Abstract
Background
Developing resistance towards existing anti-malarial therapies emphasize the urgent need for new therapeutic options. Additionally, many malaria drugs in use today have high toxicity and low therapeutic indices. Gradient Biomodeling, LLC has developed a quantum-model search technology that uses quantum similarity and does not depend explicitly on chemical structure, as molecules are rigorously described in fundamental quantum attributes related to individual pharmacological properties. Therapeutic activity, as well as toxicity and other essential properties can be analysed and optimized simultaneously, independently of one another. Such methodology is suitable for a search of novel, non-toxic, active anti-malarial compounds.
Methods
A set of innovative algorithms is used for the fast calculation and interpretation of electron-density attributes of molecular structures at the quantum level for rapid discovery of prospective pharmaceuticals. Potency and efficacy, as well as additional physicochemical, metabolic, pharmacokinetic, safety, permeability and other properties were characterized by the procedure. Once quantum models are developed and experimentally validated, the methodology provides a straightforward implementation for lead discovery, compound optimizzation and de novo molecular design.
Results
Starting with a diverse training set of 26 well-known anti-malarial agents combined with 1730 moderately active and inactive molecules, novel compounds that have strong anti-malarial activity, low cytotoxicity and structural dissimilarity from the training set were discovered and experimentally validated. Twelve compounds were identified in silico and tested in vitro; eight of them showed anti-malarial activity (IC50 ≤ 10 μM), with six being very effective (IC50 ≤ 1 μM), and four exhibiting low nanomolar potency. The most active compounds were also tested for mammalian cytotoxicity and found to be non-toxic, with a therapeutic index of more than 6,900 for the most active compound.
Conclusions
Gradient's metric modelling approach and electron-density molecular representations can be powerful tools in the discovery and design of novel anti-malarial compounds. Since the quantum models are agnostic of the particular biological target, the technology can account for different mechanisms of action and be used for de novo design of small molecules with activity against not only the asexual phase of the malaria parasite, but also against the liver stage of the parasite development, which may lead to true causal prophylaxis.
Background
Malaria is one of the most widespread infectious diseases of our time. Even though the global malaria map has been shrinking over the past 50 years, more people are at risk of suffering from malaria today than at any other time in history-close to 40% of the world's population live in countries where the disease is endemic and nearly 247 million people suffer from the disease every year [1]. Malaria is caused by protozoan parasites of the genus Plasmodium that infect and destroy red blood cells, leading to fever, severe anaemia, cerebral malaria and, if untreated, death. P. falciparum is the dominant species in sub-Saharan Africa, and is responsible for almost one million deaths each year. The disease burden is heaviest in African pregnant women and children under five years of age, who have frequent attacks and weak immunological protection. The global fight to control malaria requires a multi-faceted approach [2]. At present, a wide range of effective tools exists, including insecticide and larvicide spraying, the use of insecticide-impregnated bed nets to protect against infection by mosquitoes, and medicines to both treat the infection and prevent it in pregnant women and in young children [3-5]. However, long-term prophylaxis by vaccination has been especially challenging as the parasite has various sophisticated mechanisms to avoid the host immune system and no approved vaccine is currently available [6]. Even with all available strategies combined, a substantial number of patients will suffer from this disease over the coming decades. Due to emerging drug resistance, new medicines are needed to treat malaria episodes, mainly targeting the asexual blood stages of P. falciparum [7]. Blocking the transmission of the parasite by the mosquito vector and, in the case of P. vivax infections, targeting the dormant liver stage of the parasite, are other important steps towards eradication of the disease [8,9].
Traditional experimental methods for high-throughput screening and identification of novel compounds with activity against malaria targets are rarely effective for new therapeutics reaching clinical use. Despite considerable investments of resources and continuous improvements of these methods [10], many false positive hits arise and require further effort to triage and validate the results [11]. Increased size and complexity of molecular screening libraries often reduce the chance of finding leads among randomly chosen ligands [12] because of practical limitations associated with synthesis and testing of additional compounds with low probability of matching the requirements of the pharmaceutical target. Therefore, it is not surprising that high-throughput screening efforts frequently fail to identify suitable hit compounds, particularly against targets have not been studied previously. In attempts to improve hit rates, screening efforts are often enhanced by traditional structure-based computational methods [13,14], without much success [15-17].
