CIIA mediates the TGF-β–induced activation of SOS1–Rac1 signaling and cell migration.
Abstract
Son of sevenless 1 (SOS1) is a dual guanine nucleotide exchange factor (GEF) that activates the guanosine triphosphatases Rac1 and Ras, which mediate signaling initiated by peptide growth factors. In this paper, we show that CIIA is a new binding partner of SOS1. CIIA promoted the SOS1–Rac1 interaction and inhibited the SOS1–Ras interaction. Furthermore, CIIA promoted the formation of an SOS1–EPS8 complex and SOS1-mediated Rac1 activation, whereas it inhibited SOS1-mediated activation of Ras. Transforming growth factor β (TGF-β) up-regulated the expression of CIIA and thereby promoted the association between CIIA and SOS1 in A549 human lung adenocarcinoma cells. Depletion of CIIA in these cells by ribonucleic acid interference inhibited the TGF-β–induced interaction between SOS1 and EPS8, activation of Rac1, and cell migration. Together, these results suggest that CIIA mediates the TGF-β–induced activation of SOS1–Rac1 signaling and cell migration in A549 cells. They further show that CIIA functions as a molecular switch for the GEF activity of SOS1, directing this activity toward Rac1.
Introduction
Signal transduction initiated by receptor tyrosine kinases (RTKs) plays a pivotal role in the regulation of a variety of cellular functions, including proliferation and migration (Schlessinger, 2000). Ligand-activated RTKs initiate such signaling in part by activating small GTPases, such as Ras and Rac1, a process that is mediated by guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GTPase-bound GDP for GTP (Jaffe and Hall, 2005). Son of sevenless 1 (SOS1) is a dual GEF for Ras and Rac1 (Nimnual et al., 1998; Nimnual and Bar-Sagi, 2002). SOS1 interacts with the adaptor protein Grb2 (Bar-Sagi, 1994). The Grb2–SOS1 complex is recruited to phosphotyrosine residues of ligand-activated RTKs through the SH2 domain of Grb2 (Bar-Sagi, 1994). RTK activation thus results in the translocation of SOS1 to the plasma membrane, where Ras is present, thereby facilitating SOS1-mediated Ras activation. The Ras-specific GEF activity of SOS1 is conferred by the Cdc25 domain in the central region of the protein, which also contains a Ras-binding region designated the Ras exchanger motif (REM; Bar-Sagi, 1994).
The N-terminal region of SOS1 contains a diffuse B cell lymphoma homology (DH) domain and a pleckstrin homology (PH) domain. The DH domain is responsible for Rac1-specific GEF activity of the protein, whereas the PH domain contributes to the recruitment of SOS1 to the plasma membrane (Han et al., 1998; Nimnual et al., 1998; Das et al., 2000). SOS1 forms a complex with EPS8 and E3B1 (also known as Abi1) that mediates Rac1 activation on the basis of its GEF activity (Innocenti et al., 2002). Activated Rac1 promotes actin polymerization in lamellipodia and cell migration (Jaffe and Hall, 2005).
CIIA was initially identified as an antiapoptotic protein (Cho et al., 2003; Kim et al., 2010). It was subsequently found to be identical to mammalian Vsp28, which plays a role in endocytosis. We recently showed that CIIA promotes the epithelial–mesenchymal transition and cell migration (Han et al., 2009). We now show that CIIA is a previously unrecognized binding partner of SOS1. CIIA facilitates the SOS1-dependent activation of Rac1 while concomitantly repressing the SOS1-induced activation of Ras. Our results suggest that CIIA functions as a molecular switch of SOS1, directing its GEF activity toward the Rac1 signaling axis.
Results and discussion
CIIA physically associates with SOS1
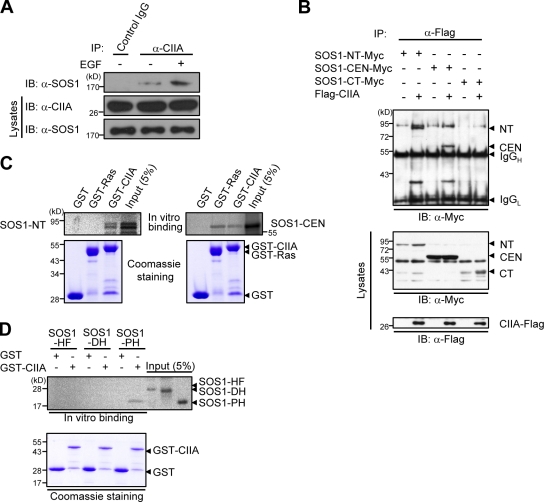
To provide further insight into the cellular function of CIIA, we searched for a CIIA-interacting protein by using a GST pull-down assay. We detected one candidate protein (170 kD), which mass spectrometric analysis identified as SOS1 (Fig. S1 A). We confirmed the physical association between CIIA and SOS1 in HeLa cells by coimmunoprecipitation (Fig. 1 A). The extent of this association was increased by EGF treatment.
Figure 1.
CIIA physically interacts with SOS1. (A) HeLa cells were deprived of serum for 16 h, incubated without or with 100 ng/ml EGF for 5 min, lysed, and subjected to immunoprecipitation (IP) with the anti-CIIA antibody or with control preimmune IgG. The resulting precipitates as well as the lysates were subjected to immunoblotting (IB) with antibodies to SOS1 or CIIA. (B) 293T cells were transfected for 48 h with various combinations of expression vectors for the indicated proteins. Cell lysates were immunoprecipitated with the anti-Flag antibody, and the resulting precipitates were examined by immunoblot analysis with the anti-Myc antibody. Cell lysates were also immunoblotted with antibodies to Flag or to Myc. IgGH and IgGL indicate heavy and light chains, respectively, of IgG. (C and D) In vitro–translated 35S-labeled SOS1 variants were pulled down with GST or GST-fused proteins immobilized on glutathione–agarose beads. The bead-bound 35S-labeled proteins were analyzed by SDS-PAGE and autoradiography. The gels were also stained with Coomassie brilliant blue. A fraction (5%) of the 35S-labeled protein input to the binding reaction is also shown. NT, N-terminal domain; CEN, central domain; CT, C-terminal domain.
