Abstract
PDZ motifs are modular protein–protein interaction domains, consisting of 80–120 amino acid residues, whose function appears to be the direction of intracellular proteins to multiprotein complexes. In skeletal muscle, there are a few known PDZ-domain proteins, which include neuronal nitric oxide synthase and syntrophin, both of which are components of the dystrophin complex, and actinin-associated LIM protein, which binds to the spectrin-like repeats of α-actinin-2. Here, we report the identification and characterization of a new skeletal muscle protein containing a PDZ domain that binds to the COOH-terminal region of α-actinin-2. This novel 31-kD protein is specifically expressed in heart and skeletal muscle. Using antibodies produced to a fragment of the protein, we can show its location in the sarcomere at the level of the Z-band by immunoelectron microscopy. At least two proteins, 32 kD and 78 kD, can be detected by Western blot analysis of both heart and skeletal muscle, suggesting the existence of alternative forms of the protein. In fact, several forms were found that appear to be the result of alternative splicing. The transcript coding for this Z-band alternatively spliced PDZ motif (ZASP) protein maps on chromosome 10q22.3-10q23.2, near the locus for infantile-onset spinocerebellar ataxia.
Keywords: skeletal muscle, sarcomeres, muscle proteins, immunoelectron microscopy, alternative splicing
Pdz motifs (Kennedy 1995), previously known as GLGF repeats (Cho et al. 1992) or DHR domains (Woods and Bryant 1991), are protein–protein interaction domains (Sheng 1996; Brenman and Bredt 1997; Kornau et al. 1997; Xia et al. 1997) composed of 80–120 amino acid residues, which can be present as single or multiple copies (Cho et al. 1992). These domains were first identified in, and named after, the three members of the membrane-associated guanylate kinase homologues (MAGUKs): PSD-95, Dlg, and ZO-1. PDZ domains can be involved in different types of interactions. NH2-terminal PDZ domains can bind to the COOH-terminal of target proteins (Kim et al. 1995, Kornau et al. 1995; Sato et al. 1995). For example, the NH2-terminal of two PDZ domains of PSD-95 and other MAGUK family members can bind to the COOH-terminal (consensus sequence T/SXV) of Shaker-type K+ channels (Kim et al. 1995) and of the modulatory subunits (NR2) of N-methyl-D-aspartate (NMDA) type-glutamate receptor ion channels (Kornau et al. 1995; Niethammer et al. 1996). In this type of interaction, PDZ domains would appear to be involved in the targeting and clustering of membrane proteins. In another type of interaction, PDZ domains can also bind internal peptides. This is demonstrated by the binding of the PDZ motif of inactivation no afterpotential D (INAD) to an internal peptide of the transient-receptor potential (TRP) calcium channel (Shieh and Zhu 1996). In a third type of interaction, PDZ domains can interact with each other in homomeric PDZ–PDZ interactions, as exemplified by the binding of the second PDZ domain of PSD-95 and that of syntrophin to the NH2-terminal PDZ domain of nNOS (Brenman et al. 1996). It is noteworthy that both the second PDZ domain of PSD-95 and the PDZ domain of neural nitric oxide synthase (nNOS) also can bind COOH-terminal sequences (Schepens et al. 1997; Stricker et al. 1997), these interactions being mediated by distinct regions of the same PDZ domain. Another type of interaction is portrayed by the PDZ domain of the actinin-associated LIM protein (ALP)1 binding to the spectrin-like repeats of α-actinin-2 (Xia et al. 1997). Recently, a novel type of PDZ interaction has been noted with the LIM domain, where the second PDZ domain of protein tyrosine phosphate-BL (PTP-BL) and the PDZ domain of reversion induced LIM (RIL) can bind to the LIM domain of RIL (Cuppen et al. 1998).
A general function of PDZ domains seems to be directing cellular proteins to multiprotein complexes. In skeletal muscle there a few PDZ domain proteins, these include neuronal nitric oxide synthase and the family of syntrophins (Ponting and Phillips 1995), both of which are components of the dystrophin complex (Adams et al. 1993; Brenman et al. 1995), and ALP, which binds to the spectrin-like repeats of α-actinin-2 (Xia et al. 1997). In this paper, we describe a new alternatively spliced skeletal muscle protein of 31 kD that has an NH2-terminal PDZ domain for which we propose the name ZASP: Z-band alternatively spliced PDZ motif protein.
Materials and Methods
Antibodies
A cDNA fragment of human ZASP lacking 165 bp from the 5′ end of the coding sequence was inserted into the His-tag prokaryote expression vector pQE9 (QIAGEN Inc.) and sequenced to confirm that there were no significant changes from the original transcript. Human α-actin cDNA was obtained by reverse transcriptase PCR using primers based on the sequence data present in the Genbank/EMBL/DDBJ database. The resulting full-length cDNA was inserted into the pQE9 vector and sequenced to detect any changes from the known sequence. Both actin and ZASP were expressed as recombinant proteins in Escherichia coli and purified by affinity chromatography using nickel-nitrilotriacetic acid resin as specified by the manufacturer (QIAGEN Inc.). The recombinant ZASP protein contains a 12-amino acid residue tag plus 228 amino acids of the ZASP protein (81% of the full-length protein) with an estimated molecular weight of 26,664 D. The human recombinant α-actin protein contains 12-amino acid residues of the tag plus the 377-amino acid residues of the full-length α-actin with an estimated molecular weight of 43,449 D. The α-actin and the ZASP recombinant proteins were used to immunize rabbits and mice for the production of polyclonal (pAb) and monoclonal (mAb) antibodies.
Construction and Screening of Human and Mouse Skeletal Muscle cDNA Libraries
Human cDNA libraries suitable for the identification of full-length transcripts were produced using a kit obtained from Invitrogen Corp. The procedure was given in detail in Valle et al. 1997. A mouse diaphragm cDNA library Uni-ZAP™XR vector (cat. 937303) used for isolation of the mouse ZASP was purchased from Stratagene. The screening was done by PCR and the transcript-specific primers were designed on mouse expressed sequence tags (ESTs) similar to the human transcript. DNA sequencing was carried out directly on 2 μl of the PCR reactions using either Dye-deoxy-terminator chemistry or Dye-primer chemistry (PE Applied Biosystems) and run on an ABI377 DNA sequencer (PE Applied Biosystems).
Culture of Primary Myoblasts
Primary human myoblasts (CHQ5B) were isolated from the quadriceps of a newborn child (5-d postnatal), without any indication of neuromuscular disease and the protocols used for this work were in full agreement with the current legislation on ethical rules. The number of myoblast divisions noted are from the isolation of the cells. These primary cells can achieve 55–60 divisions before reaching proliferative senescence. The proliferation medium used was F10-Ham (GIBCO BRL) supplemented with 20% FCS (GIBCO) and 50 μg/ml gentamycin. To obtain differentiated cells, the growth medium was replaced with DME (GIBCO BRL) without serum plus 10 μg/ml of insulin (I-5500; Sigma Chemical Co.) and 100 μg/ml of transferrin (T-2036; Sigma Chemical Co.). Myotubes can be detected two days after the addition of this medium and continue to develop for at least another four days.
Genomic Mapping
The genomic mapping was performed by PCR with the radiation hybrids method, using the GeneBridge 4 whole-genome radiation hybrid panel (Research Genetics Inc.) consisting of 93 genomic DNA preparations from human-on-hamster somatic cell lines (Walter et al. 1994). The screening results were processed by the RHMAPPER software program, available from the Whitehead Institute/MIT Center for Genomic Research (Cambridge, MA).