In contrast, Gradient's methodology did not use any prior, explicit knowledge of the malaria protein targets. Metric modelling considers the compounds of interest as quantum objects, without explicit dependence on their particular chemical structure. On a theoretical level, these quantum properties serve as powerful descriptors for molecular modelling, compound identification, optimization and de novo design. The computational platform determines essential, rigorous, easily computable molecular attributes related to chemical activity. These attributes are derived from a special representation of quantum fields. Their well-defined mathematical characteristics afford systematic theoretical treatment and property prediction with methods that would otherwise be computationally impossible. Specialized machine-learning algorithms with fuzzy decision-making protocols are applied to identify both active compounds and the corresponding quantum features of chemical and biological interest. Combined with the underlying modelling architecture, the algorithms also provide mechanistic hypothesis for the modelled interactions. Since structurally dissimilar compounds can be similar on a quantum level, this process is particularly good at identifying chemically novel compounds that have significant potency against the target.
Methods
Platform implementation for identification of existing molecules with anti-malarial activity
The first step for the generation of a model is the creation of a set of predictive quantum filters. A quantum filter is a software module that uses quantum attributes of molecules to predict their activity, or any other observable property. These filters are created by the use of a training set of compounds with known activity against the target, a crystal structure of the target, or other data related to the desired property. The structural data is first used to compute the quantum components of the involved molecular systems. Then, fuzzy machine-learning algorithms are used to classify the target property with respect to these pre-computed quantum objects. Quantum components controlling the target properties are then used for property prediction of prospective molecules. The resulting quantum filter is experimentally validated by virtual high-throughput screening of a comprehensive database of pre-computed commercially available compounds, identification of potential active molecules and testing them in vitro for activity in the appropriate biological assay.
Structure representation-localized electron-density descriptors for molecular modeling
Well-defined chemical subsystems, together with their associated local, spatially-resolved properties, are very useful in drug discovery [18]. On a theoretical level, these properties serve as powerful descriptors for molecular modelling and design. Notions from Density Functional Theory and Topological Theory of Atoms in Molecules can be combined to rigorously define and compute a complete set of such localized, electron-density descriptors.
In general, Non-Relativistic Quantum Mechanics (QM) provides the proper level of physical theory for treatment of molecular and bio-molecular systems [19,20]. However, many intuitive chemical concepts are not directly related to the corresponding wave function [21], a state-vector in Hilbert space, which is difficult to partition into chemically meaningful subsystems [22-25].
Density Functional Theory (DFT) [26,27] provides a systematic framework for inferring chemistry-related information from QM calculations. This is achieved through the use of the electron density, ρ(r), a real, nonnegative Cartesian function connected to the N-electron molecular wave function ψ by
where x = {s, r} is the four-dimensional spin-spatial coordinate. As the famous Hohenberg-Kohn theorem [28] shows, ρ(r) determines all ground-state properties of the entire system, including its chemical and biochemical features.
Furthermore, the Topological Theory of Atoms in Molecules (AIM) [29,30] uses ρ(r) to partition molecules into precise atomic subsystems. These atomic subsystems are bounded by zero-flux surfaces S, which obey the equation
where n(r) is the vector normal to S at r and ρ(r) is the corresponding electron density.
It is natural to combine DFT and AIM, together with their respective computational algorithms, in a single formalism for studying local molecular properties from first principles. This formalism has yielded meaningful interpretations of many general chemical concepts, such as energy partitioning [31], atomic softness [32], electronegativity equalization [33], atomic reactivity indices [34], etc. Augmented with the electrostatic potential, this electron density-based methodology has been applied to quantitative structure-activity relationship studies [35,36]. It also produced the molecular descriptors employed in the modelling effort described here. Importantly, when applied as descriptors, these electron-density transforms define a proper metric (molecular similarity measure) in the modelling space and allow the use of rigorous mathematical techniques.
Modelling architecture-fuzzy decision networks
Molecular modelling is a multi-step process:
The starting point, {Si, Pi}, called a training set, is a set of molecular structures Sifor which a particular property of interest P has been measured. In the first step, descriptor calculation, every structure is reduced to some form, typically a list of real numbers {Dj}, which can be modelled statistically. The second step, actual modelling, attempts to find a model-a general mapping between property P and structure S through descriptors D. If successful, the model would have predictive power that can be applied to structures for which no measurement exists. Naturally, the predictive power of the model depends on the quality (accuracy, diversity, etc.) of the training set as well as descriptor properties and modelling architecture.
Both powerful descriptors and proper modelling architecture are crucial for successful molecular modelling and compound discovery. Ideally, the modelling architecture should be chosen in accordance with the underlying fundamental processes of the system [37], and not with the type of available numerical data. Complex biochemical interactions involve local attributes of distinct and diverse molecular structures, which are best modelled with discrete combinatorial methods rather than continuous multivariate techniques. Still, inherent weaknesses of traditional molecular descriptors require the use of such continuous multivariate techniques [38]. As sophisticated as some of these techniques are, they cannot always compensate for the shortcomings of the underlying molecular-structure representations.