We next examined which region of SOS1 is responsible for its association with CIIA. SOS1 is a multidomain protein that includes the DH, PH, REM, Cdc25, and proline-rich domains (Fig. S1 B; Margarit et al., 2003; Sondermann et al., 2004). The DH and PH domains contribute to the activation of Rac1 (Nimnual et al., 1998; Soisson et al., 1998; Nimnual and Bar-Sagi, 2002), whereas the REM and Cdc25 domains are required for Ras-specific GEF activity (Boriack-Sjodin et al., 1998). We transfected 293T cells with a vector encoding Flag epitope–tagged CIIA together with a vector for Myc epitope–tagged various fragments of SOS1 (N-terminal [SOS1-NT], central [SOS1-CEN], or C-terminal [SOS1-CT] fragments). SOS1-NT contains the DH and PH domains, SOS1-CEN contains the REM and Cdc25 domains, and SOS1-CT contains the proline-rich domain that includes the binding sites for Grb2 and E3B1 (Fig. S1 B). Coimmunoprecipitation analysis revealed that CIIA-Flag physically associated with SOS1-NT and SOS1-CEN as well as SOS1 but not with SOS1-CT (Fig. 1 B). In vitro binding analysis also revealed that a GST fusion protein of CIIA bound to both 35S-labeled SOS1-NT and SOS1-CEN (Fig. 1 C). We also confirmed that GST-Ras bound to SOS1-CEN but not to SOS1-NT. Further in vitro binding analysis revealed that the PH domain of SOS1-NT was responsible for the interaction with CIIA (Fig. 1 D). A separate binding experiment using recombinant proteins confirmed the direct binding of GST-CIIA to SOS1-NT and SOS1-CEN but not to SOS1-CT (Fig. S1 C).
CIIA inhibits the SOS1–Ras–MAPK signaling axis
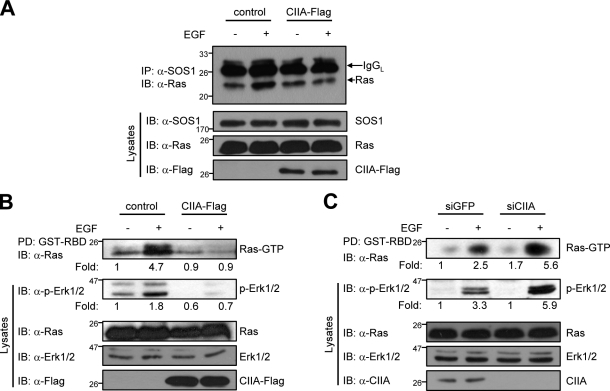
Given that CIIA bound to SOS1-CEN, which includes the binding region for Ras (Boriack-Sjodin et al., 1998), we investigated whether CIIA might affect the association between SOS1 and Ras in MDCK cells stably expressing CIIA-Flag (MDCK/CIIA-Flag cells) or MDCK/control cells. Coimmunoprecipitation analysis revealed that EGF stimulation increased the interaction between SOS1 and Ras in MDCK/control cells but not in MDCK/CIIA-Flag cells (Fig. 2 A), suggesting that CIIA indeed inhibits the EGF-induced association of SOS1 and Ras.
Figure 2.
CIIA regulates EGF-induced Ras and Erk activation. (A) MDCK/control or MDCK/CIIA-Flag cells were deprived of serum for 16 h and incubated for 5 min with or without 100 ng/ml EGF. Cell lysates were immunoprecipitated (IP) with anti-SOS1 antibody, and the pellets were immunoblotted (IB) with anti-Ras antibody. Cell lysates were also examined directly by immunoblotting with antibodies to SOS1, Ras, or Flag. (B and C) MDCK/CIIA-Flag and MDCK/control cells (B) or HeLa cells stably expressing either GFP (control) or CIIA siRNA (C) were deprived of serum for 16 h and then incubated for 5 min with or without 100 ng/ml EGF. Cell lysates were subjected to pull-down (PD) with GST-RBD and assayed for Ras activity.
SOS1 mediates the EGF-induced activation of Ras, which stimulates the Erk1/2 pathway (Buday and Downward, 1993; Schlessinger, 2000). We therefore investigated the effect of CIIA on the EGF-induced activation of Ras and Erk1/2. Activated Ras (Ras-GTP) was detected on the basis of its ability to bind to a GST fusion protein containing the Ras-GTP binding domain (RBD) of Raf1. EGF induced the activation of Ras in MDCK/control cells but not in MDCK/CIIA-Flag cells (Fig. 2 B). Forced expression of CIIA-Flag also inhibited the activation of Ras by SOS1-CEN–Myc (Fig. S2 A). The EGF-induced activation of Erk1/2 was also apparent in MDCK/control cells but not in MDCK/CIIA-Flag cells (Fig. 2 B). To investigate the role of endogenous CIIA in the activation of Ras and Erk1/2 signaling, we established HeLa cells stably expressing siRNA for CIIA or GFP (control). RNAi-mediated depletion of CIIA potentiated the EGF-induced activation of both Ras and Erk1/2 compared with that apparent in cells expressing GFP siRNA (Fig. 2 C). Similar results were obtained in experiments with a second CIIA siRNA whose nucleotide sequence did not overlap with that of the first (Fig. S2 B). Together, these results suggested that CIIA inhibits the EGF-induced SOS1–Ras–Erk1/2 signaling axis.
The central region of SOS1 comprises the REM and Cdc25 domains. The Cdc25 domain is responsible for the Ras-GEF catalytic activity of SOS1, whereas the REM domain positively regulates this activity of Cdc25 through an allosteric mechanism (Margarit et al., 2003). The binding of CIIA to the central region of SOS1 may thus be key to the molecular mechanism by which CIIA inhibits the interaction between SOS1 and Ras as well as the SOS1-mediated activation of Ras. In this regard, it is noteworthy that CIIA inhibited the GEF activity of SOS1-CEN on Ras in vitro (Fig. S2 C). Binding of CIIA to the PH domain of SOS1 may also contribute to the inhibition of SOS1-mediated Ras activation by CIIA, given that this domain has been shown to modulate the Ras-GEF activity of SOS1 (Qian et al., 1998; Zhao et al., 2007). The importance of the PH domain for the Ras-GEF activity of SOS1 has been also suggested by the findings that mutations in the PH domain are responsible for Noonan syndrome (Roberts et al., 2006; Tartaglia et al., 2006).