Immunoelectron Microscopy
Heart and skeletal muscle fibers were stretched, fixed for 2 h in 4% paraformaldehyde plus 0.05% glutaraldehyde, and then dehydrated. The temperature was decreased stepwise while simultaneously increasing the concentration of ethanol to minimize the formation of aggregates and the dislocation of cellular components during dehydration. Then, the samples were embedded in lowicryl resin K4M (Sigma Chemical Co.) and ultrathin sections (0.1 μm) of lowicryl embedded samples were cut and processed. The muscle sections were blocked in 1% BSA plus 0.05% Tween-20 for 1 h, and then treated with mouse pAb to the recombinant ZASP protein used at a 1/25 dilution for heart and a 1/50 dilution for skeletal muscle samples. The secondary antibody, anti-mouse IgG whole molecule conjugated with 5-nm gold particles (G7527; Sigma Chemical Co.), was used at a 1/20 dilution. The sections were counterstained with 3% uranyl acetate for 5 min, washed, and then stained with lead citrate for 45 s. After further washing, the sections were visualized using a transmission electron microscope (Zeiss 255/230).
Immunofluorescence Microscopy
Frozen sections (∼5-μm thick) were prepared from human skeletal muscle (Vastus) and mouse heart and skeletal muscle using a Leica Jung/CM/1800 cryostat. These sections were used for indirect immunofluorescence experiments by fixing in acetone for 5 min and then blocking in PBS containing 1% BSA and 0.05% Tween-20 for 1 h. Then, they were incubated at room temperature for 1 h in mouse anti-ZASP antibody and/or a rabbit anti–α-actin antibody (A2668; Sigma Chemical Co.) used at 1/30- and 1/40-fold dilutions, respectively. The sections were then washed five times with buffer (PBS, 0.1% BSA, 0.05% Tween-20). TRITC-labeled goat anti–mouse immunoglobulin (T7657; Sigma Chemical Co.) and/or an FITC-labeled goat anti–rabbit immunoglobulin (F0511; Sigma Chemical Co.) were used as second antibodies. Then the slides were incubated in the second antibody for 1 h at room temperature, washed extensively, and mounted. Photographs were taken at 40×.
Primary human myoblasts were grown on collagen-coated coverslips and undifferentiated and differentiated cells were fixed with paraformaldehyde (3%), then treated with 0.1 M glycine, and permeabilized with 0.05% Tween-20 for 30 min. The cells were treated with 1% BSA to block any nonspecific binding. All wash and dilution buffers contained PBS, BSA 1%, and 0.05% Tween-20. For these experiments, the secondary antibody was FITC-conjugated anti-mouse immunoglobulin (F4018; Sigma Chemical Co.). All commercial immunochemicals were diluted as recommended by the suppliers. The cells were mounted using Vectashield mounting medium H-1000 (Vector Laboratories). An Axiovert 35 fluorescence microscope (Zeiss) was used at 40× to view and photograph the slides of the cell cultures.
Immunoprecipitation
Polyadenylated mRNAs (D6098-15, D6098-25, D6064-15, and D6064-25; Invitrogen) of adult and fetal heart, as well as adult and fetal skeletal muscle, were translated in vitro using a reticulocyte lysate system (Promega Corp.) and labeled with [35S]methionine (Nycomed Amersham Inc.). Equal amounts of labeled proteins were mixed with the appropriate antibody and immunoprecipitated using protein A–Sepharose (Nycomed Amersham Inc.) in a buffer containing 50 mM Hepes, pH 8.0, 250 mM NaCl, 0.1% NP-40. The resulting immunoprecipitated samples were run on 12 (see Fig. 5 C) or 15% (see Fig. 5 B) SDS-polyacrylamide gels. After running, the gels were dried and put against super resolution type SR Packard phosphor screens. The screens were analyzed on a Packard Cyclone phosphor imager. Rainbow 14C-methylated protein molecular weight marker (Nycomed Amersham Inc.), with proteins ranging from 220 to 14.3 kD, were used in all the immunoprecipitation experiments. The mouse anti-ZASP antibody, preimmune sera, and the generic myosin antibody (MF 20) were used at a dilution of 1/75 for the immunoprecipitation experiments.
Figure 5.
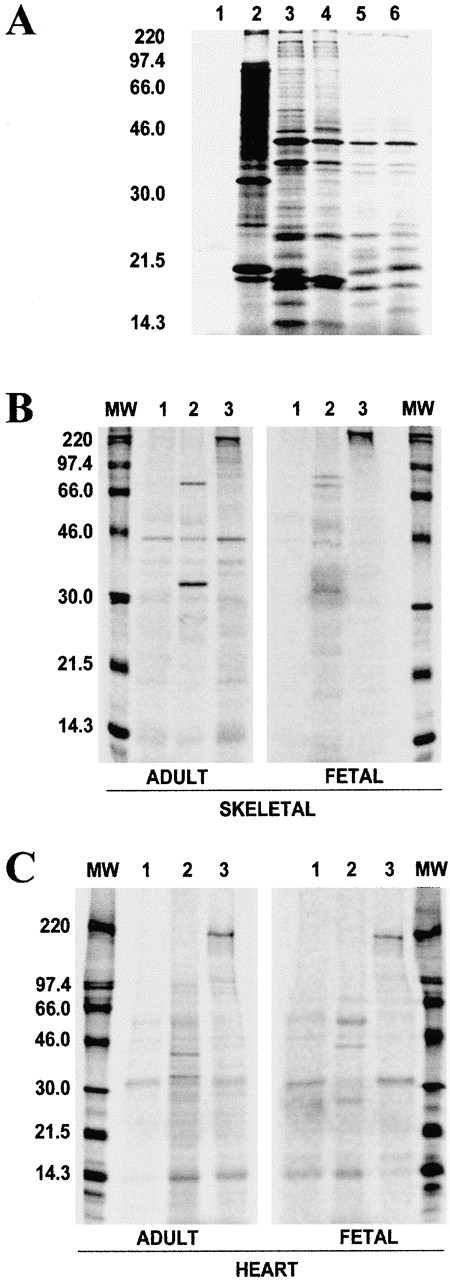
(A) Total protein obtained from in vitro translation of muscle and control mRNAs using the rabbit reticulocyte lysate system (Promega Corp.). Lanes: 1, negative control sample with water instead of mRNA; 2, positive control, BMV mRNA; 3, human adult skeletal muscle mRNA; 4, human fetal skeletal muscle mRNA; 5, human adult heart mRNA; and 6, human fetal heart mRNA. (B) Immunoprecipitation of in vitro translated human adult and fetal skeletal muscle proteins. (C) Immunoprecipitation of in vitro translated human adult and fetal heart. Equal amounts of [35S]methionine-labeled proteins were mixed with the appropriate antibody and immunoprecipitated using protein A–Sepharose, and were then run on SDS-polyacrylamide gels. In B and C, the total in vitro translated human heart and muscle proteins were immunoprecipitated with the antibodies denoted by the corresponding lane numbers: 1, preimmune sera; 2, antibody to ZASP; and 3, antibody to myosin. Numbers on the left of the figures indicate rainbow 14C-methylated protein molecular weight markers, in kD.
Northern Blot Analysis
The Northern blot analysis of ZASP transcript expression was performed on human and mouse filters supplied by CLONTECH Laboratories, Inc. The following filters were used: 7765-1, containing 2 μg/lane of mRNA from the following eight human muscle tissues: skeletal muscle, uterus (no endometrium), colon (no mucosa), small intestine, bladder, heart, stomach, and prostate; 7760-1, containing 2 μg/lane of mRNA from the following eight human tissues: heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas; 7762-1, containing 2 μg/lane of mRNA from the following mouse tissues: testis, kidney, skeletal muscle, liver, lung, spleen, brain, and heart; and 7780-1, containing 1 μg/lane of mRNA from the following twelve human tissues: brain, heart, skeletal muscle, colon, thymus, spleen, kidney, liver, small intestine, placenta, lung, peripheral blood, and leukocytes.
