7.
Anaphase/mitotic exit Brr6 regulates nuclear integrity. (A and B) Cell cycle progression of brr6+ (A) and brr6.ts8 (B) cells harboring the int::GFP-β-Gal NLS nuclear integrity marker and pREP81atb2.Cherry to mark nuclear integrity and microtubules, respectively. (C and D) The temperature was shifted from 25°C to 36°C 0.5 h before filming. Cell cycle progression of nda3.KM311 (β-tubulin mutation) cells harboring the sid4.Tom SPB marker (red) and the int::GFP-β-Gal NLS nuclear integrity marker (green) were arrested by incubation at 19°C for 9 h before the temperature was raised to 36°C to restore tubulin function. A–D show frames from Videos 1–4, respectively. The integrity of the nuclear envelope is retained during mitotic commitment (A) and anaphase B/mitotic exit of the wild-type (C) but not the mutant cells (B and D). Asterisks indicate nuclei for which the ability to retain the nuclear marker is severely compromised while the bars under the times series in C and D highlight anaphase B nuclei. (E) Cultures of the indicated strains were either shifted from 25°C to 36°C or had DMSO added at the same time as 20 µg/ml NBD-cholesterol. 1 h later, fluorescence and DIC images were captured. Bars, 10 µm.
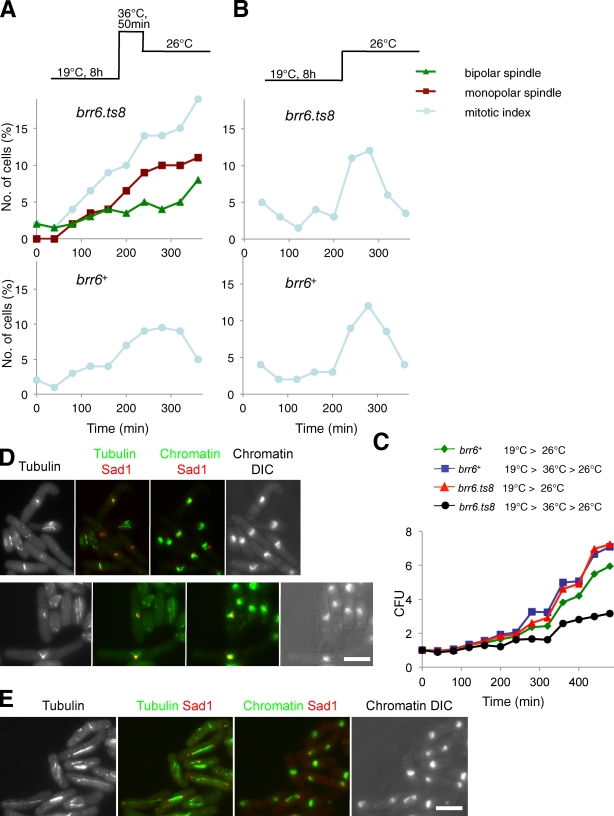
To address the integrity of the nuclear envelope during anaphase as the SPB is extruded from the nuclear envelope once more, we introduced the GFP–β-Gal NLS fusion protein into the nda3.KM311 arrest release approach (Fig. 4) to monitor nuclear envelope integrity during anaphase, when Brr6 function is compromised by temperature inactivation of the brr6.ts8 gene product. As before, cells were arrested in prophase at 20°C before warming to 36°C to inactivate Brr6 function. Although GFP–β-Gal NLS was retained within the envelope of control nda3.KM311 brr6+ cells (Fig. 7 C and Video 3), it transiently flooded out of nda3.KM311 brr6.ts8 nuclei during anaphase B/mitotic exit, which indicates that the integrity of the nuclear envelope at this time was compromised by the brr6.ts8 mutation (Fig. 7 D, asterisks; and Video 4). If this disruption of envelope integrity arose from errors in SPB/membrane topology during extrusion, the fidelity of SPB insertion in the subsequent mitosis could be compromised, even when Brr6 function is restored for the duration of the intervening G2 phase. We therefore monitored cell viability and spindle integrity in brr6+ nda3.KM311 and brr6.ts8 nda3.KM311 cells after transient incubation at 36°C (50 min) before a return to the brr6.ts8 permissive temperature of 26°C (Fig. 8 A). Control cultures were warmed directly from 19°C to 26°C (Fig. 8 B). The colony-forming ability of the brr6.ts8 mutant cells was reduced if they underwent division at 36°C, but not if they did not experience Brr6 inactivation during mitosis (Fig. 8 C). Importantly, mitotic defects were apparent in the subsequent mitosis at the permissive temperature, when brr6.ts8 prophase cells had been transiently raised to 36°C to undergo mitosis, and at the restrictive temperature before being returned to the permissive temperature for the duration of interphase (Fig. 8 D). In contrast, no mitotic defects were seen if cells had not undergone mitosis at the restrictive temperature, but merely shifted back to the permissive temperature of 26°C immediately after the prophase arrest (Fig. 8 E). These data suggest that recruitment of Brr6 to anaphase B SPBs facilitates the resolution of SPB/nuclear envelope topology during SPB extrusion.
Figure 8.
Anaphase/mitotic exit Brr6 regulates nuclear integrity and spindle formation in the next cell cycle. (A and B) brr6+ and brr6.ts8 strains containing the nda3.KM311 β-tubulin mutation were treated as shown above each of the two graphs, and aliquots were processed for cell viability assay (C) or immunofluorescence microscopy (D and E). The mitotic index and spindle morphology was scored and plotted, as indicated, in the graphs in A and B. (C) The appearance of the defective spindles in the cycle after anaphase at the restrictive temperature is accompanied by a loss in colony forming units (CFU). (D and E) Representative images of the defective brr6.ts8 or normal, brr6+ spindles from the 280-min time point are shown in D and E, respectively. Bars, 10 µm.