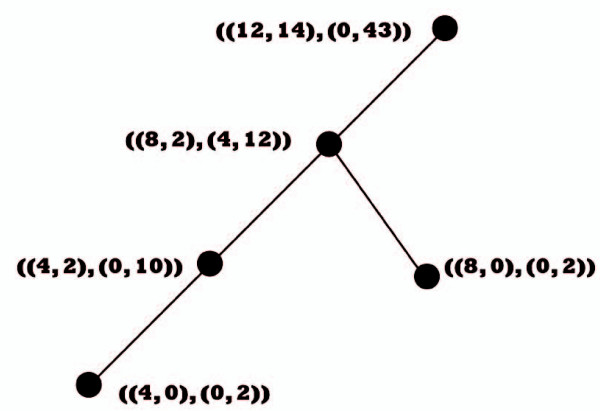
A straightforward machine-learning algorithm using fuzzy-logic decisions easily discovers the relationship between quantum components and specific interaction patterns. In its simplest implementation, the modelling algorithm produces a model (Figure 1) in the form of a fuzzy decision tree [39,40]. Each tree node corresponds to a single descriptor (interaction constraint). In a fully resolved decision tree, terminal nodes contain only either active or inactive molecules. Furthermore, each terminal node is fully characterized statistically-if a molecule belongs to it, the prediction is qualified by associated confidence intervals and other statistical parameters. A model in the form of a decision tree is easy to interpret. Each tree path that contains an active terminal node also contains a set of nodes (quantum components) that define the interaction pattern common to all training-set molecules belonging to this terminal. The fuzzy decision tree formalism can be generalized [41] to more powerful fuzzy decision algorithms. Given a diverse training set of structures with known inhibition, the modelling effort produces a decision network characterizing all present interaction patterns in terms of activity-controlling descriptors, which can be visualized [36].
Figure 1.
Fuzzy decisions tree and network. Quantum attributes are used to represent molecular systems. These attributes define a specific, relevant metric in the modelling structure space and thus allow the proper use of rigorous mathematical techniques. Also, they have strong relationships with chemical features controlling molecular interactions, and well-defined physical properties are often related to a single attribute. In its simplest implementation, the modelling algorithm produces a fuzzy decision tree, which can be generalized to a more powerful fuzzy decision network. Each tree node corresponds to a single attribute (interaction constraint). The attribute explaining most data variance occupies the highest node. In a fully resolved decision tree, terminal nodes contain only either active or inactive molecules. Further, each terminal node is fully characterized statistically by associated confidence intervals and other parameters. A decision network provides a complete characterization of the interaction patterns found within the modelling data.
Modelling data and in silico filters
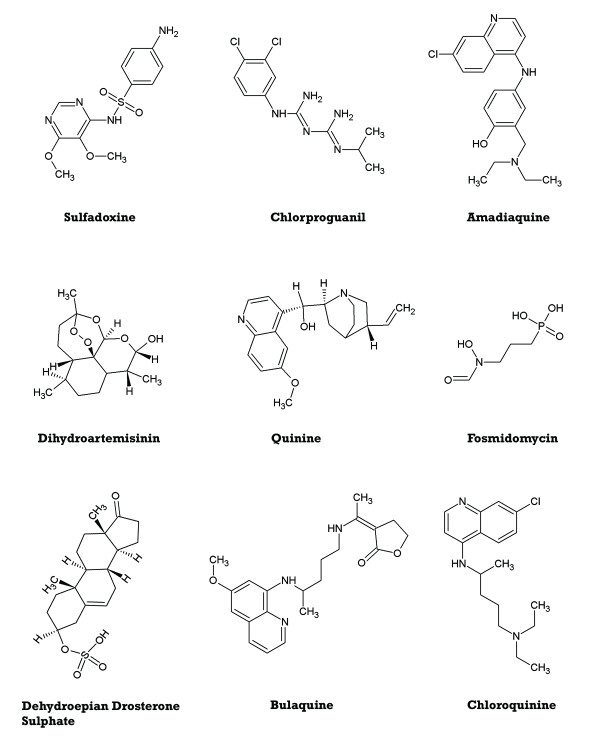
Three separate in silico models were created and then applied as screens for anti-malarial compound identification. The anti-malarial model was based on a training set consisting of 26 known anti-malarial compounds (Table 1, Figure 2) and in vitro data generated at the Johns Hopkins Malaria Research Institute of 1,730 FDA drugs. The in vitro data consisted of single-point measurements of anti-malarial percent inhibition at 10 μM concentration. Since the modelling resolution is directly related to the input data accuracy, the predictive threshold of the model was predetermined by the training set at the same 10 μM value. The training set included multiple compounds with no P. falciparum inhibition, which is important to define negative interaction constraints in quantum terms. The chemical-diversity screen was developed and applied to assure that the identified compounds are novel and chemically different than the 26 known anti-malarials. A number of commonly accepted theoretical measures of molecular similarity [42] were considered to estimate how novel the proposed compounds are. These include Tanimoto coefficients [43] based on pharmacological functional groups or compound fragments [44,45], as well as chemical diversity measures derived from electron density considerations [39]. Once computed, these indices are used to create point-to-set distance metrics [46], which determine the dissimilarity of the considered structure from the 26 known active molecules. Finally, to create filters accounting for low cytotoxicity, the publicly available data from the National Center for Computational Toxicology [47] and other sources was used [48].