CIIA promotes SOS1-mediated Rac1 activation
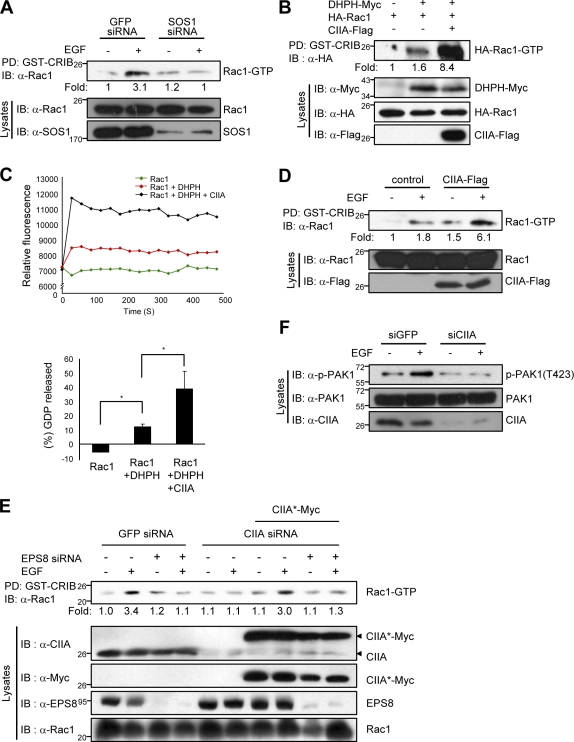
SOS1 physically interacts with Rac1 through its N-terminal region containing the DH and PH domains, resulting in Rac1 activation (Nimnual et al., 1998). RNAi-mediated depletion of SOS1 suppressed the EGF-induced activation of Rac1 in HeLa cells (Fig. 3 A), showing that SOS1 mediates Rac1 activation by this growth factor. Rac1 activation was detected by the binding of GTP-bound (active) Rac1 to a GST fusion protein containing the Cdc42 and Rac1 interactive binding (CRIB) domain of PAK1 (p21-activated kinase 1; Manser et al., 1994). Given that CIIA physically associated with the PH domain of SOS1 (Fig. 1 D), we examined the effect of CIIA on Rac1 activation induced by SOS1-DHPH, which possesses the Rac1-specific GEF activity. Transfection experiments with 293T cells showed that CIIA potentiated the binding between Rac1 and SOS1-DHPH (Fig. S3 A) and the activation of Rac1 by SOS1-DHPH (Fig. 3 B). Furthermore, in a Rac1-GEF assay, the fluorescence intensity increased when SOS1-DHPH was added to GST-Rac1, and this increase was further enhanced by CIIA (Fig. 3 C), indicating that CIIA increased the GEF activity of SOS1-DHPH on Rac1. Next, we examined the actions of CIIA variants on EGF-induced Rac1 activation in 239T cells that had been transfected with vectors for HA-SOS1 and Flag-tagged CIIA variants (CIIA, CIIA-ΔC, and CIIA-ΔN). CIIA-ΔC and CIIA-ΔN are the deletion mutants of CIIA containing amino acids 1–174 and 79–221, respectively. CIIA-ΔC, but not CIIA-ΔN, associated with SOS1 in the coimmunoprecipitation analysis (Fig. S3 B). CIIA and CIIA-ΔC potentiated the EGF-induced stimulation of Rac1 activity, whereas CIIA-ΔN did not (Fig. S3 C). Additionally, CIIA and CIIA-ΔC, but not CIIA-ΔN, inhibited EGF-stimulated Ras activation (Fig. S3 C). We also showed that the extent of EGF-induced Rac1 activation in MDCK/CIIA-Flag cells was markedly greater than that in MDCK/control cells (Fig. 3 D). Consistent with these results, RNAi-mediated depletion of CIIA blocked EGF-induced Rac1 activation in HeLa cells, and this effect was reversed by ectopic expression of CIIA*-Myc (Fig. 3 E), which contains three silent point mutations within the region targeted by the CIIA siRNA. This Rac1 activation by CIIA*-Myc in the cells was abrogated by RNAi-mediated depletion of EPS8, suggesting that a positive effect of CIIA on EGF-induced Rac1 activation requires EPS8. Moreover, immunoblot analysis with antibodies to phospho(Thr423)-PAK1 revealed that the RNAi-mediated knockdown of CIIA inhibited the EGF-induced activation of PAK1 (Fig. 3 F), a downstream target of Rac1 (Bokoch, 2003). Collectively, these results suggested that CIIA promoted the activation of Rac1 by EGF.
Figure 3.
CIIA promotes EGF-induced SOS1–Rac1 signaling. (A) HeLa cells transfected with GFP or SOS1 siRNA for 12 h were deprived of serum for 16 h and then incubated for 5 min with or without 100 ng/ml EGF. Cell lysates were pulled down (PD) with GST-CRIB and assayed for Rac1 activity. (B) 293T cells were transfected for 48 h with vectors for the indicated proteins, after which cell lysates were subjected to a pull-down assay with GST-CRIB as in A. (C) The GEF activity of SOS1-DHPH on Rac1 was determined in vitro in the absence or presence of CIIA by fluorescence spectroscopy with the use of the GEF exchange assay kit as described in Materials and methods. (top) Relative fluorescence intensity data from one representative experiment. (bottom) Data are the means ± SD of three independent experiments. Results are expressed as the mant-GTP incorporation into GST-Rac1 after 480 s relative to time 0 after adding His6-SOS1-DHPH. *, P < 0.05. (D) MDCK/CIIA-Flag and MDCK/control cells were deprived of serum for 16 h and incubated for 5 min with or without 100 ng/ml EGF. Cell lysates were subjected to Rac1 activity assay as in A. (E) HeLa cells were transfected for 24 h with the indicated combinations of GFP, CIIA, or E3B1 siRNA and a vector encoding CIIA*-Myc. Then, the cells were deprived of serum for 16 h and incubated for 5 min in the presence or absence of 100 ng/ml EGF. Cell lysates were assayed for Rac1 activity as in A. (F) HeLa-siGFP or HeLa-siCIIA cells were deprived of serum for 16 h and then incubated for 5 min with or without 100 ng/ml EGF. Cell lysates were examined by immunoblotting (IB) with antibodies to phospho(Thr423)-PAK1, PAK1, or CIIA.