Brr6 does not associate with nuclear pores
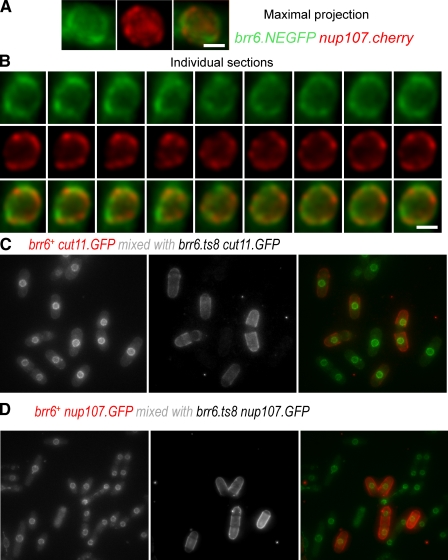
The association of Brr6 with the nuclear periphery throughout the cell cycle prompted us to ask whether this distribution reflected an association of Brr6 with nuclear pores; however, we did not detect a marked registration between the signals for Brr6.NEGFP and the NPC marker Nup107.cherry (Fig. 9, A and B). Furthermore, imaging either the Nup107.GFP or a Cut11.GFP signal in mixed brr6+/brr6.ts8 cultures at the restrictive temperature did not reveal any alterations in the distribution or quantity of nuclear pores in S. pombe brr6.ts8 mutants after 1 h at the restrictive temperature (Fig. 9, C and D).
Figure 9.
Brr6 shows no functional or physical association with nuclear pores. (A and B) Optical sections were taken through nuclei of brr6.NEGFP nup107.cherry cells. Images in A show a maximal projection of the individual 0.2-µm step size z sections shown in B. There is very little registry between the Nup107 and the Brr6 signals. (C) brr6+ cut11.GFP and brr6.ts8 cut11.GFP cells were grown to early log phase in EMM2 before being shifted to the restrictive temperature of 36°C for 1 h. brr6+ cut11.GFP cells were isolated by filtration, stained with TRITC-conjugated lectin, mixed with an equal number of brr6.ts8 cut11.GFP cells, and mounted for live cell imaging at 36°C. Maximal projections of series of optical sections in the z axis show that the intensity of the Cut11.GFP signal is not affected by the inactivation of Brr6. (D) The same as for C, except that Nup107.GFP is used in place of Cut11.GFP. Again, there is no impact of the brr6.ts8 deficiency upon the distribution of the nuclear pore component. Bars: (A and B) 2 µm; (C and D) 10 µm.
Cooperation between Apq12 and Brr6
apq12+ was isolated as a multicopy suppressor of brr6.ds1 (Fig. S5 A). The intimate genetic and functional relationship between budding yeast Apq12 and Brr6 (Hodge et al., 2010) prompted us to assess the relationship between the fission yeast homologues. Although apq12+ is not an essential gene (Decottignies et al., 2003), the addition of 4% DMSO to apq12.Δ cells did compromise spindle formation (Fig. S5 B), which suggests that it may indeed act in partnership with Brr6. This view was consolidated by the synthetic lethality arising from combining apq12.Δ cells with either brr6.ds1 or brr6.ts8 (Fig. S5 C and not depicted), and the striking concordance of the genetic relationships between apq12.Δ and mutations in SPB components and those displayed between the same SPB mutants and brr6.ts8/brr6.ds1 (Fig. S5 D and not depicted). Apq12.tdTom did not associate with SPBs at any point in the cell cycle but exhibited a more general membrane reminiscent of the distribution of the endoplasmic reticulum (Fig. S5 E; Broughton et al., 1997).
The sensitivity of S. cerevisiae brr6 and apq12 mutants to reagents that increase membrane fluidity led to the conclusion that they modulate lipid homeostasis to facilitate the assembly of the outer components of the nuclear pore on the nuclear envelope (Hodge et al., 2010). Strikingly, S. pombe brr6 and apq12 mutants also had a marked impact upon the tolerance of these agents (Fig. S5 F). Moreover, mutation of either brr6 or apq12 compromised the incorporation of NBD cholesterol into membranes (Fig. 7 E), which is indicative of an impact upon sterol metabolism.
Discussion
We show that Brr6 is required for the correct insertion of the fission yeast SPB during mitotic commitment. It is also required for nuclear envelope integrity during anaphase B/mitotic exit. This latter requirement coincides with the time at which the interphase separation of the nuclear and cytoplasmic SPB components is reestablished (Ding et al., 1997). Consistently, Brr6 is transiently recruited to the SPB at mitotic commitment and again during anaphase B.
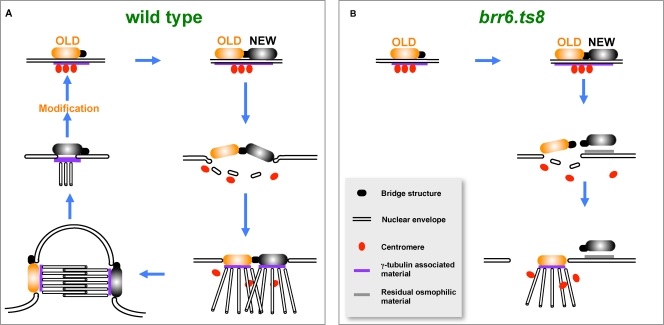
Ablation of Brr6 function with the brr6.ts8 mutation blocked the dispersal of the nuclear envelope at the new SPB during mitotic commitment (Fig. 10). The retention of osmophilic material on the outer face of the envelope beneath the inactive cytoplasmic SPB component and the consistency in the distance separating it from the envelope both suggest that the new SPB retains a degree of attachment to the nuclear envelope. Integration of the active, old, SPB into the envelope was accompanied by the formation of a large hole next to it, indicating that this integration event was also defective. These data suggest that, unlike Ndc1/Cut11 that anchors the SPB within the nuclear envelope (West et al., 1998), Brr6 coordinates the SPB insertion cycle with the generation of the pore within the envelope.
Figure 10.
A diagram summarizing the SPB insertion defect of brr6.ts8 cells.