The hybridization protocol and solutions were provided by CLONTECH Laboratories, Inc. (ExpressHyb Hybridization Solution, cat. S0910). Probes were obtained either by labeling PCR fragments by random priming (DECAprimeII™ DNA labeling kit, Ambicon, cat. 1455) with 32P-dCTP, or by SP6 transcription (Strip-EZ™ RNA, Ambicon, cat. 1366) with 32P-UTP.
Sequence Analysis
Sequence similarity searches were performed using the programs BLASTN, BLASTP, and TBLASTN 2.0.6 (Altschul et al. 1997) with nonredundant nucleotide and protein databases, as well as with human and mouse EST databases, running on the NCBI BLAST server (Bethesda, MD). The FASTA program (Pearson and Lipman 1988) was run using EMBL and SwissProt databases. The protein sequences of ZASP and KIAA0613 were used to search the PROSITE database (Hofmann et al. 1999) using ScanProsite software (ExPASy Molecular Biology World Wide Web server, Swiss Institute of Bioinformatics, Geneva, Switzerland). These protein sequences were also employed in searching various databases of functional profiles, using ProfileScan (World Wide Web server, Bioinformatics Group, ISREC, Switzerland), SMART (Schultz et al. 1998), and Pfam (Bateman et al. 1999).
Western Blotting and Quantitation
Human muscle and heart extracts used in Western blot experiments were obtained by homogenizing fragments of frozen tissue under liquid nitrogen using a mortar and pestle. The resulting frozen powder was solubilized in a urea buffer (8 M urea) and then centrifuged to remove any insoluble material. The extracts were run on 15% SDS-polyacrylamide gels (10 μg of total protein per lane). Proteins from different human tissues (brain, heart, kidney, lung, skeletal muscle, liver, placenta, ovary, testis, and spleen) were obtained from CLONTECH Laboratories Inc. (cat. 7800–7808 and 7813) and used at various protein concentrations (10–60 μg of total protein). The ZASP mAbs and pAbs were used undiluted and at various dilutions (1/200 and 1/20,000, respectively). The myosin mAb MF 20 developed by Dr. D.A. Fischman was obtained from the Developmental Studies Hybridoma bank maintained by the University of Iowa's Department of Biological Sciences (Iowa City, IA). MF 20 is a generic myosin antibody obtained as an ascites fluid, which was used at a 1/10,000 dilution. Goat anti–mouse immunoglobulin conjugated with alkaline phosphatase (A3562; Sigma Chemical Co.) was used as the second antibody. As detailed previously (Valle et al. 1997), the intensity of the signal obtained from Western blot analysis can be used to estimate the relative amount of a specific protein in heart and skeletal muscle extracts. This procedure was used to estimate the amount of human α-actin (see Fig. 4 A) and ZASP protein (see Fig. 4 B) present in total heart and skeletal muscle. The mouse pAbs to ZASP and α-actin were used at dilutions of 1/20,000 and 1/4,000, respectively. A prestained wide-range color molecular weight marker (C3437; Sigma Chemical Co.) was used in all the Western blot experiments. The batch used had the following colors associated to the molecular weight marker: 205 kD, blue; 126 kD, turquoise; 83 kD, pink; 48 kD, yellow; 28 kD, orange; 22 kD, green; 15 kD, purple; and 9.5 kD, blue.
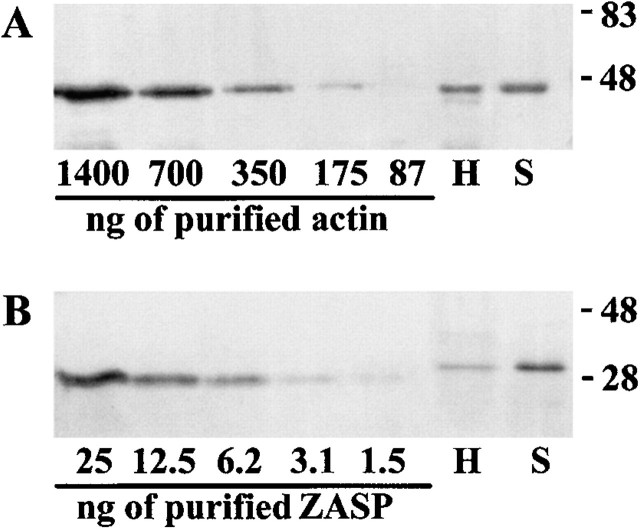
Figure 4.
(A) Western blot analysis of known amounts of recombinant human α-actin (left) and known amounts of total protein (2.5 μg) from human heart and skeletal muscle tissue (right). Actin was detected using a mouse pAb specific for human α-actin. (B) Western blot analysis of known amounts of recombinant ZASP protein (left) and known amounts of total protein (10 μg) from human heart and skeletal muscle tissue (right). The recombinant protein lacks some amino acids from the NH2-terminal end of the ZASP sequence, therefore it is slightly smaller in size than the native muscle protein. Mouse pAb to ZASP was used to detect the protein. From densitometric analysis of the bands, it can be calculated that the actin signal from 2.5 μg of total muscle proteins (both heart or skeletal muscle) is approximately equivalent to 500 ng of purified α-actin, whereas for ZASP, there is less protein present in heart than skeletal muscle tissue: 4.5 ng, as opposed to 18 ng in 10 μg of total muscle proteins.
Yeast Two-hybrid Experiments
Unless otherwise specified, the recipes and protocols used for yeast culture were obtained from Ausubel et al. 1994. The cDNA fragment encoding the NH2 terminus (amino acid residues 1–107) of the human ZASP protein was amplified by PCR using a forward (KpnI 686 PDZ-FOR GGGGTACCCCGGATGTCTTACAGTGTGACCCTGA) and a reverse (SalI 686 PDZ-REV ACGCGTCGACGTTCTGGTGAGGGATCACCG) oligonucleotide incorporating restriction sites KpnI and SalI, respectively. The amplified product was digested and cloned into the KpnI–SalI cut vector pHybLex/Zeo (Invitrogen) to create a hybrid protein between the LexA DNA binding domain and the PDZ domain of ZASP. This construct was verified by DNA sequencing before it was used to transform (Agatep et al. 1998) the yeast strain L40 (genotype MATa his3Δ200 trp1-901 leu2-3112 ade2 LYS2:(4lexAop-HIS3) URA3::(8lexAop-lacZ) GAL4). The background, due to histidine leakage, was measured by plating the transformed yeast on YC-HUK plates containing 300 μg/ml of Zeocin (Z300) and a range of 3-Aminotriazole (0, 1, 3, and 5 mM). No histidine expression could be detected after 5 d at 30°C from plates with 1, 3, and 5 mM 3-Aminotriazole. Several transformants were tested for the expression of the bait by Western blot assay using both anti-ZASP pAb and mAb, as well as anti-Lex antibody (Santa Cruz Biotechnology). Yeast cells from the best clone were transformed with three different libraries: human heart and skeletal muscle cDNA libraries fused to the GAL4 activation domain (CLONTECH Laboratories Inc.) and the third library of human heart cDNA fused to the B42 activation domain (Display System Biotech). The transformants made with the GAL4 libraries were plated onto 50 YC-LHUK Z300 plates (150-mm diam) and those obtained with the B42 were plated onto 50 YC-WHUK Z300 plates (150-mm diam). The growing clones were recovered from three to ten days at 30°C and the lacZ expression measured by the β-galactosidase filter assay. Positive clones were selected, the activating insert was amplified, and the PCR product was sequenced.
Results
Molecular Cloning and Characterization of ZASP mRNA
Many novel genes have been discovered from the systematic sequencing of human skeletal muscle ESTs carried out in one of our laboratories (Lanfranchi et al. 1996), including telethonin (Valle et al. 1997), which is bound, as well as phosphorylated, by titin and located at the level of the Z-band (Mayans et al. 1998; Mues et al. 1998). Currently, >30,000 ESTs have been identified, representing well over 4,000 different independent transcripts. Amongst these, transcript HSPD00686, hereafter named ZASP, appeared to be particularly interesting, as it was found at a moderately high frequency (0.06%) and, from a preliminary study based on reverse transcriptase-PCR, seemed to be expressed only in heart and skeletal muscle.