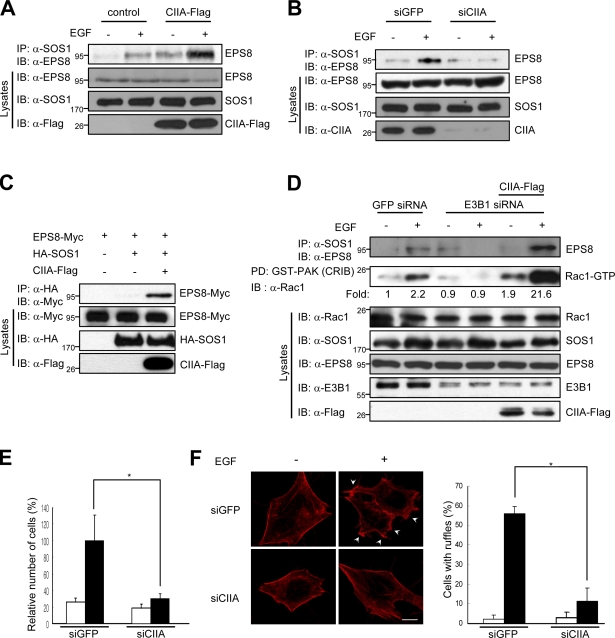
We next examined whether CIIA might affect the physical association between SOS1 and EPS8. EGF induced the association of SOS1 and of EPS8 in MDCK/control cells, and this effect was enhanced in MDCK/CIIA-Flag cells (Fig. 4 A). Conversely, silencing of CIIA inhibited the EGF-induced interaction between SOS1 and EPS8 in HeLa cells (Fig. 4 B). These results thus suggested that CIIA promotes the interaction between SOS1 and EPS8. Given that E3B1 is required for the interaction between SOS1 and EPS8 in formation of the SOS1–EPS8–E3B1 tripartite complex (Scita et al., 1999), we examined whether CIIA exerts an E3B1-like function as a scaffold for SOS1 and EPS8. Indeed, CIIA physically associated with EPS8, but not with E3B1, in transfected 293T cells (Fig. S3 D). Furthermore, coimmunoprecipitation analysis of transfected 293T cells revealed a physical interaction between HA-SOS1 and EPS8-Myc only in cells coexpressing CIIA-Flag (Fig. 4 C). In addition, RNAi-mediated depletion of E3B1 abolished the EGF-induced binding between SOS1 and EPS8 as well as Rac1 activation in HeLa cells, whereas ectopic expression of CIIA in the E3B1-depleted cells restored these effects of EGF (Fig. 4 D). These results suggested that CIIA facilitates the interaction between SOS1 and EPS8 as well as SOS1-mediated Rac1 activation in a manner independent of E3B1. CIIA thus appears to function as a scaffold that supports the interaction between SOS1 and EPS8. This scaffold function might be an integral component of the mechanism by which CIIA promotes the SOS1-mediated activation of Rac1.
Figure 4.
CIIA promotes SOS1-mediated activation of Rac1. (A and B) MDCK/CIIA-Flag and MDCK/control cells (A) or HeLa cells expressing GFP (control) or CIIA siRNA (B) were deprived of serum for 16 h, incubated for 5 min with or without 100 ng/ml EGF, and lysed. Cell lysates were immunoprecipitated (IP) with the anti-SOS1 antibody, and the resulting precipitates were immunoblotted (IB) with the anti-EPS8 antibody. (C) 293T cells were transfected for 48 h with vectors for EPS8-Myc, HA-SOS1, and CIIA-Flag as indicated. Cell lysates were immunoprecipitated with the anti-HA antibody, and the precipitates were immunoblotted with the anti-Myc antibody. (D) HeLa cells were transfected with either GFP or E3B1 siRNA alone or together with a vector encoding CIIA-Flag. After 8 h of transfection, the cells were deprived of serum for 16 h and then incubated for 5 min with or without 100 ng/ml EGF. Cell lysates were immunoprecipitated with the anti-SOS1 antibody, and the resulting precipitates were immunoblotted with the anti-EPS8 antibody. Cell lysates were also subjected to pull-down (PD) with GST-CRIB and assayed for Rac1 activity. (E) HeLa cells stably expressing GFP or CIIA siRNA were deprived of serum for 16 h, transferred to the upper chambers of a Transwell plate, incubated for 24 h with (black bars) or without (white bars) 100 ng/ml EGF in the lower chambers, and analyzed for migration. (F) HeLa cells stably expressing GFP or CIIA siRNA were serum starved for 16 h, incubated for 5 min with (black bars) or without (white bars) 100 ng/ml EGF, and stained with Alexa Fluor red–conjugated phalloidin to detect F-actin. Arrowheads indicate F-actin–enriched membrane ruffles. The number of cells with ruffles were counted and expressed as the percentages relative to the total number of cells. Data are means ± SD from three independent experiments. *, P < 0.01. Bar, 10 µm.
CIIA promotes SOS1- and Rac1-dependent cell migration
Rac1 mediates EGF-induced cell migration (Russell et al., 2003; Katso et al., 2006; Yamaguchi et al., 2006). Indeed, a dominant-negative Rac1 mutant (Rac1N17) markedly inhibited EGF-induced migration of HeLa cells (Fig. S3 E). RNAi-mediated knockdown of SOS1 also blocked EGF-induced HeLa cell migration (Fig. S3 F). These results suggested that SOS1–Rac1 signaling mediates EGF-induced migration of HeLa cells. Given that CIIA facilitated SOS1-mediated Rac1 activation (Fig. 4), we investigated the possible effect of CIIA on EGF-induced HeLa cell migration. The stimulatory effect of EGF on cell migration was markedly attenuated in cells expressing CIIA siRNA (Fig. 4 E), suggesting that CIIA facilitates the migration of HeLa cells dependent on the EGF–SOS1–Rac1 signaling axis. It is also noteworthy that depletion of CIIA by RNAi abrogated the EGF-induced formation of actin-based membrane ruffles (Fig. 4 F).