The loss of nuclear integrity during mitotic commitment in cut11 mutants could indicate that Cut11 anchorage must be tightly coordinated with the Brr6-driven integration. However, nuclear integrity is also compromised during mitotic commitment of cut12.1 mutants. In contrast to Cut11 and Brr6, the sequence and relationship between cut12 and cdc25 (for example, the ability to suppress the nuclear integrity defect by elevation of Cdc25 levels) suggest that Cut12 regulates SPB activation and mitotic commitment rather than providing a physical interface with the envelope (Tallada et al., 2009). Thus, loss of envelope integrity may reflect a generic “Achilles heel” of spindle formation in fission yeast that arises from the need to transiently generate a hole large enough to accept the SPBs. In this context, it is notable that compromised nuclear envelope integrity has not been reported for Ndc1, or any budding yeast SPB insertion mutants (Adams and Kilmartin, 2000). Thus, the route of wholesale fenestration to accept both SPBs simultaneously may place more of a strain upon a system than one in which a second SPB is “pulled” into the envelope via interactions with its embedded sister.
The failure to integrate the new SPB while an adjacent, active, old SPB borders a hole in the envelope also mirrors the consequences of mutating the core SPB component Cut12 and the Ndc1 orthologue Cut11 (West et al., 1998; Tallada et al., 2009). As Cut11 and Brr6 recruitment to the SPB are both confined to mitosis, the functional asymmetry between the SPBs most likely reflects an inherent difference between the two SPBs by themselves rather than a differential impact of the mutation in these molecules upon SPB structure. This relationship between SPB age and nuclear envelope dynamics extends to more global organization. Reducing membrane incorporation into the mitotic envelope results in asymmetric nuclear separation; a larger nucleus separates from a smaller one (Gonzalez et al., 2009). Fascinatingly, the large nucleus is associated with the old SPB when the membrane deficiency arises from mutation of the RAN pathway; however, there is no correlation with SPB age when the membrane deficiency arises from a simple reduction in membrane biogenesis (Gonzalez et al., 2009). Furthermore, SPB age dictates function at other cell cycle stages; the molecules of the SIN are recruited to the new SPB in anaphase B, Kms2 associates with one of the two interphase SPBs in ima1.Δ cells, and only one of the two SPBs expands after overproduction of the MPF inhibitor Rum1 (Sohrmann et al., 1998; Grallert et al., 2004; Uzawa et al., 2004; King et al., 2008). Thus, there are many inherent functional differences between the old and new SPBs; the old SPB appears to be more robust, as it tolerates deficiencies in constituent or interacting molecules that render the new SPB defective. The molecular basis for these distinctions between the old and new SPBs remains to be established.
Fission yeast Brr6 is named after the budding yeast homologue BRR6 (Bad Response to Refrigeration). BRR6 and BRL1 (BRR6 related 1) are essential components of the nuclear periphery that are required for efficient nuclear transport (de Bruyn Kops and Guthrie, 2001; Saitoh et al., 2005). Loss of either can be complemented by S. pombe or Pneumocystis carinii brr6+ (Saitoh et al., 2005; Lo Presti et al., 2007). BRL1 and BRR6 are thought to have arisen from a genome duplication event that is specific to the Saccharomycotina subphylum of the Ascomycota. As brr6+ genes outside this subphylum are represented once in their genomes, it has been proposed that Brr6 function was originally executed by a single isoform in budding yeast ancestors before the duplication event (Lo Presti et al., 2007).
S. cerevisiae Brl1 was identified as a multicopy suppressor of a mutation in the XPO1/exportin gene CRM1 (Saitoh et al., 2005). brl1 mutants are compromised for protein export, and Brl1 associates with the NPC outer ring component Nup145 (Alber et al., 2007) in two-hybrid assays (Ito et al., 2001). However, Brl1 does not behave as an integral NPC component; it is not found in NPC complexes (Alber et al., 2007), and no Brl1/Brr6 homologues exist in higher eukaryotes whose NPCs mirror yeast NPCs in composition and structure (Lo Presti et al., 2007). Furthermore, its distribution around the budding yeast nuclear periphery is not altered by the nup133.Δ mutation that clusters nuclear pores (Saitoh et al., 2005). The transport defects exhibited by brl1 mutants may therefore arise from altered nuclear envelope organization, as NPC distribution is disrupted in such cells (Saitoh et al., 2005). Similarly, although S. pombe Brr6 neither associates with NPCs nor impacts upon NPC number or distribution, defects in the ability to sequester a β-Gal NLS fusion protein indicate that S. pombe Brr6 function does have an impact on nuclear envelope physiology (unpublished data).
Altered assimilation of sterols and sensitivity to agents that alter membrane fluidity in both S. pombe and S. cerevisiae brr6 mutants suggests that a principle function for Brr6-related molecules is to alter membrane composition. In the case of fission yeast Brr6, its recruitment to SPBs during SPB integration and extrusion suggests that the physical association of Brr6 with the SPB at this time facilitates changes in SPB association with the envelope because it physically alters the lipid composition of the nuclear envelope in the neighborhood of the SPB at these times. Strikingly, defects in both fission and budding yeast affect events on the cytoplasmic face of the nuclear envelope (new SPB integration in S. pombe and assembly of NPC components in S. cerevisiae; Hodge et al., 2010), raising the question as to whether Brr6-related molecules specifically modify this outer membrane?
The membrane-spanning Pfam motif PF10104, which defines a Brr6 molecule, is found in fungi and protists that generate a polar fenestra in the nuclear envelope to enable an MTOC to organize both cytoplasmic and nuclear microtubules within a closed mitosis (Heath, 1980); e.g., Dictyostelium discoideum (Moens, 1976). We believe that it is highly significant that PF10104 is not found in microbes that do not need a fenestra to form within the envelope, either because their anastral spindles are organized by an intranuclear MTOC as in Trypanosoma brucei or no MTOC at all, as in Tetrahymena (Fujiu and Numata, 1999; Ogbadoyi et al., 2000). This striking distribution of PF10104 and the clear demonstration that fission yeast Brr6 mediates SPB insertion and exit lead us to propose that Brr6-related molecules modulate similar interactions between MTOCs and the nuclear envelope in other species.