ZASP was initially identified in our laboratory as a cluster of muscle ESTs corresponding to the 3′ terminus of the mRNA; then we isolated and sequenced the entire human and mouse ZASP transcripts by screening full-length muscle cDNA libraries and performing 5′ RACE experiments on muscle mRNA. The total length of the nucleotide sequence shown in Fig. 1 is 1,607 bases for human and 1,469 bases for mouse. The translation of the human sequence reveals an open reading frame of 849 bases, encoding a putative protein of 283 amino acids with a molecular weight of 30,998 D, whereas the open reading frame of mouse ZASP encodes 288 amino acids and has a molecular weight of 31,426 D. The human and mouse coding sequences are very similar; there is a single insertion of 15 bases (five amino acid residues: Ala, Ser, Pro, Leu, and Ala) in mouse, and 69 base substitutions between mouse and human, resulting in eight changes in amino acid residues, as can be seen in Fig. 1 (Val→Ile, Thr→Ser, Val→Ala, Ala→Val, Ile→Val, Ser→Thr, Asn→Ser, Phe→Tyr). Thus, the identity of human and mouse ZASP is 92% at the nucleotide level and 97% at the amino acid level.
Figure 1.
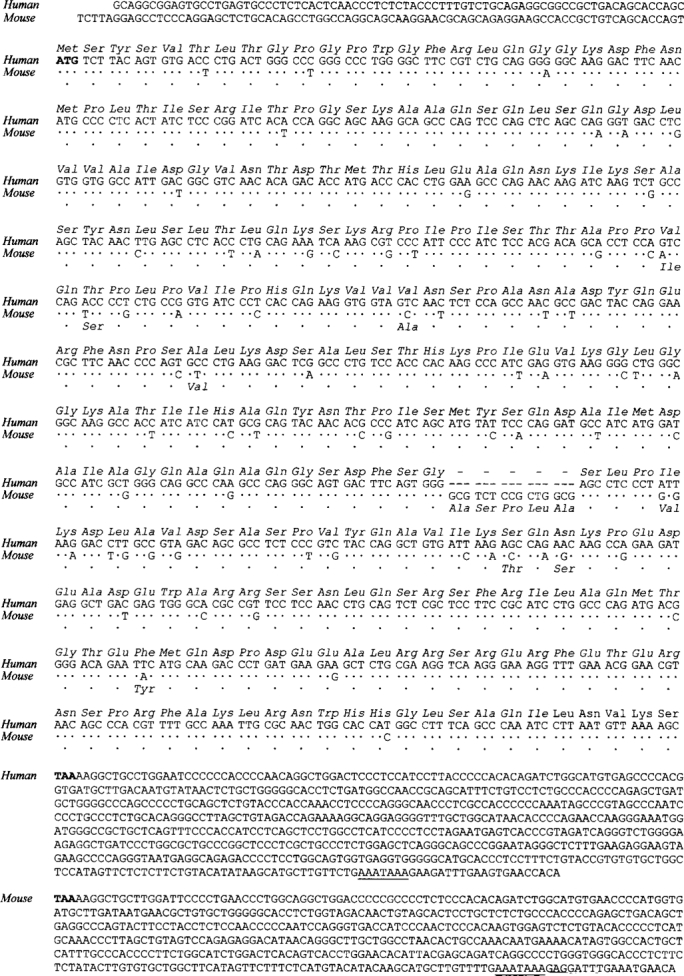
cDNA and amino acid sequences of human and mouse ZASP. The start and stop codons are in bold characters and the polyadenylation sites are underlined. In the coding part of the mouse sequence, the conserved nucleotides and amino acids are represented by dots. Dashes indicate gaps that have been inserted to align the two sequences. The sequence data are available from Genbank/EMBL/DDBJ under the accession number AJ133766 and AJ005621, respectively, for human and mouse. The two variant forms of ZASP, described in the other sections of this paper, are available under accession number AJ133768 and AJ133767, as well as at http://grup. bio.unipd.it/muscle.
An extensive similarity search that was done using the coding sequences (nucleotide and amino acid) of human ZASP revealed a significant similarity to KIAA0613, both at the nucleotide and amino acid level. The KIAA0613 sequence (GenBank/EMBL/DDBJ accession number AB014513) was found as a cDNA clone from brain as part of a systematic sequencing project (Ishikawa et al. 1998). A PDZ domain was detected at the NH2-terminal of ZASP, from amino acid 1 to 85 by the ProfileScan, SMART, and Pfam programs as described in Materials and Methods. PredictProtein (Rost et al. 1994), Psort (Nakai and Kanehisa 1992), and SMART programs did not detect any transmembrane domain in the ZASP protein.
Genomic mapping was done using the radiation hybrid technique, as described in Materials and Methods, revealing that the ZASP gene maps near the locus for infantile-onset spinocerebellar ataxia (OMIM 271245) on the human chromosome region 10q22.3-23.2, with a significant lod score of 17.
ZASP Is Primarily Expressed in Heart and Skeletal Muscle
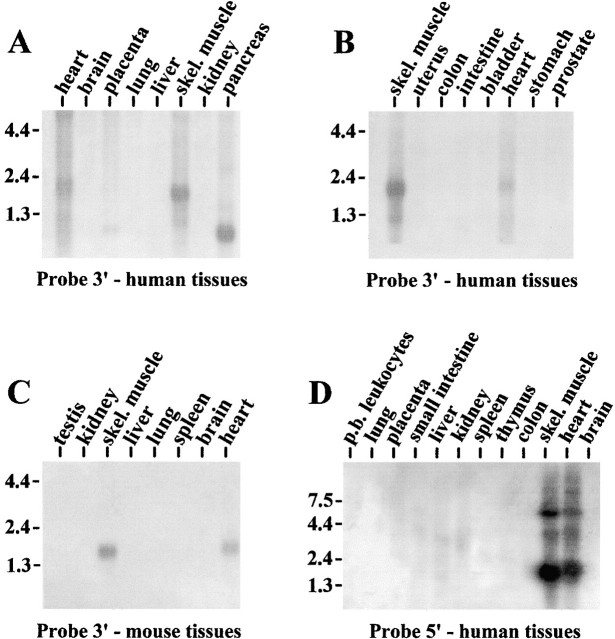
Northern blot analysis of different tissues using the 3′ untranslated region of ZASP as a probe revealed that skeletal muscle is the major site of expression of this gene (Fig. 2A and Fig. B). It is also expressed in heart, but to a lesser degree. A major band was detected at 1.9 kb in human heart and skeletal muscle, and at ∼1.6 kb in mouse heart and skeletal muscle (Fig. 2 C). Smaller transcripts could also be seen in pancreas and placenta at ∼1 kb, the signal in pancreas being quite strong. When Northern blot analysis of different human tissues was done using as a probe at the 5′ end of ZASP (Fig. 2 D), three transcripts of 1.9 kb, 4 kb, and 5.4 kb were detected both in heart and skeletal muscle. A weak signal could also be detected in brain, at ∼6 kb.
Figure 2.
Northern blot analysis of human (A, B, and D) and mouse (C) tissues demonstrating patterns of expression of ZASP mRNAs. Blots containing Poly(A)+ RNA from a variety of human and mouse tissues were probed with 3′ untranslated region of ZASP (A–C) or with the 5′ region (D) as indicated. The ZASP transcript is expressed primarily in skeletal muscle and heart. The numbers on the side indicate size in kb.