Table 1.
Anti-malarial training set-full list of molecules
| sulphadoxine | amodiaquine | atovaquone | mefloquine |
|---|---|---|---|
| pyrimethamine | artesunate | proguanil | pyronaridine |
| chlorproguanil | dihydroartemisinine | azithromycin | artemether |
| Dapsone | piperaquine | quinine | lumefantrine |
| Dihydroartemisinin | chloroquine | bulaquine | tafenoquine |
| Trimethoprim | sulphamethoxazole | fosmidomycin | clindamycin |
| Artemotil | dehydroepian drosterone sulphate | - | - |
Figure 2.
Anti-malarial training set example structures. The core of the training set used to create the anti-malarial filter consists of 26 known anti-malarial compounds (full list in Table 1).
The database
A compound database of commercially available molecules was compiled for the virtual screening part of this experiment. A total of about 5.8 million structures were included by incorporating compounds from multiple sources like Enamine, ChemBridge, LifeChemicals, ChemDiv, TimTec, the National Cancer Institute, etc. All molecules were pre-computed and stored in quantum binary form for virtual screening purposes.
In vitro anti-malarial activity assay
The in vitro anti-malarial activity was measured using the [3H]-hypoxanthine incorporation assay [49] with various strains of P. falciparum (Roche). Results were expressed as the concentration corresponding to 50% inhibition. The anti-malarial assays were performed at the Swiss Tropical Institute, Basel, Switzerland.
Toxicity assay
Toxicity was determined by using the colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) assay [50]. The toxicity assays were performed at the Swiss Tropical Institute, Basel, Switzerland.
Results and Discussion
Conventional drug discovery implies a slow, incremental search for chemicals structurally similar to known active compounds. This process is inherently limited and rarely finds molecules having at once all the required properties of a successful drug. Chemical structure alone does not provide adequate description of molecular interactions, which are quantum in nature. Without quantum science any understanding of molecular interactions is incomplete. In theory, chemistry and biology can be fully derived from quantum mechanics [19,20]. It was hypothesized that quantum representation of a small, diverse set of known anti-malarial compounds can be used identify in silico novel, non-toxic molecules that inhibit P. falciparum. Starting with the training set of the 26 known anti-malarials and the 1730 FDA drugs with malaria activity screened at 10 μM, a quantum anti-malarial model was constructed. The model produced 12 quantum components positively correlated to activity. Each of these quantum components can be expressed by multiple chemical substructures. In addition to the 12 active quantum components, the model produced more than 20 quantum components with negative correlation to malaria action. This translated to detrimental chemical substructures which were also used in the subsequent in silico screening process. Additional filters generated from the data from the National Center for Computational Toxicology were used to ensure that the selected compounds were non-toxic.
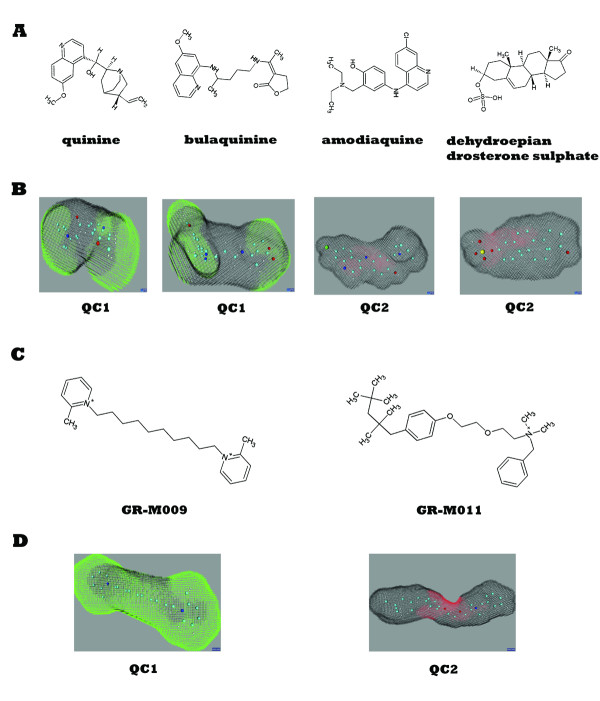
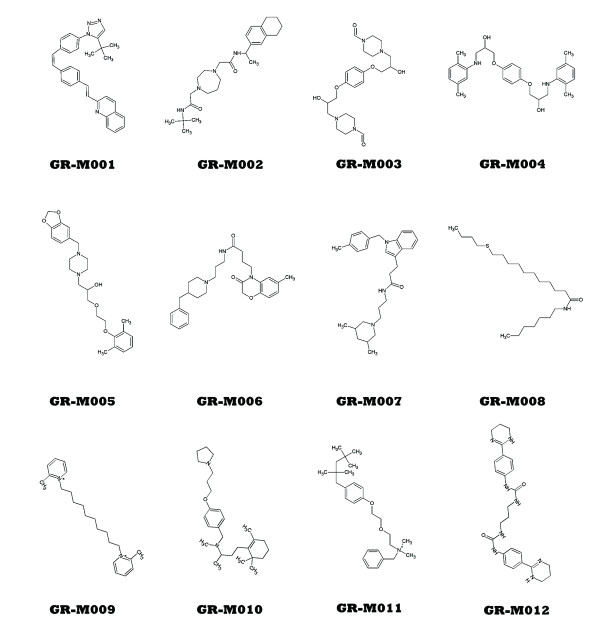