CIIA mediates TGF-β–induced cell migration
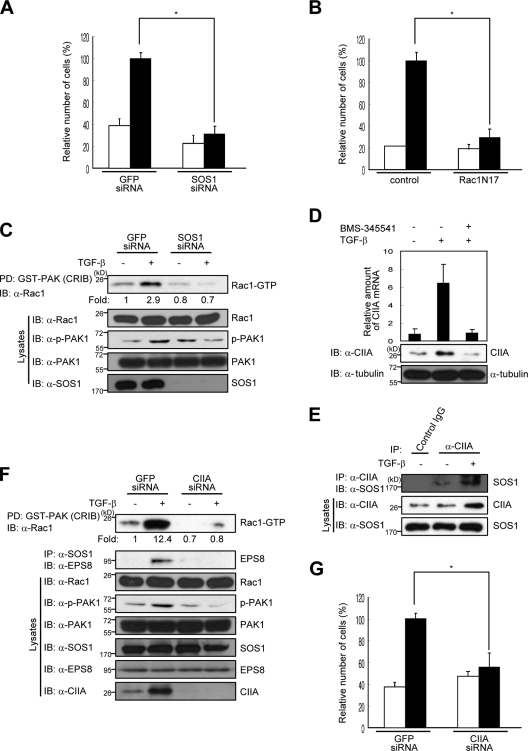
TGF-β induces cell motility in various types of tumor cells (Massagué, 2008). Rac1 contributes in a manner that is independent of Smad signaling to the mechanism by which TGF-β induces such cell migration (Ng, 2008; Zhang, 2009). Indeed, TGF-β promoted the migration of A549 human lung adenocarcinoma cells, and this effect was blocked either by RNAi-mediated depletion of SOS1 (Fig. 5 A) or by expression of the dominant-negative Rac1 mutant Rac1N17 (Fig. 5 B), suggesting that SOS1-Rac1 signaling mediates the stimulatory effect of TGF-β on A549 cell migration. Furthermore, TGF-β induced activation of Rac1 and PAK1 in A549 cells transfected with a control siRNA but not in those transfected with SOS1 siRNA (Fig. 5 C). TGF-β increased the expression of CIIA at both the mRNA and protein levels in A549 cells, and this effect was blocked by the IκB kinase inhibitor BMS-345541 (Fig. 5 D), suggesting that TGF-β induces the expression of CIIA through the NF-kB signaling pathway. TGF-β also increased the interaction between CIIA and SOS1 in these cells (Fig. 5 E). We therefore examined the possible role of CIIA in TGF-β–induced SOS1-Rac1 signaling and cell migration in A549 cells. RNAi-mediated depletion of CIIA inhibited the TGF-β–induced association between SOS1 and EPS8, activation of Rac1, phosphorylation of PAK1 (Fig. 5 F), and cell migration (Fig. 5 G). Together, these results suggested that CIIA mediates SOS1-dependent Rac1 activation initiated by TGF-β and that the CIIA–SOS1–Rac1 signaling axis is important for TGF-β–induced cell migration. Given that TGF-β is implicated in the migration and invasion of tumorigenic cells (Welch et al., 1990; Breuhahn et al., 2006; Jakowlew, 2006; Fransvea et al., 2008), clarification of the relations among TGF-β, SOS1, and CIIA in tumors may provide insight into the molecular mechanism of tumor progression.
Figure 5.
CIIA mediates TGF-β–induced migration of A549 cells. (A, B, and G) A549 cells were transfected either with GFP (control), SOS1 (A), or CIIA (G) siRNA oligonucleotides or with a vector for Rac1N17 or the corresponding empty vector (B). After 8 h of transfection, the cells were deprived of serum for 16 h, transferred to the upper chambers of a Transwell plate, incubated for 48 h with (black bars) or without (white bars) 2 ng/ml TGF-β in the lower chambers, and analyzed for migration. Data are means ± SD from three independent experiments. *, P < 0.01. (C) A549 cells were transfected with GFP or SOS1 siRNA for 16 h, deprived of serum for 10 h, and incubated with or without 2 ng/ml TGF-β for 16 h. Cell lysates were then subjected to pull-down (PD) with GST-CRIB and assayed for Rac1 activity. (D) A549 cells were incubated with or without 10 µM BMS345541 for 1 h and then in the presence or absence of 2 ng/ml TGF-β for 6 h (for quantitative RT-PCR analysis of CIIA mRNA) or for 16 h (for immunoblot [IB] with antibodies to CIIA or α-tubulin [loading control]). RT-PCR data are means ± SD of triplicates from a representative experiment. (E) A549 cells were left untreated or treated with 2 ng/ml TGF-β for 16 h, lysed, and immunoprecipitated (IP) with the anti-CIIA antibody or rabbit preimmune IgG. The resulting precipitates were immunoblotted with the anti-SOS1 antibody. (F) A549 cells were transfected for 12 h with GFP or CIIA siRNA oligonucleotides, deprived of serum for 16 h, and incubated with or without 2 ng/ml TGF-β for 16 h. Cell lysates were then subjected to Rac1 activity assay as in C. Cell lysates were also immunoprecipitated with the anti-SOS1 antibody, and the resulting precipitates were immunoblotted with the anti-EPS8 antibody.
Materials and methods
DNA constructs and antibodies
The cDNAs of human SOS1 variants, including the full-length protein, SOS1-NT (amino acid residues 1–550), histone-fold domain of SOS1 (SOS1-HF; residues 1–198), SOS1-DH (residues 199–404), SOS1-PH (residues 442–550), SOS1-DHPH (residues 199–550), SOS1-CEN (residues 551–1,050), and SOS1-CT (residues 1,051–1,333), were generated by PCR with pMT2-HA-hSOS1 (provided by P.P. Di Fiore, University of Milan, Milan, Italy) as a template and were inserted into pcDNA6-His-Myc-A vector (Invitrogen). Mouse CIIA cDNA was inserted into pcDNA6-Myc-A or pcDNA3-puro for expression of Myc epitope–tagged or Flag-tagged proteins, respectively. An expression vector for Myc-tagged Rac1N17 was provided by A. Hall (Memorial Sloan-Kettering Cancer Center, New York, NY). A bacterial expression vector encoding a GST fusion protein of CRIB was constructed by subcloning cDNA corresponding to the CRIB region of rat α-PAK1 into pGEX-4T2. A pGEX-2T vector encoding GST-fused RBD of Raf1 was provided by C. Herrmann (Ruhr University Bochum, Bochum, Germany). Vectors encoding Myc-tagged human E3B1 (pcDNA3-E3B1-Myc) and Myc-tagged human EPS8 (pCR-Myc-EPS8) were provided by D. Park (Seoul National University, Seoul, Korea) and G. Scita (Universitá degli Studi di Milano, Milan, Italy), respectively.