Western blot analysis was done to determine the electrophoretic mobility and tissue distribution of ZASP. From the results using human tissue (Fig. 3 A), it can be seen that ZASP is present to a lesser extent in heart than in skeletal muscle tissue. ZASP has the same pattern of distribution in mouse tissue as in human, that is, it is predominantly found in skeletal muscle and, to a lesser extent, in heart (data not shown). When the ZASP antibody is used at high dilutions (1/20,000) it detects two bands (Fig. 3 A). A prominent lower band corresponds to a molecular weight of ∼32 kD and an upper band of ∼78 kD. However, on using twofold higher amounts of total human protein (20 μg per lane) and 100-fold more concentrated antibody (1/200 dilution), extra bands can be detected in both heart and skeletal muscle (Fig. 3 B) with apparent molecular weights of 22, 27, and 67 kD. Also, there are two extra proteins that can be detected only in heart (43 and 83 kD). Several human tissues (brain, heart, kidney, lung, skeletal muscle, liver, placenta, ovary, testis, and spleen) were screened by Western blotting using a high concentration of ZASP antibody (1/200 dilution) as the probe. To detect bands in tissues other than heart and skeletal muscle, six times more protein had to be used (60 μg), as well as high concentration of antibody. Under these conditions, bands could be detected in brain and placenta (Fig. 3 B). Four bands corresponding to proteins of ∼198, 175, 38, and 20 kD could be detected in brain, and one band corresponding to a protein of 43 kD in placenta.
Figure 3.
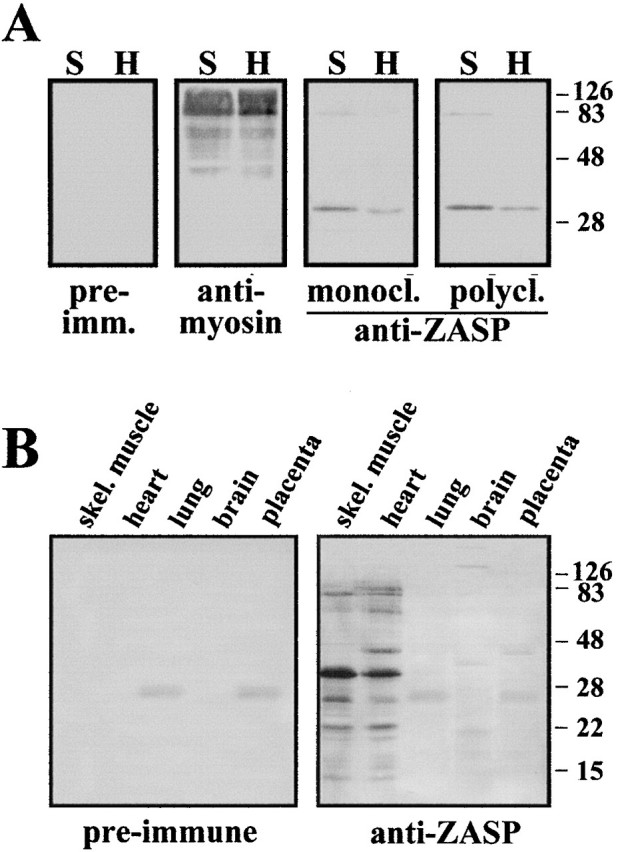
(A) Western blot analysis of heart and skeletal muscle tissue with antibodies to myosin, ZASP, and preimmune sera. Equal amounts of proteins were run in each lane (10 μg) on a 15% SDS-polyacrylamide gel and then blotted onto Immobilon-P membrane. The ZASP mAb was used undiluted, whereas the pAb was used at 1/20,00 dilution, as were the preimmune and myosin sera. (B) Tissue distribution of ZASP as demonstrated by Western blotting. Protein extracts from human heart and skeletal muscle (10 μg), as well as from brain, lung, and placenta (60 μg) were loaded in each lane, run on a 15% SDS-polyacrylamide gel, and then blotted. The membrane was probed with mouse pAb specific for ZASP and preimmune mouse sera, both used at 1/200 dilution. Sigma Chemical Co. color molecular weight markers were used.
The bands seen at low dilutions (1/200) with the polyclonal ZASP antibody do not appear to be cross-reactions to bacterial proteins, as these bands were not removed by preadsorption of the antiserum with an acetone powder of the bacteria used for the recombinant protein production (data not shown). It is possible that the extra bands seen at low dilutions could be due to cross-reactions with alternative forms of the ZASP protein, or with other PDZ-containing proteins present in muscle, e.g., ALP (39 kD) and the syntrophins (58–60 kD). It is interesting to note that the complex pattern of proteins seen in Western blotting with low dilutions of polyclonal antiserum (Fig. 3 B) was also seen with ZASP mAb (data not shown).
Since ZASP does not contain any cysteine residues, the higher forms, seen when using more protein and a higher concentration of antisera, are unlikely to be due to homodimer formation. Also, the proteins were run on SDS-PAGE under denaturing conditions in the presence of high amounts of DTT. The adsorption of ZASP antisera with either the 32 or 78 kD protein had only the effect of reducing the affinity of the preadsorbed antisera for both proteins, not one in particular (data not shown), with the ratio of the proteins remaining the same, thus suggesting that the anti-ZASP antibody recognizes an epitope, which is also present in the 78-kD protein.
To have an estimate of the level of ZASP present in muscle tissue, we employed a method (Valle et al. 1997) based on the intensity of the signal obtained from Western blot analysis to determine the relative amount of a specific protein in tissue extracts. This procedure was used to estimate the amount of human α-actin (Fig. 4 A) and ZASP (Fig. 4 B) present in 2.5 and 10 μg of total heart and skeletal muscle protein, respectively. There is less ZASP present in heart than in skeletal muscle tissue: 4.5 ng as opposed to 18 ng in 10 μg of total proteins, which is 0.05 and 0.18%, respectively. From densitometric analysis of the bands, it can be calculated that the actin signal from 2.5 μg of total muscle proteins is approximately equivalent to 500 ng of recombinant α-actin. Therefore, using this method the percentage of actin present in heart and skeletal muscle would be ∼20%, which is in agreement with the percentage (19%) previously found in adult rabbit muscle (Pollard 1981). This method only gives an estimate of the amount of protein present in muscle tissues, based on two main assumptions: that proteins of the same size, blotted under the same conditions have the same rate of blotting; and that the antibody used has the same affinity for the native and recombinant proteins. However, within these limits it gives a reasonable approximation of the percentage of an unknown recombinant protein present in muscle tissue.
The pattern of expression of ZASP and its ability to coimmunoprecipitate other muscle proteins was studied using immunoprecipitation of total human muscle proteins obtained by in vitro translation of muscle mRNA. In Fig. 5B and Fig. C, for both adult and fetal proteins, immunoprecipitation using preimmune sera is shown in lane 1, anti-ZASP antibody in lane 2, and antimyosin antibody in lane 3. Both the 32 and 78 kD proteins could be immunoprecipitated from in vitro translated proteins of adult and fetal skeletal muscle using the ZASP antibody (Fig. 5 B, lane 2). Also, from in vitro translated fetal skeletal muscle, three other proteins of ∼43, 50, and 72 kD could be immunoprecipitated (Fig. 5 B, lane 2). The 32-kD ZASP protein was immunoprecipitated, along with proteins of 14.3, 40, and 50 kD, from total adult heart proteins using anti-ZASP antibody (Fig. 5 C, lane 2), whereas in fetal heart (Fig. 5 C, lane 2) only proteins of 26, 40, and 50 kD could be detected. Therefore, it would appear that the 32-kD protein is not detectable in fetal heart. It is clear that there is more ZASP present in skeletal muscle than in heart, this data being confirmed by both Western blot analysis and these immunoprecipitation experiments. The amount of ZASP immunoprecipitated from in vitro translated fetal skeletal muscle was much less than that from adult, which would indicate that ZASP is present at very low levels in fetal tissue. This variation was not due to poor translation of fetal mRNAs, as can be seen by the total protein obtained from in vitro translation (Fig. 5 A), for both heart and skeletal muscle.