Since the modelling procedure employed this diverse training set of structures with known activity (Table 1, Figure 2), it produced a model in the form of a decision network characterising multiple anti-malaria interaction patterns. These interactions were defined in terms of activity-controlling quantum attributes, which were visualized by projection on corresponding Cartesian molecular surfaces (Figure 3). Importantly, the same quantum attribute can be found on chemically dissimilar molecules, which enables discovery of novel molecules with anti-malarial activity. Quantum attributes as shown in Figure 3 were used to construct the malaria-specific in silico filter. Together with the quantum toxicity filter and the chemical diversity filter described in the Materials and Methods, it was used in a virtual search for novel, non-toxic anti-malarial compounds. The virtual screen identified a number of molecules from the quantum database and rank-ordered them according to their quantum anti-malarial attributes. Based on commercial availability, twelve of the top 25 rank-ordered compounds were obtained for in vitro validation in both anti- P. falciparum and mammalian cytotoxicity assays. The in vitro results are presented at Table 2. All tested molecules are novel, with low structural similarity (average Tanimoto coefficient < 0.2) to the known drugs used in our training set (Figure 4). Eight of the twelve showed anti-malarial activity at or below the modelling threshold (IC50 = 10 μM), with six being very effective (IC50 ≤ 1 μM). Four compounds exhibited potency in the low nanomolar range (IC50s of 27nM, 185nM, 328nM and 332nM, respectively). The toxicity of the six most active molecules was also measured (Table 2), and their respective therapeutic indices (ratios of anti-malarial activity to mammalian cytotoxicity) were calculated. The most potent of the tested compounds has an index greater than 6900. In contrast, most malaria drugs in use today have much lower therapeutic indices [51]. These results exceed conventional state-of-the-art computational methods and are not subject to the limitations of popular docking programs [52]. For example, these findings substantially outperform a recent malaria study [17,53] not only in success rate of compound discovery (75%), but in speed and need for computational resources as well. The research described here confirms the ability of the quantum-similarity platform to generate anti-malarial compounds that are simultaneously active, novel and non-toxic-the three most important characteristics of an effective therapy for this disease. Once validated, the quantum anti-malarial components discovered by this approach can be employed in straightforward de novo design of new chemical entities possessing these three features as well as all other ADME/Tox properties required for successful anti-malarial therapeutics.
Figure 3.
Quantum anti-malarial model. Starting with known anti-malarial drugs (A), quantum components (QCs) that control anti-malarial activity were identified (red and green shaded areas). Dissimilar nuclear arrangements in active molecules can have similar anti-malarial electron density transforms (EDTs). For example, even though the red-shaded area illustrated as QC2 in panel A is comprised of different atoms in a different chemical substructure, the algorithms calculated their anti-malarial EDTs to be similar to one another. The QCs (B) were calculated and visualized as described in Materials and Methods. The subsequent virtual screen identified novel compounds (C) predicted to be active against P. falciparum based on these pre-computed anti-malarial QCs, i.e., it discovered molecules with novel nuclear arrangements that carry the same anti-malarial QCs (D). Containing two symmetrical quantum components (2 × QC1) which encompass the entire molecule, GR-M009 is the most active novel compound, while the less active GR-M011 contains only one quantum component (QC2). Red dots represent oxygen atoms, dark blue dots represent nitrogen atoms, light blue dots represent carbon atoms, and yellow dots represent sulphur atoms.
Table 2.