Rabbit polyclonal antibodies to CIIA, SOS1, and EPS8 were obtained from Santa Cruz Biotechnology, Inc. Rabbit polyclonal antibodies to CIIA were purchased from LabFrontier, and those to E3B1 (Abi1) were purchased from Sigma-Aldrich. Mouse monoclonal antibodies to Ras and Rac1 were obtained from BD, and those to the HA epitope and Flag were obtained from Sigma-Aldrich. Rabbit polyclonal antibodies to Erk1/2, phosphorylated Erk1/2, PAK1, and phosphorylated PAK1 as well as mouse monoclonal antibodies to the Myc epitope were obtained from Cell Signaling Technology.
Cell culture and DNA transfection
Human embryonic kidney 293T, MDCK, HeLa, and A549 cells were routinely maintained under a humidified atmosphere of 5% CO2 at 37°C in DME (Hyclone) supplemented with 10% heat-inactivated FBS. Cultured cells were transfected with expression vectors either by the calcium phosphate method or with the use of Lipofectamine (Invitrogen). For stable transfection of MDCK cells, the cells were transfected with pcDNA3-puro-CIIA-Flag or with the empty vector (pcDNA3-puro), and puromycin-resistant cells were selected in complete medium containing 1.7 µg/ml puromycin. MDCK cells stably expressing CIIA-Flag and the corresponding control cells were named as MDCK/CIIA-Flag and MDCK/control cells, respectively.
In vitro binding assay
35S-labeled SOS1 and deletion mutants thereof were produced in vitro with the use of the reticulocyte lysate system (TnT; Promega). The 35S-labeled proteins were incubated at 4°C for 2 h in a binding buffer (Cho et al., 2003) with GST alone or GST fusion proteins immobilized on glutathione–agarose beads. The beads were then washed, and bead-bound 35S-labeled proteins were separated by SDS-PAGE and analyzed with a phosphoimager (BAS-2500; Fujifilm).
Coimmunoprecipitation and immunoblot analysis
Cell lysates were incubated at 4°C first overnight with the appropriate antibodies and then for an additional 1 h with protein G–agarose beads (GE Healthcare). The beads were washed three times with cell lysis buffer (Cho et al., 2003), and bead-bound proteins were subjected to SDS-PAGE followed by immunoblot analysis. For immunoblot analysis, proteins were transferred electrophoretically from the SDS-PAGE gel to a polyvinylidene fluoride membrane, which was then incubated for 1 h at room temperature with 5% nonfat dry milk to block nonspecific sites. The membrane was probed with primary antibodies, washed three times, and incubated with horseradish peroxidase–conjugated secondary antibodies. Immune complexes were visualized with the use of an enhanced chemiluminescence kit (SuperSignal West Pico; Thermo Fisher Scientific).
Assay for Ras and Rac1 activities
Activation of Ras and Rac1 was measured on the basis of the ability of the activated proteins to bind GST-RBD and GST-CRIB, respectively. GST-RBD is a GST fusion protein containing a fragment of Raf that includes the binding domain for Ras-GTP (de Rooij and Bos, 1997), whereas GST-CRIB is a GST fusion protein containing a fragment of α-PAK1 that includes the CRIB region, which specifically binds GTP-bound Rac1 (Manser et al., 1994). Cell lysates were centrifuged at 12,000 g for 15 min at 4°C, and the resulting soluble fraction was incubated for 2 h at 4°C with GST-RBD or GST-CRIB immobilized on glutathione–agarose beads. The bead-bound proteins were collected by centrifugation, and Ras-GTP or Rac1-GTP was detected by immunoblot analysis with antibodies to Ras or to Rac1, respectively. The intensities of bands corresponding to Ras-GTP or Rac1-GTP were determined by densitometry and expressed as fold increases as indicated.
RNAi
HeLa cells were stably transfected with pSuper-retro vector (Oligoengine) encoding either an siRNA specific for human CIIA mRNA or a control (GFP) siRNA (Hamar et al., 2004) and named HeLa-siCIIA or HeLa-siGFP cells, respectively. Stable transfectants were selected in the presence of 0.2 µg/ml puromycin. The target sequences for the CIIA and GFP siRNAs were 5′-CCTGGGAACAAGCCGGAGCTGTATGAGGA-3′ and 5′-GCTGGAGTACAACTACAACAGCCACAACG-3′, respectively. Where indicated, siRNA oligonucleotides for GFP, CIIA, SOS1 (Boykevisch et al., 2006), or E3B1 (Innocenti et al., 2004) mRNAs were obtained from Invitrogen and introduced into cells by transfection (electroporation) with the use of a microporator (Neon Transfection System; Invitrogen). The sequences of the siRNA oligonucleotides for human SOS1 and E3B1 were 5′-GACAGTGTTGTAATGAATT-3′ and 5′-ATGCATTGGCCAACAATGT-3′, respectively. CIIA*-Myc is a Myc epitope-tagged full-length version of CIIA that is resistant to the CIIA siRNA as a result of three silent point mutations (AAG → AAA, GAG → GAA, and TAT → TAC) within the region targeted by the siRNA.
Quantitative real-time RT-PCR
1 µg total RNA isolated from A549 cells with the use of a purification kit (RNeasy Mini; QIAGEN) was subjected to reverse transcription in a final volume of 20 µl with an oligo(dT) primer and Moloney murine leukemia virus reverse transcription (Invitrogen) as described previously (Wang et al., 2002). Real-time PCR was performed with 5 µl of a 1:5 dilution of the resulting cDNA, 10 µM of each primer, and 10 µl of 2× SYBR Green Supermix (iQ; Bio-Rad Laboratories) in a final volume of 20 µl. The reaction was performed in a multicolor real-time PCR detection system (iQ5; Bio-Rad Laboratories), and data were analyzed with iQ5 optical system software (Bio-Rad Laboratories). The PCR primers (forward and reverse, respectively) were 5′-CAACCTCAACCGCTGCAT-3′ and 5′-ATGGTCTCCATCAGCTCTCG-3′ for human CIIA and 5′-GAGTCCACTGGCGTCTTCAC-3′ and 5′-GTTCACACCCATGACGAACA-3′ for glyceraldehyde-3-phosphate dehydrogenase. Reactions were performed in triplicate, and the amount of CIIA mRNA was normalized by the corresponding amount of glyceraldehyde-3-phosphate dehydrogenase.