Although we propose a conserved function for Brr6-related proteins, elaborations around a core PF10104 function could account for the diversity of solutions that have been adopted to cope with the challenges of segregating genomes within an enclosed nuclear envelope (Kubai, 1976; Heath, 1980). For example, it is possible that the need to retain the SPB within the envelope throughout the cell cycle (Adams and Kilmartin, 2000) has set different hierarchies for SPB incorporation in budding and fission yeasts. As discussed earlier, the insertion event in S. cerevisiae in which the duplication plaque is pulled into the envelope alongside its integrated parental SPB differs considerably from insertion into a fenestra within the S. pombe nuclear envelope. Maybe these distinct topological demands explain the absence from S. pombe of the Nbp1 molecule that associates with Ndc1 and Mps2 to drive the insertion of the S. cerevisiae duplication plaque (Araki et al., 2006). In this context, it would be interesting to determine the contributions of Brl1 and Brr6 to SPB integration in strains in which a modified SESA network facilitates life without Mps2 (Sezen et al., 2009).
There is currently no link between Brr6 or Brl1 and SPB insertion in S. cerevisiae. Perhaps this reflects a need to simultaneously mutate both loci to reveal an SPB phenotype. However, the functional data that links Brr6 with NPC assembly (Hodge et al., 2010) does challenge our proposal that the primary requirement for the PF10104 domain is to generate polar fenestrae. It may be important to consider the generic evolutionary conservation of NPC architecture and the restriction of PF10104 distribution; although NPC architecture is conserved, many eukaryotes do not need a PF10104 protein for NPC assembly. So why does S. cerevisiae need one? Perhaps this requirement reflects a coevolutionary adaptation to the coexistence of NPCs and PF10104 domains; i.e., although PF10104 domain proteins are not required for the assembly of a generic eukaryotic NPC, if one is present, NPC components adapt to the alterations in membrane composition they promote, leading to an “addiction” to PF10104-driven modifications for NPC assembly. Thus, if a PF10104 molecule is modifying membranes, it is understandable that resident membrane proteins would adapt to accommodate, or take advantage of, the modified membrane properties.
In summary, we believe that the characterization of S. pombe Brr6 presented here and the evolutionary distribution of the PF10104 domain suggests that the Brr6 family enables a significant subset of lower eukaryotes to deal with one of the significant distinctions between many higher and lower eukaryotes: genome segregation within a closed mitosis.
Materials and methods
Cell culture and strains
Strains are listed in Table I. Cells were cultured in YE5S or appropriately supplemented Edinburgh Minimal Medium number 2 (EMM2; Moreno et al., 1991). Size selection for cell cycle synchronization used an elutriator rotor (JE-5.0; Beckman Coulter; Mitchison and Carter, 1975). The screen of Broek et al. (1991) was modified by the inclusion of cut12.NEGFP to identify conditional mutations that condensed chromosomes and exhibited defects in SPB separation in the presence of 4% DMSO (Craven et al., 1998; Poloni and Simanis, 2002). The pURB1 and pURB2 libraries were used to clone plasmids that complement the brr6.ds1 mutation according to standard procedures (Moreno et al., 1991; Barbet et al., 1992).
Table I.
Strains used in this study
| Strain number | Genotype | Source |
| IH304 | h− nda3.KM311 leu1.32 | Toda et al., 1983 |
| IH531 | h− leu1.32 | Laboratory stock |
| IH654 | h− cut11.1 ura4.d18 leu1.32 | West et al., 1998 |
| IH754 | h− cut12.1 ade6.704 ura4.D18 | Bridge et al., 1998 |
| IH759 | h− cdc7.HA:ura4+ ura4.d18 | Sohrmann et al., 1998 |
| IH1389 | h− cut11.GFP ade6.704 ura4.d18 leu1.32 | West et al., 1998 |
| IH1740 | h− cdc7.A20 spg1.B8 leu1.32 ura4.d18 | Tanaka et al., 2001 |
| IH2466 | h− cdc31.1 ura4.d18 leu1.32 | This paper |
| IH2584 | h+ brr6.ds1 leu1.32 his2 ade6.M210 | This paper |
| IH2586 | h− brr6.ds1 leu1.32 ura4.d18 | This paper |
| IH3317 | nmt81gfpatb2:kanR pcp1.cRFP:kanR leu1.32 | Grallert et al., 2004 |
| IH3365 | h+/h− brr6+/brr6+ ade6.M210/ade6.M216 leu1.32/leu1.32 | This paper |
| IH3366 | h+/h− brr6::kanR/brr6+ ade6.M210/ade6.M216 leu1.32/leu1.32 | This paper |
| IH3412 | h− brr6.C.kanR ade6.M210 leu1.32 ura4.d18 | This paper |
| IH3442 | h+ apq12::kanR ade6 leu1.32 | This paper |
| IH3487 | h− brr6::kanR ade6.M210 leu1.32 pREP81brr6+ | This paper |
| IH3508 | h− leu1::ura4+ brr6.N3Pk ura4.d18 | This paper |
| IH3587 | h− brr6.C.ura4+ ade6.M210 leu1.32 ura4.d18 | This paper |
| IH3620 | h− brr6.ts8:ura4+ ura4.d18 | This paper |
| IH3781 | h− brr6.N3Pk:ura4+ ade6.M210 leu1.32 ura4.d18 | This paper |
| IH3957 | h− brr6.ts8:ura4+ cut11.GFP ade6.M210 ura4.d18 leu1.32 | This paper |
| IH3991 | h− cut12.1 brr6.ts8:ura4+ ura4.d18 | This paper |
| IH4081 | h− sad1.2 leu1.32 | This paper |
| IH4165 | h+ cut11.1 brr6.ts8:ura4+ ura4.d18 | This paper |
| IH4167 | h− sad1.2 brr6.ts8:ura4+ leu1.32 ura4.d18 | This paper |
| IH4194 | h+ apq12::kanR cut11.1 leu1.32 | This paper |