Summary of anti-malarial activity and cytotoxicity
| Compound | Average Anti-malarial Activity**(ng/ml) |
IC50 (μM) |
Average Cytotoxicity** (ng/ml) |
Therapeutic Index | Average Tanimoto Coefficient |
|---|---|---|---|---|---|
| GR-M001 | > 10'000* | > 20.0 | ND | NA | 0.170 |
| GR-M002 | 5381 | 12.54 | ND | NA | 0.162 |
| GR-M003 | > 10'000 | > 20.0 | ND | NA | 0.148 |
| GR-M004 | 4778 | 10.28 | ND | NA | 0.175 |
| GR-M005 | 9736 | 18.91 | ND | NA | 0.147 |
| GR-M006 | 1634 | 3.520 | ND | NA | 0.175 |
| GR-M007 | 446 | 0.999 | 10420 | 23 | 0.186 |
| GR-M008 | 382 | 1.027 | > 90'000 | > 236 | 0.124 |
| GR-M009 | 13 | 0.027 | > 90'000 | > 6923 | 0.159 |
| GR-M010 | 172 | 0.332 | 4860 | 28 | 0.183 |
| GR-M011 | 86 | 0.185 | 3250 | 38 | 0.159 |
| GR-M012 | 168* | 0.328 | 89770 | 533 | 0.145 |
| Anti-malarial control (chloroquine) | 4.2 | 0.013 | NA | ~2 | ND |
| Cytotoxicity control (Podophyllotoxin) | NA | NA | 9.5 | NA | NA |
*Not fully soluble in DMSO
**Data from two independent assays
ND: not determined
NA: not applicable
Figure 4.
Novel anti-malarial compounds identified through quantum metric modelling.
Given the growing resistance of the malaria parasite, the ability to discover new classes of active, safe molecules will be essential in the search for anti-malarial agents. Furthermore, since quantum features can be defined for compounds known to impact different stages of the parasite life cycle, the methodology could provide, for instance, an opportunity to identify alternative P. vivax hypnozoitocidals by the use of primaquine, tafenoquine and pamaquine in the training set [7,9]. This is an extremely difficult and demanding area of biology and there is a great need for alternative radical cures without side effects. Finally, this computational platform opens the possibility of exploring novel chemical spaces and specifically and accurately targeting elusive, hard to modulate protein-protein interactions previously considered unapproachable by current discovery methods.
Conclusions
To summarize, after starting from a training set of 26 known anti-malarial drugs and a collection of 1730 FDA drugs, several novel, chemically different molecules with high potency and low toxicity were identified and experimentally validated by testing only 12 compounds. The computational work was performed in less than a month on a single computer. Together with the experimental validation, the whole process took less than four months and required significantly smaller resources than similar drug discovery efforts. Gradient's innovative approach significantly reduces the time and cost needed to generate pre-clinical drug candidates and greatly improves the chances to discover and develop a true causal anti-malarial prophylaxis therapy.
Competing interests
The core quantum technologies being used by Gradient Biomodeling, LLC, were developed and are owned by Martin N. Martinov, PhD, the chief scientist and managing partner of Gradient.
Authors' contributions
MNM and DS conceived and designed the study. MNM performed the quantum analysis. MM, NK and DS analysed the data. MNM and NK produced figures and tables. MNM, NK and DS drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
David J Sullivan, Jr, Email: dsulliva@jhsph.edu.
Nikola Kaludov, Email: kaludov.n@gradientbiomodeling.com.
Martin N Martinov, Email: martinov.mn@gradientbiomodeling.com.
Acknowledgements
We would like to thank Roy Crandall from Park City, Utah, for his help in initiating and coordinating the project. We would like to thank Christian Scheurer and Sergio Wittlin, PhD from the Swiss Tropical & Public Health Institute, Switzerland, for performing the in vitro anti-malarial and cytotoxicity assays. The drug library work was supported the Johns Hopkins Malaria Research Institute and the Bloomberg Family Foundation. The work was supported by Gradient Biomodeling, LLC.