Phalloidin staining
HeLa-siGFP or HeLa-siCIIA cells seeded on glass coverslips were deprived of serum for 16 h, treated with 100 ng/ml EGF for 5 min where indicated, fixed with 3.0% formaldehyde, permeabilized in 0.1% Triton X-100, and then stained with Alexa Fluor red–conjugated phalloidin. Coverslips were mounted on a microscope slide with gold antifade reagent containing DAPI (ProLong; Invitrogen). F-actin images in the cells were acquired at room temperature with the use of a confocal microscope (LSM 510 Meta; Carl Zeiss) with C-Aprochromat 40×/1.2 W corr objective lens (Carl Zeiss) and a 2× zoom in magnification. Data were acquired with LSM Image Examiner software (Carl Zeiss) and imported into Photoshop (Adobe).
In vitro GEF assay
Recombinant proteins of His6-SOS1-DHPH, His6-SOS1-CEN, His6-CIIA, GST-Rac1, and GST-Ras were expressed in and purified from Escherichia coli. In vitro GEF assays were performed with the use of a GEF exchange assay (Biochem Kit; Cytoskeleton) according to the manufacturer’s protocol. In brief, N-methylanthraniloyl (mant)–GTP incorporation into GST-Rac1 or GST-Ras was analyzed by fluorescence spectroscopy with the use of a multilabel plate reader (fluorescence/luminescence; Victor3; PerkinElmer). Either 0.2 µg GST-Rac1 or 0.2 µg GST-Ras was added without or with 0.2 µg His6-CIIA in 15 µl reaction buffer containing 20 mM Tris, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 50 µg/ml BSA, and 0.75 µM mant-GTP. The exchange reaction mixtures were placed into a 384-well black-colored plate (Corning), and the fluorescence of each sample was measured at 20°C every 30 s with excitation and emission wavelengths of 360 and 440 nm, respectively. After 150 s, the exchange reaction was initiated by adding His6-SOS1-DHPH or His6-SOS1-CEN into the Rac1 or Ras reaction mixtures, respectively, and the relative mant fluorescence was monitored for 55 readings.
Cell migration assay
For Transwell migration assays, 104 cells were deprived of serum for 16 h and then added in 0.2 ml culture medium containing 0.1% (instead of 10%) FBS to the upper chambers of a 24-well Transwell plate (pore size of 8 µm; Corning). The lower chambers of the plate were loaded with 0.6 ml of the same medium. Where indicated, 100 ng/ml EGF or 2 ng/ml TGF-β was added to the lower chambers. After incubation with EGF for 24 h or with TGF-β for 48 h, cells on the upper surface of each membrane were removed by scraping with a cotton swab. Motile cells on the underside of the membrane were fixed and stained with a stain kit (Diff Quik; Sysmex). For each experiment, the stained cells in three random fields on the underside of the membrane were quantified by light microscopy (40× objective). Data are means ± SD of values from three independent experiments.
Statistical analysis
Data are presented as means ± SD and were analyzed by analysis of variance. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the identification of SOS1 as a CIIA-binding protein. Fig. S2 shows that CIIA inhibits the SOS1-mediated activation of Ras. Fig. S3 shows that CIIA promotes the binding of Rac1 to SOS1-DHPH and that a dominant-negative Rac1 mutant Rac1N17 inhibits EGF-induced migration of HeLa cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106138/DC1.
Acknowledgments
We thank P.P. Di Fiore, A. Hall, C. Herrmann, G. Scita, and Dongeun Park for providing HA-SOS1, Myc-Rac1N17, GST-RBD, EPS8-Myc, and E3B1-Myc cDNA clones, respectively.
This work was supported by the National Research Foundation grant 2006-0093855 and National Research Foundation grants 20090081488 and 2010-0001197 funded by the Ministry of Education, Science and Technology of South Korea (to E.-J. Choi).
Footnotes
Abbreviations used in this paper:
- CRIB
- Cdc42 and Rac1 interactive binding
- DH
- diffuse B cell lymphoma homology
- GEF
- guanine nucleotide exchange factor
- mant
- N-methylanthraniloyl
- PH
- pleckstrin homology
- RBD
- Ras-GTP binding domain
- REM
- Ras exchanger motif
- RTK
- receptor tyrosine kinase
- SOS1
- son of sevenless 1
References
- Bar-Sagi D. 1994. The Sos (Son of sevenless) protein. Trends Endocrinol. Metab. 5:165–169 10.1016/1043-2760(94)90014-0 [DOI] [PubMed] [Google Scholar]
- Bokoch G.M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72:743–781 10.1146/annurev.biochem.72.121801.161742 [DOI] [PubMed] [Google Scholar]
- Boriack-Sjodin P.A., Margarit S.M., Bar-Sagi D., Kuriyan J. 1998. The structural basis of the activation of Ras by Sos. Nature. 394:337–343 10.1038/28548 [DOI] [PubMed] [Google Scholar]
- Boykevisch S., Zhao C., Sondermann H., Philippidou P., Halegoua S., Kuriyan J., Bar-Sagi D. 2006. Regulation of ras signaling dynamics by Sos-mediated positive feedback. Curr. Biol. 16:2173–2179 10.1016/j.cub.2006.09.033 [DOI] [PubMed] [Google Scholar]
- Breuhahn K., Longerich T., Schirmacher P. 2006. Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene. 25:3787–3800 10.1038/sj.onc.1209556 [DOI] [PubMed] [Google Scholar]
- Buday L., Downward J. 1993. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 73:611–620 10.1016/0092-8674(93)90146-H [DOI] [PubMed] [Google Scholar]
- Cho S.G., Kim J.W., Lee Y.H., Hwang H.S., Kim M.S., Ryoo K., Kim M.J., Noh K.T., Kim E.K., Cho J.H., et al. 2003. Identification of a novel antiapoptotic protein that antagonizes ASK1 and CAD activities. J. Cell Biol. 163:71–81 10.1083/jcb.200303003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B., Shu X., Day G.J., Han J., Krishna U.M., Falck J.R., Broek D. 2000. Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sos1 regulates Rac binding. J. Biol. Chem. 275:15074–15081 10.1074/jbc.M907269199 [DOI] [PubMed] [Google Scholar]
- de Rooij J., Bos J.L. 1997. Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene. 14:623–625 10.1038/sj.onc.1201005 [DOI] [PubMed] [Google Scholar]
- Fransvea E., Angelotti U., Antonaci S., Giannelli G. 2008. Blocking transforming growth factor-beta up-regulates E-cadherin and reduces migration and invasion of hepatocellular carcinoma cells. Hepatology. 47:1557–1566 10.1002/hep.22201 [DOI] [PubMed] [Google Scholar]