| IH4202 | h− kms1::ura4+ leu1.32 ura4.d18 | Miki et al., 2004 |
| IH4206 | h− kms2::ura4+ leu1.32 ura4.d18 | Miki et al., 2004 |
| IH4282 | h+ kms1::ura4+ brr6.ds1 ura4.d18 leu1.32 | This paper |
| IH4292 | h+ cut11.1 brr6.ds1 leu1.32 his2 | This paper |
| IH4293 | h− apq12::kanR sad1.2 leu1.32 | This paper |
| IH4295 | h− apq12::kanR cut12.1 ade6. | This paper |
| IH4297 | h+ apq12::kanR kms2::ura4+ leu1.32 ura4.d18 ade6. | This paper |
| IH4323 | h− cdc31.1 brr6.ds1 ura4.d18 leu1.32 | This paper |
| IH4326 | h+ apq12::kanR cdc31.1 leu1.32 | This paper |
| IH4333 | h+ nup107.cGFP:kanR ade6.M216 ura4.d18 leu1.32 | Baï et al., 2004 |
| IH4358 | h+ brr6.ts8:ura4+ kms2::ura4+ ura4.d18 | This paper |
| IH4381 | h− brr6.nEGFP:ura4+ ura4.d18 | This paper |
| IH4423 | h− brr6.ts8:ura4+ nup107.cGFP:kanR ura4.d18 leu1.32 ade6.M216 | This paper |
| IH4600 | h−/h+ ade6.M210/ade6.M216 leu1+/leu1.32 brr6.ts8:ura4+/brr6+ ura4.d18/ura4+ | This paper |
| IH4750 | brr6.ts8:ura4+ nmt81gfpatb2:kanR pcp1.cRFP:kanR ura4.d18 | This paper |
| IH5076 | h− brr6.nEGFP:ura4+ nup107.cCherry:kanR ade6.M210 leu1.32 ura4.d18 | This paper |
| IH5113 | brr6.ts8:ura4+ nda3.KM311 cdc7.HA:ura4+ | This paper |
| IH5253 | leu1::nmt4x-SV40NLS.GFP-LacZ:ura4+ ade6.M216 leu1.32 ura4.D18 | Yoshida and Sazer, 2004 |
| IH5254 | brr6.ts8:ura4+ leu1::nmt4x-SV40NLS.GFP-LacZ:ura4+ ade6.M216 leu1.32 ura4.D18 | This paper |
| IH5155 | brr6.ts8:ura4+ nda3.KM311 | This paper |
| IH5974 | h− 972 | Laboratory stock |
| IH6083 | brr6.ts8:ura4+ wee1.50 ura4.d18 | This paper |
| IH6091 | cut11.GFP sid4.tdTom:hphR leu1.32 ura4.d18 brr6.ts8:ura | This paper |
| IH6177 | h+ apq12.tdTom:hphR pcp1.gfp his2 leu1.32 ura4.d18 | This paper |
| IH7609 | h− brr6.nEGFP:ura4+ cdc25.22 ura4.d18 | This paper |
| IH8062 | brr6.ts8:ura4+ nda3.KM311 leu1::nmt4x-SV40NLS.GFP-LacZ:ura4+ sid4.Tdtom:hphR ura4.d18 | This paper |
| IH8088 | nda3.KM311 leu1::nmt4x-SV40NLS.GFP-LacZ:ura4+ ura4.d18 sid4.Tdtom:hphR | This paper |
| IH9993 | brr6.ts8:ura4+ sid4.Tdtom:hphR cut12.NEGFP leu1.32 ura4.d18 h- | This paper |
| IH9997 | brr6.ts8:ura4+ sid4.Tdtom:hphR leu1.32 ura4.d18 sad1.CFP | This paper |
Master strains containing sad1.2, kms1::ura4+, kms2::ura4+, nup107.cGFP:kanR were provided by K. Sheldrick (University of Manchester, Manchester, England, UK), O. Niwa (Kazusa DNA Research Institute, Kisarazu, Chiba, Japan), and V. Doye (Institut Curie, Paris, France), respectively.
Isolation of apq12+ and brr6+
Transformation of IH2586 with the pURB2 library (Barbet et al., 1992) generated 3 × 105 transformants. Plasmid rescue, retransformation, and subcloning established that two of the transformants are viable because the apq12+ gene on the plasmids they contained suppressed the DMSO sensitivity of brr6.ds1. Mitotic haploidization with lys1/ura1 leu1/his3 and ade6 markers (Kohli et al., 1977) established that the sdd1.ds1 mutation resided on chromosome I. After initial long-range mapping in a swi5.158 background (Schmidt, 1993), crosses between sdd1.ds1 and cdc24.M38 produced no recombinants in 13 tetrads. Subcloning cosmid SPAC8F11 (Wellcome Trust Sanger Institute) and Tn1000 transposition analysis (Morgan et al., 1996) established that sdd1+ was allelic to brr6+. The SMART RACE cDNA kit (Takara Bio Inc.) established that the 5′ untranslated region (UTR) and 3′ UTR for brr6+ were 257 nt and 232 nt, respectively.
Mutagenesis of S. pombe loci
PCR technologies were used to delete brr6+ and tag the nup107+ and apq12+ loci (Bähler et al., 1998). The temperature-sensitive brr6.ts8 and N-terminally tagged brr6.NEGFP and brr6.N3Pk alleles were generated by a marker switch approach (MacIver et al., 2003): The ADH1 terminator and kanMX6 marker cassette were integrated immediately after the brr6 ORF stop codon (IH3412; Bähler et al., 1998). This cassette was switched with a fragment in which ura4+ was inserted downstream of an altered brr6 ORF (IH3587). To create conditional alleles, the second fragment (the 3.7-kb genomic PstI–HindIII fragment from cosmid SPAC8F11) was cloned into the pUC18 to generate pUCBrr6. A BglII site was then introduced 10 nt downstream from the brr6+ ORF into which the BamHI fragment from pREP42PkN (Craven et al., 1998) was inserted to generate pUCBrr6.ura4. Mutagenic PCR using pUCBrr6.ura4 as a template generated the fragment for transformation (MacIver et al., 2003). To generate alleles in which 3Pk or EGFP were inserted at the initiator ATG, the ATG of pUCBrr6 was converted to an NdeI site, creating pUCBrr6-5NdeI. The SacI–PstI fragment of pUCBrr6-5NdeI was inserted into pGEM/T (Promega) in which the NdeI site had been destroyed generating pGEMBrr6-5NdeI. The 3Pk and EGFP NdeI cassettes (Craven et al., 1998) were inserted into the NdeI site of pGEMBrr6-5NdeI. The introduction of a BglII and the BamHI fragment from pREP42PkN as for pUCBrr6.ura4 generated pGEM-PkNBrr6.ura4 and pGEM-EGFPNBrr6.ura4. Fragments spanning from 352 bp upstream of brr6 to 189 bp downstream of ura4+ were transformed into strain IH3412. PCR analysis of strains that switched the kanR marker for ura4+ identified strains bearing the tagged alleles. The SacI–PstI fragment from pGEM-PkNBrr6 was cloned into the pINTA (Petersen et al., 2001) for integration at the leu1 locus to create IH3508.