References
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks GD, Kain KC, Keystone JS. Malaria chemoprophylaxis in the age of drug resistance. II. Drugs that may be available in the future. Clin Infect Dis. 2001;33:381–385. doi: 10.1086/321866. [DOI] [PubMed] [Google Scholar]
- Chong CR, Chen X, Shi L, Liu JO, Sullivan DJ Jr. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2:415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- Weisman JL, Liou AP, Shelat AA, Cohen FE, Guy RK, DeRisi JL. Searching for new antimalarial therapeutics amongst known drugs. ChemBiol Drug Des. 2006;67:409–416. doi: 10.1111/j.1747-0285.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri SK, Singh N. Azithromycin: antimalarial profile against blood- and sporozoite-induced infections in mice and monkeys. Exp Parasitol. 2000;94:8–14. doi: 10.1006/expr.1999.4465. [DOI] [PubMed] [Google Scholar]
- Chilengi R, Gitaka J. Is vaccine the magic bullet for malaria elimination? A reality check. Malar J. 2010;9(Suppl 3):S1. doi: 10.1186/1475-2875-9-S3-S1. [DOI] [PubMed] [Google Scholar]
- Mazier D, Rénia L, Snounou G. A pre-emptive strike against malaria's stealthy hepatic forms. Nat Rev Drug Disc. 2009;8:854–864. doi: 10.1038/nrd2960. [DOI] [PubMed] [Google Scholar]
- Wells TNC, Alonso PL, Gutteridge WE. New medicines to improve control and contribute to the eradication of malaria. Nat Rev Drug Disc. 2009;8:879–891. doi: 10.1038/nrd2972. [DOI] [PubMed] [Google Scholar]
- Mahmoudi N, Garcia-Domenech R, Galvez J, Farhati K, Franetich JF, Sauerwein R, Hannoun L, Derouin D, Danis M, Mazier D. New active drugs against liver stages of Plasmodium predicted by Molecular Topology. Antimicrob Agents Chemother. 2008;52:1215–1220. doi: 10.1128/AAC.01043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macarron R. Critical review of the role of HTS in drug discovery. Drug Discov Today. 2006;11:277–279. doi: 10.1016/j.drudis.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Pellecchia M, Bertini I, Cowburn D, Dalvit C, Giralt E, Jahnke W, James TL, Homans SW, Kessler H, Luchinat C, Meyer B, Oschkinat H, Peng J, Schwalbe H, Siegal G. Perspectives on NMR in drug discovery: A technique comes of age. Nat Rev Drug Discov. 2008;7:738–745. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- Bleicher KH, Böhm HJ, Müller K, Alanine AI. Hit and lead generation: beyond high-throughput screening. Nat Rev Drug Discov. 2003;5:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- Boehm HJ, Schneider G. Virtual screening for bioactive molecules. Methods and Principles in Medicinal Chemistry, Germany: John Wiley & Sons, Inc; 2000. [Google Scholar]
- Ojha PK, Roy K. Chemometric modeling, docking and in silico design of triazolopyrimidine-based dihydroorotatedehydrogenase inhibitors as antimalarials. Eur J Med Chem. 2010;45:4645–4656. doi: 10.1016/j.ejmech.2010.07.034. [DOI] [PubMed] [Google Scholar]
- Adane L, Patel DS, Bharatam PV. Shape- and chemical feature-based 3D-pharmacophore model generation and virtual screening: identification of potential leads for P. falciparum DHFR enzyme inhibition. ChemBiol Drug Des. 2010;75:115–126. doi: 10.1111/j.1747-0285.2009.00908.x. [DOI] [PubMed] [Google Scholar]
- Kasam V, Salzemann J, Botha M, Dacosta A, Degliesposti G, Isea R, Kim D, Maass A, Kenyon C, Rastelli G, Hofmann-Apitius M, Breton V. WISDOM-II: screening against multiple targets implicated in malaria using computational grid infrastructures. Malar J. 2009;8:88. doi: 10.1186/1475-2875-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zartler ER, Shapiro MJ, eds. Fragment-Based Drug Discovery. A Practical Approach. John Wiley & Sons; 2008. [Google Scholar]
- Erwin Schrodinger. What is Life? Macmillan. 1946.
- Abbott D, Davies PCW, Pati AK, Eds. Quantum Aspects of Life. Imperial College Press; 2008. [Google Scholar]
- Mulliken RS. Molecular science: some reminiscences. J Chem Phys. 1965;43:S2–11. doi: 10.1063/1.1701510. [DOI] [Google Scholar]
- Morokuma K. Why do molecules interact? The origin of electron donor-acceptor complexes, hydrogen bonding and proton affinity. Acc Chem Res. 1977;10:294–300. doi: 10.1021/ar50116a004. [DOI] [Google Scholar]
- Glendenning ED, Streitwieser A. Natural energy decomposition analysis: An energy partitioning procedure for molecular interactions with application to weak hydrogen bonding, strong ionic, and moderate donor-acceptor interactions. J Chem Phys. 1994;100:2900–2909. doi: 10.1063/1.466432. [DOI] [Google Scholar]