Myosin mAb was used in the immunoprecipitation experiments as a positive control. It immunoprecipitated a protein of ∼220 kD from the in vitro translated adult and fetal skeletal muscle proteins, as well as from adult and fetal heart proteins. Both myosin and ZASP antibodies immunoprecipitated a protein of 14.3 kD from adult heart proteins.
Localization of ZASP Protein in Human Muscle Cells
Immunofluorescence experiments were undertaken in primary human myoblasts (Fig. 6 A) and myotubes (Fig. 6 B), as well as skeletal (Fig. 6 C) and heart muscle tissues, with the scope of detecting the intracellular localization of the ZASP protein. A fluorescence signal can be detected in some, but not all, of the primary undifferentiated muscle cells incubated with ZASP antibodies. The fluorescence is usually restricted to the pseudopodia and to the area of the cytoplasm around the nuclei (Fig. 6 A), where it can be seen as strongly fluorescing dots. The fluorescence intensity in individual cells may be similar in differentiated and undifferentiated cells, but the strong fluorescence seen in the undifferentiated cells is restricted to a small percentage of the total cells (5–10%), whereas in differentiated cells, the percentage is much higher (90%). In differentiated cells incubated with antibodies to ZASP, a fluorescent signal can be detected in nearly all of the cells and it is especially strong throughout the myotubes (Fig. 6 B). In these cells, cross-striations can be seen that are reminiscent to those seen in tissue sections incubated with ZASP antibodies (Fig. 6 C). However, there are also cells that show a pattern of strongly fluorescing dots similar to those seen in undifferentiated cells, and these may in fact be cells in the early stages of differentiation. A weak fluorescent signal can be detected in undifferentiated and differentiated cells incubated with preimmune serum (Fig. 6A and Fig. B), as well as undifferentiated cells incubated with myosin (Fig. 6 A). However, in differentiated cells incubated with myosin (Fig. 6 B), strong fluorescence can be detected in the cytoplasm near the nuclei and as cross-striations throughout the myotubes.
Figure 6.
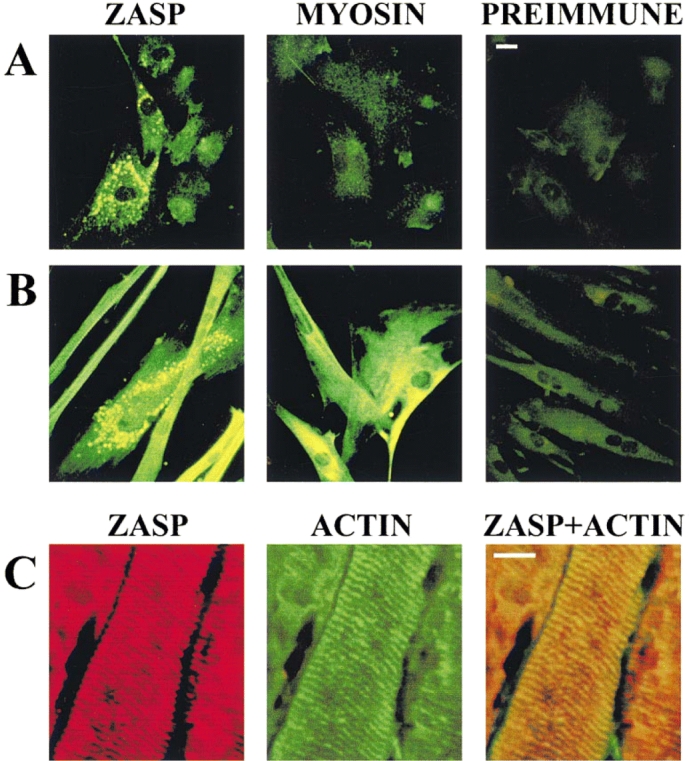
Indirect immunofluorescence of undifferentiated human myoblasts (A) and differentiated human myotubes (B). Preimmune sera and ZASP pAb were used at a dilution of 1/50; myosin mAb (MF 20) was used at 1/100 dilution. FITC-conjugated anti-mouse immunoglobulin (Sigma Chemical Co.) was used as the second antibody. (C) Indirect immunofluorescence of skeletal muscle tissue sections using mouse pAb to ZASP (1/30 dilution) and rabbit antiactin antibody (1/40). FITC-labeled goat anti–rabbit Ig (green) and TRITC-labeled goat anti–mouse Ig (red) were used as second antibodies. Bars, 10 μm.
In tissue sections of human heart (not shown) and skeletal muscle (Fig. 6 C), an alternate banding pattern could be detected by indirect immunofluorescence experiments using antibodies to ZASP (red) and actin (green). From double fluorescence experiments, the ZASP and actin signals seem to be coincident, as seen in Fig. 6 C, which would suggest that the ZASP protein is present in the I-band.
Immunoelectron microscopy of heart and skeletal muscle tissue sections demonstrated that ZASP is located within the Z-band, as can be seen in Fig. 7A and Fig. B, the latter showing a higher magnification of the same section. Therefore, ZASP would appear to be present throughout the Z-band.
Figure 7.
Localization of ZASP in human skeletal muscle by immunoelectron microscopy. Immunoelectron microscopy with ZASP pAb as the primary antibody and anti-mouse IgG whole molecule conjugated with 5-nm gold particles (Sigma Chemical Co.; G7527) as the secondary antibody. (A) Low magnification of a section of skeletal muscle. The gold particles were detected in the Z-band. (B) Higher magnification of the same image, rotated 90°. The ZASP protein is seen throughout the Z-band. Bars, 0.1 μm.
Characterization of Alternative Forms of ZASP
The full-length cDNA sequence of ZASP was used to search for similar sequences in the Genbank/EMBL/DDBJ databases. Two regions of ZASP (322 and 203 bp) were found to be identical to KIAA0613, as shown in Fig. 8. As mentioned previously, KIAA0613 is a sequence obtained from systematic sequencing of a brain library (Ishikawa et al. 1998). Interestingly, the 3′ end region of KIAA0613 matches perfectly with a cluster of ESTs from the 3′ end skeletal muscle catalogue mentioned above. This cluster is referred to as HSPD1333 and contains four ESTs, which are found with a frequency of 0.012% (about five times less than ZASP). Therefore, we decided to investigate further the following two points: is the KIAA0613 actually expressed in muscle, as the 3′ end tag would indicate? And, are ZASP and KIAA0613 two alternatively spliced forms encoded by the same gene?
Figure 8.
Schematic representation of the ZASP transcript, two alternative muscle variants, and brain transcript KIAA0613. Boxes (not always in scale) represent the coding region and the numbers inside each box indicate the length in bases. The 3′ and 5′ untranslated regions are represented by the terminal lines. The original KIAA0613 sequence begins just a few bases in front of the first ATG starting codon. However, the box has been marked as 322 bases long to facilitate the interpretation.
To verify whether the entire KIAA0613 is actually expressed in muscle, we screened by PCR our full-length cDNA library of skeletal muscle, using two primers designed respectively on the 5′ and 3′ end of the KIAA0613 sequence. As a result, we obtained two variant bands that were sequenced, neither of which corresponded to the KIAA0613 sequence (Fig. 8). An identical result was obtained from a heart library. Therefore, we do not have any evidence that KIAA0613 is expressed in skeletal muscle or in heart. However, the two variant transcripts that were identified give further support and complexity to the idea of alternative splicing. In the schematic view presented in Fig. 8, it can be seen that the four transcripts are composed of different combinations of ten fragments. The perfect identity of these fragments in the four transcripts, and the way that they are assorted, is compatible with the hypothesis of alternative splicing.