Antibody production
DNA fragments corresponding to codons of 1–79 (Brr6-Nt) and 253–297 (Brr6-Ct) of Brr6 were cloned into pGEX-6P-1 (GE Healthcare). GST fusion proteins were expressed in Escherichia coli BL21(DE3)RIL and purified on glutathione Sepharose 4B. A mix of GST-Brr6-Nt and GST-Brr6-Ct were used to generate and affinity purify separate rabbit polyclonal antibodies (Scottish Antibody Production Unit).
Light microscopy
Indirect immunofluorescence of S. pombe to stain microtubules (using TAT1; a generous gift from K. Gull, Oxford University, England, UK) and Sad1 has been described previously (Hagan and Yanagida, 1995). For Brr6 and Cdc7.HA staining, cells were processed after fixation in 3.6% formaldehyde for 10 min. A DeltaVision system (Applied Precision) fitted with a 100× 1.45 NA α-plan Fluor objective lens (Carl Zeiss) was used for live cell imaging in conjunction with SoftWorX and Imaris (Bitplane) software. A UApo TIRFM 150×/1.45 NA lens (Olympus) was used with a microscope (Deltavision) for imaging the registry of Nup107.cherry and Brr6.NEGFP signals. The cells were mounted in a Bioptechs chamber. Bioptechs controls regulated the temperature of the chamber and objective while the stage was maintained in a temperature-controlled environmental chamber (Solent Scientific). Fig. 6 B, Fig. 9, Fig. S4 C, and Fig. S5 E show maximal projections of 14 sections, 0.2 µm between the slices. Fig. 7 (A–D) and Videos 1–4 show maximal projections of 14 sections, 0.3 µm between the slices. Images were taken every 3 min.
Electron microscopy
Cells were prepared for electron microscopy using high-pressure freezing and freeze substitution as described previously (Murray, 2008; Tallada et al., 2009). Wild-type brr6+ cells were filtered and fixed from mid-log phase asynchronous cultures. brr6.ts8 cells were synchronized with respect to cell cycle progression by centrifugal elutriation (Mitchison and Carter, 1975) using an elutriator rotor (JE-5.0; Beckman Coulter). After filtration onto 0.45-µm membrane filters (Millipore), cells were loaded into interlocking brass hats (Swiss Precision Instruments) and fixed by high-pressure freezing with a high-pressure freezer (HPM010; Bal-Tec). Freeze substitution into 2% OsO4 + 0.1% uranyl acetate in anhydrous acetone was conducted using an automatic freeze substitution chamber unit (AFS; Leica) at 90°C for 72 h with a 5°C h−1 slope to raise the temperature to −20°C, at which point cells were held for 2 h before a final increase to 4°C for 4 h at a rate of 5°C h−1. After infiltration with Spurr’s resin, blocks were trimmed and serial sections were prepared with a diamond knife, mounted on Formvar/carbon-coated single-slot grids, stained with Reynolds’s solution for 5 min, and imaged on a transmission electron microscope (model 1220; JEOL) at 80 kV. 12 and 15 complete serial thin section images of SPBs were analyzed for brr6.ts8 and wild-type cells, respectively.
Online supplemental material
Fig. S1 shows wild-type controls for anti-tubulin/Sad1 immunofluorescence and electron microscopy alongside Pcp1.RFP/Atb1.GFP images of old pole activation in brr6.ts8. Fig. S2 shows the phenotype arising from complete loss of Brr6 and that brr6.ts8 is a recessive allele. Fig. S3 shows further electron micrographs of brr6.ts8 cells at the restrictive temperature. Fig. S4 shows the genetic interactions between brr6 mutant alleles and strains harboring mutations in SPB components. Fig. S5 shows the characterization of apq12.Δ phenotype and genetic interactions with SPB components and brr6 alleles. Fig. 5 also shows the sensitivity of brr6 alleles and apq12.Δ to reagents that alter membrane fluidity. Video 1 shows the maintenance of nuclear integrity during mitotic commitment of wild-type cells, whereas Video 2 shows that nuclear integrity is transiently breached during mitotic commitment of brr6.ts8 cells. Video 3 shows that the integrity of the nucleus is maintained during anaphase of nda3.KM311 brr6+ cells, whereas Video 4 shows that this integrity is breached, sometimes repeatedly, toward the end of anaphase B/mitotic exit of brr6.ts8 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106076/DC1.
Acknowledgments
We thank Osami Niwa (Kazusa DNA Research Institute, Japan), Valerie Doye (Institut Curie, France), and Katherine Sheldrick for strains; Stephen Murray and Sandra Rutherford for technical assistance; and Steve Bagley, the Paterson advanced imaging facility for technical support, and Keith Gull (Oxford University, UK) for the TAT1 antibody.
This work was supported by Cancer Research UK (CRUK) grant C147/A6058.