- Cybulski SM, Scheiner S. Comparison of Morokuma and Perturbation Theory Approaches to Decomposition of Molecular Interaction Energy. (NH4)+...NH3. Chem Phys Lett. 1990;166:57–64. doi: 10.1016/0009-2614(90)87050-2. [DOI] [Google Scholar]
- Frey R, Davidson ER. Energy Partitioning of the SCF Interaction Energy of ScCO. J Chem Phys. 1989;90:5555–5562. doi: 10.1063/1.456408. [DOI] [Google Scholar]
- Parr RG, Yang W. Density-Functional Theory of Atoms and Molecules. Oxford University Press; 1989. [Google Scholar]
- Sholl D, Steckel JA. Density Functional Theory: A Practical Introduction. Wiley-Interscience; 2009. [Google Scholar]
- Hohenberg P, Kohn W. Inhomogeneous electron Gas. Phys Rev. 1964;136:B864–871. doi: 10.1103/PhysRev.136.B864. [DOI] [Google Scholar]
- Bader RFW. Atoms in Molecules. Clarendon Press, Oxford; 1990. [Google Scholar]
- Matta CF, Boyd RJ, Eds. The Quantum Theory of Atoms in Molecules. Wiley-VCH; 2007. [Google Scholar]
- Martinov MN, Cioslowski J. A rigorous energy partitioning scheme for analysis of molecular interactions. Mol Phys. 1995;85:121–125. doi: 10.1080/00268979500100981. [DOI] [Google Scholar]
- Cioslowski J, Martinov M. The atomic softness matrix. J Chem Phys. 1994;101:366–370. doi: 10.1063/1.468143. [DOI] [Google Scholar]
- Cioslowski J, Martinov M. Electronegativity equalization in polyyne carbon chains. J Phys Chem. 1996;100:6156–6160. doi: 10.1021/jp952814l. [DOI] [Google Scholar]
- Cioslowski J, Martinov M, Mixon ST. Atomic Fukui indexes from the topological theory of atoms in molecules applied to Hartree-Fock and correlated electron densities. J Phys Chem. 1993;97:10948–10951. doi: 10.1021/j100144a008. [DOI] [Google Scholar]
- Breneman CM, Martinov M. In: Molecular Electrostatic Potentials: Concepts and Applications, Elsevier. Murray JS, Sen K, editor. 1996. The use of electrostatic potential fields in QSAR and QSPR; pp. 143–180. [Google Scholar]
- Breneman CM, Martinov M. In: The Amide Linkage: Structural Significance in Chemistry, Biochemistry, and Materials Science. Greenberg A, Breneman CM, Liebman JF, editor. John Wiley and Sons, Inc; 2000. The electron density distribution of amides and related compounds; pp. 1–33. [Google Scholar]
- Gershenfeld NA. The Nature of Mathematical Modeling. Cambridge University Press; 1999. [Google Scholar]
- Martin YC. Quantitative Drug Design: A Critical Introduction. Second. CRC Press; 2010. [Google Scholar]
- Yuana Y, Shawb MJ. Induction of fuzzy decision trees. Fuzzy Sets and Systems. 1995;69:125–139. doi: 10.1016/0165-0114(94)00229-Z. [DOI] [Google Scholar]
- Janikow CZ. Fuzzy Decision Trees: Issues and Methods. IEEE Transactions on Man, Systems, and Cybernetics. 1998;28:1–14. doi: 10.1109/3477.658573. [DOI] [PubMed] [Google Scholar]
- Janikow CZ. Fuzzy Decision Forest. Fuzzy Information Processing Society, NAFIPS; 2003. pp. 480–483. [Google Scholar]
- Carbó R, Gironés X, Mezey Paul G, eds. Fundamentals of Molecular Similarity. Plenum Pub Corp; 2001. [Google Scholar]
- Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discovery Today. 2006;11:1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner JC, Willighagen E. The blue obelisk-interoperability in chemical informatics. J Chem Inf Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Open Babel Package, version 2.0.1. http://openbabel.sourceforge.net
- Deza M, Deza E. Encyclopedia of Distances. Springer-Verlag; 2009. [Google Scholar]
- DSSTox Computational Toxicology Research Program (Comptox) http://www.epa.gov/ncct/dsstox/DataFiles.html
- Cheminformatics. http://www.cheminformatics.org/datasets
- Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. Quantitative assessment of antimalarial activity in vitro by a semiautomatedmicrodilution technique. Antimicrob Agents Chemother. 1979;16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Meth. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Carraz M, Jossang A, Franetich JF, Siau A, Ciceron L, Hannoun L, Sauerwein R, Frappier F, Rasoanaivo P, Snounou G, Mazier D. A plant-derived morphinan as a novel lead compound active against malaria liver stages. PLoS Med. 2006;3:e513. doi: 10.1371/journal.pmed.0030513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem. 2004;47:45–55. doi: 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- Jacq N, Salzemann J, Jacq F, Legré Y, Medernach E, Montagnat J, Maaß A, Reichstadt M, Schwichtenberg H, Sridhar M, Kasam V, Zimmermann M, Hofmann M, Bretonm V. Grid-enabled virtual screening against malaria. J Grid Computing. 2008;6:29–43. doi: 10.1007/s10723-007-9085-5. [DOI] [Google Scholar]