- Hamar P., Song E., Kökény G., Chen A., Ouyang N., Lieberman J. 2004. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA. 101:14883–14888 10.1073/pnas.0406421101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Luby-Phelps K., Das B., Shu X., Xia Y., Mosteller R.D., Krishna U.M., Falck J.R., White M.A., Broek D. 1998. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 279:558–560 10.1126/science.279.5350.558 [DOI] [PubMed] [Google Scholar]
- Han S.Y., Hwang H.S., Chae J.S., Yang S.J., Yoon J.H., Yeom Y.I., Choi E.J. 2009. CIIA induces the epithelial-mesenchymal transition and cell invasion. Biochem. Biophys. Res. Commun. 387:548–552 10.1016/j.bbrc.2009.07.050 [DOI] [PubMed] [Google Scholar]
- Innocenti M., Tenca P., Frittoli E., Faretta M., Tocchetti A., Di Fiore P.P., Scita G. 2002. Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J. Cell Biol. 156:125–136 10.1083/jcb.200108035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L.B., Steffen A., Stradal T.E.B., Di Fiore P.P., Carlier M.-F., Scita G. 2004. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6:319–327 10.1038/ncb1105 [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Jakowlew S.B. 2006. Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 25:435–457 10.1007/s10555-006-9006-2 [DOI] [PubMed] [Google Scholar]
- Katso R.M., Pardo O.E., Palamidessi A., Franz C.M., Marinov M., De Laurentiis A., Downward J., Scita G., Ridley A.J., Waterfield M.D., Arcaro A. 2006. Phosphoinositide 3-Kinase C2β regulates cytoskeletal organization and cell migration via Rac-dependent mechanisms. Mol. Biol. Cell. 17:3729–3744 10.1091/mbc.E05-11-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.J., Yu J.W., Hwang H.S., Choi E.-J. 2010. CIIA is a novel regulator of detachment-induced cell death. Cancer Res. 70:6352–6358 10.1158/0008-5472.CAN-09-4394 [DOI] [PubMed] [Google Scholar]
- Manser E., Leung T., Salihuddin H., Zhao Z.S., Lim L. 1994. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 367:40–46 10.1038/367040a0 [DOI] [PubMed] [Google Scholar]
- Margarit S.M., Sondermann H., Hall B.E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., Kuriyan J. 2003. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 112:685–695 10.1016/S0092-8674(03)00149-1 [DOI] [PubMed] [Google Scholar]
- Massagué J. 2008. TGFbeta in Cancer. Cell. 134:215–230 10.1016/j.cell.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. 2008. TGF-β signals regulate axonal development through distinct Smad-independent mechanisms. Development. 135:4025–4035 10.1242/dev.028209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual A., Bar-Sagi D. 2002. The two hats of SOS. Sci. STKE. 2002:pe36 10.1126/stke.2002.145.pe36 [DOI] [PubMed] [Google Scholar]
- Nimnual A.S., Yatsula B.A., Bar-Sagi D. 1998. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 279:560–563 10.1126/science.279.5350.560 [DOI] [PubMed] [Google Scholar]
- Qian X., Vass W.C., Papageorge A.G., Anborgh P.H., Lowy D.R. 1998. N terminus of Sos1 Ras exchange factor: critical roles for the Dbl and pleckstrin homology domains. Mol. Cell. Biol. 18:771–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.E., Araki T., Swanson K.D., Montgomery K.T., Schiripo T.A., Joshi V.A., Li L., Yassin Y., Tamburino A.M., Neel B.G., Kucherlapati R.S. 2006. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 39:70–74 10.1038/ng1926 [DOI] [PubMed] [Google Scholar]
- Russell A.J., Fincher E.F., Millman L., Smith R., Vela V., Waterman E.A., Dey C.N., Guide S., Weaver V.M., Marinkovich M.P. 2003. Alpha 6 beta 4 integrin regulates keratinocyte chemotaxis through differential GTPase activation and antagonism of alpha 3 beta 1 integrin. J. Cell Sci. 116:3543–3556 10.1242/jcs.00663 [DOI] [PubMed] [Google Scholar]
- Schlessinger J. 2000. Cell signaling by receptor tyrosine kinases. Cell. 103:211–225 10.1016/S0092-8674(00)00114-8 [DOI] [PubMed] [Google Scholar]
- Scita G., Nordstrom J., Carbone R., Tenca P., Giardina G., Gutkind S., Bjarnegård M., Betsholtz C., Di Fiore P.P. 1999. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 401:290–293 10.1038/45822 [DOI] [PubMed] [Google Scholar]
- Soisson S.M., Nimnual A.S., Uy M., Bar-Sagi D., Kuriyan J. 1998. Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell. 95:259–268 10.1016/S0092-8674(00)81756-0 [DOI] [PubMed] [Google Scholar]
- Sondermann H., Soisson S.M., Boykevisch S., Yang S.S., Bar-Sagi D., Kuriyan J. 2004. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 119:393–405 10.1016/j.cell.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Tartaglia M., Pennacchio L.A., Zhao C., Yadav K.K., Fodale V., Sarkozy A., Pandit B., Oishi K., Martinelli S., Schackwitz W., et al. 2006. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 39:75–79 10.1038/ng1939 [DOI] [PubMed] [Google Scholar]
- Wang W., Wyckoff J.B., Frohlich V.C., Oleynikov Y., Hüttelmaier S., Zavadil J., Cermak L., Bottinger E.P., Singer R.H., White J.G., et al. 2002. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 62:6278–6288 [PubMed] [Google Scholar]
- Welch D.R., Fabra A., Nakajima M. 1990. Transforming growth factor β stimulates mammary adenocarcinoma cell invasion and metastatic potential. Proc. Natl. Acad. Sci. USA. 87:7678–7682 10.1073/pnas.87.19.7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Hata K., Wada T., Moriya S., Miyagi T. 2006. Epidermal growth factor-induced mobilization of a ganglioside-specific sialidase (NEU3) to membrane ruffles. Biochem. Biophys. Res. Commun. 346:484–490 10.1016/j.bbrc.2006.05.136 [DOI] [PubMed] [Google Scholar]
- Zhang Y.E. 2009. Non-Smad pathways in TGF-β signaling. Cell Res. 19:128–139 10.1038/cr.2008.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Du G., Skowronek K., Frohman M.A., Bar-Sagi D. 2007. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9:706–712 [DOI] [PubMed] [Google Scholar]