Footnotes
Abbreviations used in this paper:
- DIC
- differential interference contrast
- MTOC
- microtubule organizing center
- NPC
- nuclear pore complex
- SIN
- septum initiation network
- SPB
- spindle pole body
- SUN
- Sad1/Unc84
References
- Adams I.R., Kilmartin J.V. 2000. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10:329–335 10.1016/S0962-8924(00)01798-0 [DOI] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007. The molecular architecture of the nuclear pore complex. Nature. 450:695–701 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Araki Y., Lau C.K., Maekawa H., Jaspersen S.L., Giddings T.H., Jr, Schiebel E., Winey M. 2006. The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p. Mol. Biol. Cell. 17:1959–1970 10.1091/mbc.E05-07-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Baï S.W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., Doye V. 2004. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell. Biol. 24:6379–6392 10.1128/MCB.24.14.6379-6392.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N., Muriel W.J., Carr A.M. 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 114:59–66 10.1016/0378-1119(92)90707-V [DOI] [PubMed] [Google Scholar]
- Bridge A.J., Morphew M., Bartlett R., Hagan I.M. 1998. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 12:927–942 10.1101/gad.12.7.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D., Bartlett R., Crawford K., Nurse P. 1991. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature. 349:388–393 10.1038/349388a0 [DOI] [PubMed] [Google Scholar]
- Broughton J., Swennen D., Wilkinson B.M., Joyet P., Gaillardin C., Stirling C.J. 1997. Cloning of SEC61 homologues from Schizosaccharomyces pombe and Yarrowia lipolytica reveals the extent of functional conservation within this core component of the ER translocation machinery. J. Cell Sci. 110:2715–2727 [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial H.J., Rout M.P., Giddings T.H., Winey M. 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143:1789–1800 10.1083/jcb.143.7.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R.A., Griffiths D.J., Sheldrick K.S., Randall R.E., Hagan I.M., Carr A.M. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 221:59–68 10.1016/S0378-1119(98)00434-X [DOI] [PubMed] [Google Scholar]
- de Bruyn Kops A., Guthrie C. 2001. An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J. 20:4183–4193 10.1093/emboj/20.15.4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A., Sanchez-Perez I., Nurse P. 2003. Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 13:399–406 10.1101/gr.636103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., McDonald K.L., McIntosh J.R. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141–151 10.1083/jcb.120.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., West R.R., Morphew D.M., Oakley B.R., McIntosh J.R. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 8:1461–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Tate J., Mistry J., Coggill P.C., Sammut S.J., Hotz H.R., Ceric G., Forslund K., Eddy S.R., Sonnhammer E.L., Bateman A. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–D288 10.1093/nar/gkm960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Rodríguez-Navarro S., Pereira G., Rácz A., Schiebel E., Hurt E. 2004. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 6:840–848 10.1038/ncb1163 [DOI] [PubMed] [Google Scholar]
- Flory M.R., Morphew M.M., Joseph J.D., Means A.R., Davis T.N. 2002. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 13:47–58 [PubMed] [Google Scholar]
- Fujiu K., Numata O. 1999. Localization of microtubules during macronuclear division in Tetrahymena and possible involvement of macronuclear microtubules in ‘amitotic’ chromatin distribution. Cell Struct. Funct. 24:401–404 10.1247/csf.24.401 [DOI] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M. 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121:961–976 10.1083/jcb.121.5.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Y., Meerbrey K., Chong J., Torii Y., Padte N.N., Sazer S. 2009. Nuclear shape, growth and integrity in the closed mitosis of fission yeast depend on the Ran-GTPase system, the spindle pole body and the endoplasmic reticulum. J. Cell Sci. 122:2464–2472 10.1242/jcs.049999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto B., Okazaki K., Niwa O. 2001. Cytoplasmic microtubular system implicated in de novo formation of a Rabl-like orientation of chromosomes in fission yeast. J. Cell Sci. 114:2427–2435 [DOI] [PubMed] [Google Scholar]
- Grallert A., Krapp A., Bagley S., Simanis V., Hagan I.M. 2004. Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 18:1007–1021 10.1101/gad.296204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047 10.1083/jcb.129.4.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M., Riddle P.N., Hyams J.S. 1990. Intramitotic controls in the fission yeast Schizosaccharomyces pombe: the effect of cell size on spindle length and the timing of mitotic events. J. Cell Biol. 110:1617–1621 10.1083/jcb.110.5.1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath I.B. 1980. Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis. Int. Rev. Cytol. 64:1–80 10.1016/S0074-7696(08)60235-1 [DOI] [PubMed] [Google Scholar]
- Hodge C.A., Choudhary V., Wolyniak M.J., Scarcelli J.J., Schneiter R., Cole C.N. 2010. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 123:141–151 10.1242/jcs.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 98:4569–4574 10.1073/pnas.061034498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Giddings T.H., Jr, Winey M. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956 10.1083/jcb.200208169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Martin A.E., Glazko G., Giddings T.H., Jr, Morgan G., Mushegian A., Winey M. 2006. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174:665–675 10.1083/jcb.200601062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe T., Hiraoka Y., Tanaka K., Yanagida M. 1990. The transition of cells of the fission yeast beta-tubulin mutant nda3-311 as seen by freeze-substitution electron microscopy. Requirement of functional tubulin for spindle pole body duplication. J. Cell Sci. 96:275–282 [DOI] [PubMed] [Google Scholar]
- Kilmartin J.V. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162:1211–1221 10.1083/jcb.200307064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Drivas T.G., Blobel G. 2008. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 134:427–438 10.1016/j.cell.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B., O’Toole E., McIntosh J.R., Mellone B., Allshire R., Mengarelli S., Hultenby K., Ekwall K. 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell. 12:2767–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Hottinger H., Munz P., Strauss A., Thuriaux P. 1977. Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics. 87:471–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubai D.F. 1976. The evolution of the mitotic spindle. Int. Rev. Cytol. 43:167–227 10.1016/S0074-7696(08)60069-8 [DOI] [PubMed] [Google Scholar]