To address more specifically whether these transcripts could have originated by alternative splicing from the same gene, we amplified human genomic DNA using a forward oligo designed on box 365 (see Fig. 8), and a reverse oligo designed on the 203-bp box. As a result, a band >10,000 bases was obtained (data not shown). This band was used as a template for a PCR reaction, directed by primers specific for box 197 of ZASP, giving an amplified fragment identical to that of a control performed on genomic DNA. This result confirms the hypothesis of alternative splicing and indicates that the putative exon corresponding to box 197 of ZASP is located after the exon with box 365 of KIAA0613.
The 3′ end region of KIAA0613 was analyzed by the radiation hybrid technique and found to map at 10q22.3-10q23.2, the same position of the 3′ end region of ZASP.
The PDZ Domain of ZASP Interacts with α-Actinin-2
To identify muscle proteins, which could bind to the PDZ domain at the NH2-terminal of ZASP, three cDNA libraries were screened by the yeast two-hybrid system.
The segment consisting of the first 321 coding bases of ZASP was subcloned into the pHybridLex/Zeo vector as a bait and transformed into L40 yeast strain. Then, 2,500,000 transformants were screened from various muscle libraries: 280,000 clones from the pGAD10 human skeletal muscle library (pGAD10S), 600,000 clones from the pGAD10 human heart library (pGAD10H), and 1,750,000 clones from the pDisplayTarget human heart library (pDTH).
Growing clones were picked from the different libraries: 17 clones from the pGAD10S, 9 clones from the pGAD10H, and 87 clones from pDTH, and their interactions confirmed with the β-galactosidase filter assay. The inserts associated with the activation domains of the positive clones were directly amplified from yeast cells by PCR and 30 were sequenced. The inserts of 23 clones were identified as fragments of the α-actinin-2 gene (Beggs et al. 1992), whereas the other seven clones matched mitochondrial genes and transcription factors, typical false positives of the yeast two-hybrid system.
All the clones containing α-actinin-2 cDNA, although they start from different positions, extend to the end of the coding region, as shown in Fig. 9. The region of α-actinin-2 binding to the PDZ domain of ZASP can be inferred from the clones containing the shortest cDNA inserts that have only the final 155 amino acids of the COOH-terminal region of the α-actinin-2 protein.
Figure 9.
Schematic representation of the positive α-actinin-2 clones found using the yeast two-hybrid system. The domains of α-actinin-2 are shown at the top, whereas the remaining part of the figure shows the coding regions of the α-actinin-2, which are involved in a positive interaction with the PDZ domain of ZASP. The numbers at the side of the bars indicate the starting amino acid. The coding regions of all the α-actinin-2 clones reach the true stop codon of the protein at position 894.
Discussion
From the data presented in this paper, it is evident that in human heart and skeletal muscle there are at least three different forms of ZASP, derived by alternative splicing. A fourth form, also derived by alternative splicing, is present in brain and was previously described as KIAA0613 (Ishikawa et al. 1998). Furthermore, an EST obtained from human fetal lung (GenBank/EMBL/DDBJ accession number AI193732) indicates the presence of a transcript in which the boxes of 322 and 203 bp of Fig. 8 are joined together. The presence of more variants of this transcript in other tissues is suggested by Northern blot analysis. For instance, in Fig. 2 A, it can be seen that using the 3′ end region of ZASP as a probe, a very strong signal is also found in pancreas, producing a band smaller than that seen in muscle. A faint band of ∼1 kb can be detected also in placenta. Similarly, using a probe specific for the 322-bp box, several strong bands can be seen in muscle and heart (Fig. 2 D). After overexposure (data not shown), weak bands of different sizes can be seen in brain, small intestine, and placenta. These data suggest a complex case of alternative splicing, producing a wide variety of transcripts in different tissues, also confirmed at the protein level, as shown in Fig. 3 B.
To approach the issue of the possible function of these alternatively spliced transcripts, we tried to dissect the corresponding putative proteins on the basis of their alternatively assorted fragments, as well as on any recognizable functional domains. The only domain that is common to the different forms, that so far have been sequenced, is the PDZ domain at the NH2 terminus, which is fully encoded by the 322-base box shown in Fig. 8. As it was said in the introduction, the PDZ domain is generally engaged in protein–protein interaction. To further investigate this issue, we performed a yeast two-hybrid assay and discovered that the PDZ domain of ZASP is interacting with the region extending across 150 COOH-terminal amino acids of the Z-band protein α-actinin-2. This latter protein is known to bind other Z-band proteins such as titin (Ohtsuka et al. 1997; Sorimachi et al. 1997; Young et al. 1998) and ALP (Xia et al. 1997). Titin has been reported to bind the region spanning the final 73 COOH-terminal amino acids of α-actinin-2 (Ohtsuka et al. 1997; Sorimachi et al. 1997), therefore, both ZASP and titin seem to bind α-actinin-2 around the same region, near the COOH terminus. However, recently it has been reported that titin can also interact with the central spectrin-like repeat of α-actinin-2, as well as with the COOH-terminal domain (Young et al. 1998). The analogy between ZASP and ALP in binding α-actinin-2 is also very interesting, as the PDZ domain of ALP is one of the most similar to the PDZ domain of ZASP, sharing 53/84 identical residues. Therefore, it should not be surprising that both of these PDZ domains bind α-actinin-2. However, it is strange that the PDZ of ALP has been reported (Xia et al. 1997) to bind the spectrin-like domains 2 and 3 of α-actinin-2 (see map on Fig. 9), whereas the PDZ domain of ZASP binds to the COOH-terminal region. Although we cannot exclude that the PDZ domain of ZASP may also bind the spectrin-like domains of α-actinin-2, this observation remains puzzling.
A second fragment that can be functionally dissected is that of 1,061 bp (see Fig. 8), which encodes three LIM domains, as detected by the program SMART (Schultz et al. 1998). The LIM domains are also involved in protein–protein interaction. None of the other fragments are similar to any known functional domain.
PDZ and LIM domains are often found associated on the same proteins. In fact, using the program BLASTP (Altschul et al. 1997) we searched the NCBI nonredundant protein databases for similarity to the PDZ domain of ZASP and we found that the 15 best hits (53–63% of identities on an 80–84 amino acid overlap) are to PDZ domains belonging to proteins containing LIM domains at the COOH terminus. Some of these proteins have only a single LIM domain, such as rat CLP36 (GenBank/EMBL/DDBJ accession number U23769); human and mouse CLIM1 (GenBank/EMBL/DDBJ accession numbers U90878 and AF053367); mouse, human, and rat RIL (GenBank/EMBL/DDBJ accession numbers Y08361, X93510, and X76454); and human, mouse, and rat ALP (GenBank/EMBL/DDBJ accession numbers AF002280 to AF002283). The remaining proteins have an organization similar to the variant forms of ZASP, in which the PDZ domain is followed by three LIM domains (see Fig. 8). These proteins are the LIM protein itself (GenBank/EMBL/DDBJ accession AF061258), human and rat Enigma (GenBank/EMBL/DDBJ accession numbers L35241 and U48247), and rat LMP1 (GenBank/EMBL/DDBJ accession number AF095585).
A similar result was obtained when the sequence containing the three LIM domains of the variant forms of ZASP was used as a query for a BLASTP search. The best hits returned were the same Enigma, LIM, and LPM1 proteins seen above, showing alignments of 174–178 amino acids with a percentage of identity ranging from 58% to 67%. None of the other boxes of Fig. 8 show any significant similarity to known proteins, with the exception of the 197-bp box that shows similarity (45% identity on a 51 amino acids overlap) with the skeletal muscle isoforms of ALP. In fact, ALP is found as two isoforms derived from alternative splicing.
Another intriguing feature is the presence in the 203-bp box (which is common to all the ZASP isoforms shown in Fig. 8) of a conserved stretch of 15 amino acids (QSRSFRILAQMTGTE), which is also found in the human LIM and in rat Enigma proteins.
A suggestive speculation is that, in general, these PDZ/LIM proteins could act as some kind of adapters (Cuppen et al. 1998). The data presented in this paper give further support to this hypothesis, adding an extended degree of modularity by means of an intricate alternative splicing. In this scheme, the idea of ZASP as a truncated form of a PDZ/LIM adapter does not exclude the possibility that this protein may act as a competitor against the two other variant forms found in muscle, which have the LIM domains.
Acknowledgments
We would like to thank Francesca Dalla Vecchia (University of Padua, Italy) for her help and advice with immunoelectron microscopy experiments. Thanks are also due to Rosanna Zimbello (University of Padua, Italy) for DNA sequencing, and Mauro Sturnega and GianCarlo Lunazzi (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy) for excellent technical assistance in the immunization of animals used for antibody production.
This work was supported by Fondazione Telethon (Italy) grant number 1023 (to G. Faulkner) and grant number B.41 (to G. Lanfranchi and G. Valle).
Footnotes
1.used in this paper: ALP, actinin-associated LIM protein; EST, expressed sequence tag; pAb, polyclonal antibody; ZASP, Z-band alternatively spliced PDZ motif
Georgine Faulkner and Alberto Pallavicini contributed equally to this work.
References
- Adams M.E., Butler M.H., Dwyer T.M., Peters M.F., Murnane A.A., Froehner S.C. Two forms of mouse syntrophin, a 58-kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron. 1993;11:531–540. doi: 10.1016/0896-6273(93)90157-m. [DOI] [PubMed] [Google Scholar]
- Agatep, R., R.D. Kirkpatrick, D.L. Parchaliuk, R.A. Woods, and R.D. Gietz. 1998. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol protocol. Technical Tips Online. (http://tto.biomednet.com) 01525.
- Altschul S.F., Madden T.S., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLASTa new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience; New York: 1994. [Google Scholar]
- Bateman A., Birney E., Rubin R., Eddy S.R., Finn R.D., Sonnhammer E.L. Pfam3.11313 multiple alignments match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs A.H., Byers T.J., Knoll J.H., Boyce F.M., Bruns G.A., Kunkel L.M. Cloning and characterization of two human skeletal muscle α-actinin genes located on chromosomes 1 and 11. J. Biol. Chem. 1992;267:9281–9288. [PubMed] [Google Scholar]
- Brenman J.E., Bredt D.S. Synaptic signaling by nitric oxide. Curr. Opin. Neurobiol. 1997;7:374–378. doi: 10.1016/s0959-4388(97)80065-7. [DOI] [PubMed] [Google Scholar]
- Brenman J.E., Chao D.S., Xia H., Aldape K., Bredt D.S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- Brenman J.E., Chao D.S., Gee S.H., McGee A.W., Craven S.E., Santillano D.R., Huang F., Xia H., Peters M.F., Froehner S.C. Interaction of nitric oxide synthase with postsynaptic density protein PSD-95 and α-1-syntrophin mediated by PDZ motifs. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- Cho K.O., Hunt C.A., Kennedy M.B. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- Cuppen E., Gerrits H., Pepers B., Wieringa B., Hendriks W. PDZ motifs in PTP-BL and RIL bind to internal protein segments in the LIM domain protein RIL. Mol. Biol. Cell. 1998;9:671–683. doi: 10.1091/mbc.9.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Bucher P., Falquet L., Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K., Nagase T., Suyama M., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro . DNA Res. 1998;5:169–176. doi: 10.1093/dnares/5.3.169. [DOI] [PubMed] [Google Scholar]
- Kennedy M.B. Origin of PDZ (DHR, GLGF) domains. Trends Biochem. Sci. 1995;21:350. doi: 10.1016/s0968-0004(00)89074-x. [DOI] [PubMed] [Google Scholar]
- Kim E., Niethammer M., Rothschild A., Jan Y.N., Sheng M. Clustering of Shaker-type K+ channels by direct interaction with the PSD-95/SAP90 family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kornau H.-C., Schenker L.T., Kennedy M.B., Seeburg P.H. Domain interactions between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Kornau H.-C., Seeburg P.H., Kennedy M.B. Interaction of ion channels and receptors with PDZ domains. Curr. Opin. Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- Lanfranchi G., Muraro T., Caldara F., Pacchioni B., Pallavicini A., Pandolfo D., Toppo S., Trevisan S., Scarso S., Valle G. Identification of 4,370 expressed sequence tags from a 3′-end-specific cDNA library of human skeletal muscle by DNA sequencing and filter hybridization. Genome Res. 1996;6:35–42. doi: 10.1101/gr.6.1.35. [DOI] [PubMed] [Google Scholar]
- Mayans O., van der Ven P., Wilm M., Mues A., Young P., Fürst D.O., Wilmanns M., Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- Mues A., van der Ven P.F.M., Young P., Fürst P.D.O., Gautel M. Two immunoglobulin-like domains of the Z-disc portion of titin interact in a conformation-dependent way with telethonin. FEBS Lett. 1998;428:111–114. doi: 10.1016/s0014-5793(98)00501-8. [DOI] [PubMed] [Google Scholar]
- Nakai K., Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M., Kim E., Sheng M. Interaction between the C-terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J. Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka H., Yajima H., Maruyama K., Kimura S. The N-terminal Z repeat 5 of connectin/titin binds to the C-terminal region of α-actinin. Biochem. Biophys. Res. Commun. 1997;235:1–3. doi: 10.1006/bbrc.1997.6534. [DOI] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. Cytoplasmic contractile proteins. J. Cell Biol. 1981;91:156s–165s. doi: 10.1083/jcb.91.3.156s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Phillips C. DHR domains in syntrophins, neuronal NO synthase and other intracellular proteins. Trends Biol. Sci. 1995;20:102–103. doi: 10.1016/s0968-0004(00)88973-2. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C., Schneider R. PHDan automatic mail server for protein secondary structure prediction. CABIOS. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Sato T., Irie S., Kitada S., Reed J.C. FAP-1a protein tyrosine phosphatase that associates with FAS. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- Schepens J., Cuppen E., Wieringa B., Hendricks W. The neuronal nitric oxide synthase PDZ motif binds to −G(D < E)XV* carboxy-terminal sequences. FEBS Lett. 1997;409:53–56. doi: 10.1016/s0014-5793(97)00481-x. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research toolidentification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M. PDZs and receptor/channel clusteringrounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Shieh B.-H., Zhu M.Y. Regulation of the TRP Ca2+ channel by INAD in Drosophila photoreceptors. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Sorimachi H., Freiburg A., Kolmerer B., Ishiura S., Stier G., Gregorio C.C., Labeit D., Linke W.A., Suzuki K., Labeit S. Tissue-specific expression and α-actinin binding properties of the Z-disc titinimplications for the nature of vertebrate Z-discs. J. Mol. Biol. 1997;270:668–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- Stricker N.L., Christopherson K.S., Yi B.A., Schatz P.J., Raab R.W., Dawes G., Basset D.E., Jr., Bredt D.S., Li M. PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences as determined by in vitro selection. Nat. Biotechnol. 1997;15:336–342. doi: 10.1038/nbt0497-336. [DOI] [PubMed] [Google Scholar]
- Valle G., Faulkner G., De Antoni A., Pacchioni B., Pallavicini A., Pandolfo D., Tiso N., Toppo S., Trevisan S., Lanfranchi G. Telethonin, a novel sarcomeric protein of heart and skeletal muscle. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- Walter M.A., Spillet D.J., Thomas P., Weissenbach J., Goodfellow P.N. A method for constructing radiation hybrid maps of whole genomes. Nat. Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- Woods D.F., Bryant P.J. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- Xia H., Winnnokur S.T., Kuo W.-L., Altherr M.R., Bredt D.S. Actinin-associated LIM proteinidentification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P., Ferguson C., Bañuelos S., Gautel M. Molecular structure of the sarcomeric Z-disktwo types of titin interactions lead to an asymmetrical sorting of α-actinin. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:1614–1624. doi: 10.1093/emboj/17.6.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]