- Lo Presti L., Cockell M., Cerutti L., Simanis V., Hauser P.M. 2007. Functional characterization of Pneumocystis carinii brl1 by transspecies complementation analysis. Eukaryot. Cell. 6:2448–2452 10.1128/EC.00321-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver F.H., Glover D.M., Hagan I.M. 2003. A ‘marker switch’ approach for targeted mutagenesis of genes in Schizosaccharomyces pombe. Yeast. 20:587–594 10.1002/yea.983 [DOI] [PubMed] [Google Scholar]
- McCully E.K., Robinow C.F. 1971. Mitosis in the fission yeast Schizosaccharomyces pombe: a comparative study with light and electron microscopy. J. Cell Sci. 9:475–507 [DOI] [PubMed] [Google Scholar]
- Miao M., Ryan K.J., Wente S.R. 2006. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 172:1441–1457 10.1534/genetics.105.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F., Kurabayashi A., Tange Y., Okazaki K., Shimanuki M., Niwa O. 2004. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genomics. 270:449–461 10.1007/s00438-003-0938-8 [DOI] [PubMed] [Google Scholar]
- Mitchison J.M., Carter B.L.A. 1975. Cell cycle analysis. Methods in Cell Biology. Vol. 11 Prescott D.M., editor Academic Press, New York: 201–219 [PubMed] [Google Scholar]
- Moens P.B. 1976. Spindle and kinetochore morphology of Dictyostelium discoideum. J. Cell Biol. 68:113–122 10.1083/jcb.68.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Morgan B.A., Conlon F.L., Manzanares M., Millar J.B.A., Kanuga N., Sharpe J., Krumlauf R., Smith J.C., Sedgwick S.G. 1996. Transposon tools for recombinant DNA manipulation: characterization of transcriptional regulators from yeast, Xenopus, and mouse. Proc. Natl. Acad. Sci. USA. 93:2801–2806 10.1073/pnas.93.7.2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Centeno M.C., McBratney S., Monterrosa A., Byers B., Mann C., Winey M. 1999. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol. Biol. Cell. 10:2393–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S. 2008. High pressure freezing and freeze substitution of Schizosaccharomyces pombe and Saccharomyces cerevisiae for TEM. Methods Cell Biol. 88:3–17 10.1016/S0091-679X(08)00401-9 [DOI] [PubMed] [Google Scholar]
- Niepel M., Strambio-de-Castillia C., Fasolo J., Chait B.T., Rout M.P. 2005. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol. 170:225–235 10.1083/jcb.200504140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. 1975. Genetic control of cell size at cell division in yeast. Nature. 256:547–551 10.1038/256547a0 [DOI] [PubMed] [Google Scholar]
- Ogbadoyi E., Ersfeld K., Robinson D., Sherwin T., Gull K. 2000. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma. 108:501–513 10.1007/s004120050402 [DOI] [PubMed] [Google Scholar]
- Onischenko E., Stanton L.H., Madrid A.S., Kieselbach T., Weis K. 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 185:475–491 10.1083/jcb.200810030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A., Bordes N., Haddad R., Schwartz C.L., Chang F., Bornens M. 2003. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell. 14:2793–2808 10.1091/mbc.E02-10-0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J., Paris J., Willer M., Philippe M., Hagan I.M. 2001. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 114:4371–4384 [DOI] [PubMed] [Google Scholar]
- Poloni D., Simanis V. 2002. A DMSO-sensitive conditional mutant of the fission yeast orthologue of the Saccharomyces cerevisiae SEC13 gene is defective in septation. FEBS Lett. 511:85–89 10.1016/S0014-5793(01)03285-9 [DOI] [PubMed] [Google Scholar]
- Razafsky D., Hodzic D. 2009. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 186:461–472 10.1083/jcb.200906068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y.H., Ogawa K., Nishimoto T. 2005. Brl1p -- a novel nuclear envelope protein required for nuclear transport. Traffic. 6:502–517 10.1111/j.1600-0854.2005.00295.x [DOI] [PubMed] [Google Scholar]
- Schmidt H. 1993. Effective long range mapping in Schizosaccharomyces pombe with the help of swi5. Curr. Genet. 24:271–273 10.1007/BF00351803 [DOI] [PubMed] [Google Scholar]
- Schramm C., Elliott S., Shevchenko A., Schiebel E. 2000. The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 19:421–433 10.1093/emboj/19.3.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezen B., Seedorf M., Schiebel E. 2009. The SESA network links duplication of the yeast centrosome with the protein translation machinery. Genes Dev. 23:1559–1570 10.1101/gad.524209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M., Schmidt S., Hagan I., Simanis V. 1998. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 12:84–94 10.1101/gad.12.1.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallada V.A., Tanaka K., Yanagida M., Hagan I.M. 2009. The S. pombe mitotic regulator Cut12 promotes spindle pole body activation and integration into the nuclear envelope. J. Cell Biol. 185:875–888 10.1083/jcb.200812108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Petersen J., MacIver F.H., Mulvihill D.P., Glover D.M., Hagan I.M. 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 20:1259–1270 10.1093/emboj/20.6.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Umesono K., Hirata A., Yanagida M. 1983. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J. Mol. Biol. 168:251–270 10.1016/S0022-2836(83)80017-5 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 50:917–925 10.1016/0092-8674(87)90518-6 [DOI] [PubMed] [Google Scholar]
- Uzawa S., Li F., Jin Y., McDonald K.L., Braunfeld M.B., Agard D.A., Cande W.Z. 2004. Spindle pole body duplication in fission yeast occurs at the G1/S boundary but maturation is blocked until exit from S by an event downstream of cdc10+. Mol. Biol. Cell. 15:5219–5230 10.1091/mbc.E04-03-0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Vaisberg E.V., Ding R., Nurse P., McIntosh J.R. 1998. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell. 9:2839–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin K.L., Friederichs J.M., Cohen-Fix O., Jaspersen S.L. 2010. Changes in the nuclear envelope environment affect spindle pole body duplication in Saccharomyces cerevisiae. Genetics. 186:867–883 10.1534/genetics.110.119149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., Blobel G., Rout M.P. 1994. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 125:31–42 10.1083/jcb.125.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Sazer S. 2004. Nucleocytoplasmic transport and nuclear envelope integrity in the fission yeast Schizosaccharomyces pombe. Methods. 33:226–238 10.1016/j.ymeth.2003.11.018 [DOI] [PubMed] [Google Scholar]