Abstract
Using RNA-mediated genetic interference in a phenotypic screen, we identified a conserved nonmuscle myosin II regulatory light chain gene in Caenorhabditis elegans, which we name mlc-4. Maternally supplied mlc-4 function is required for cytokinesis during both meiosis and mitosis and for establishment of anterior-posterior (a-p) asymmetries after fertilization. Reducing the function of mlc-4 or nmy-2, a nonmuscle myosin II gene, also leads to a loss of polarized cytoplasmic flow in the C. elegans zygote, supporting models in which cytoplasmic flow may be required to establish a-p differences. Germline P granule localization at the time of cytoplasmic flow is also lost in these embryos, although P granules do become localized to the posterior pole after the first mitosis. This result suggests that a mechanism other than cytoplasmic flow or mlc-4/nmy-2 activity can generate some a-p asymmetries in the C. elegans zygote. By isolating a deletion allele, we show that removing zygotic mlc-4 function results in an elongation phenotype during embryogenesis. An mlc-4/green fluorescent protein transgene is expressed in lateral rows of hypodermal cells and these cells fail to properly change shape in mlc-4 mutant animals during elongation.
Keywords: microfilament proteins, RNA-mediated interference, nonmuscle myosin II regulatory light chain, P granules, morphogenesis
Sperm entry appears to provide the initial positional cue responsible for establishing anterior-posterior (a-p)1 polarity in the Caenorhabditis elegans zygote, possibly by directing cytoplasmic movements shortly after fertilization (Goldstein and Hird 1996). During the pronuclear stage, a polarized cytoplasmic flow occurs within the zygote based on the position of the sperm pronucleus and associated centrosomes: cytoplasm near the cortex flows away from, whereas internal cytoplasm flows towards, the paternal pronuclear–centrosomal complex. If the paternal pronucleus is located at a lateral position, this cytoplasmic flow appears able to move the paternal pronucleus to one end of the oblong zygote with its position defining the posterior pole and, therefore, the a-p axis of the embryo (Goldstein and Hird 1996). In addition, ribonucleoprotein structures called P granules, initially present throughout the oocyte, become localized to the posterior pole during this period. Treatment of embryos with cytochalasin D to disrupt actin microfilaments blocks polarized cytoplasmic flow, prevents P granule segregation, and causes other losses in a-p asymmetry (Hill and Strome 1988; Hird et al. 1996). The critical period of microfilament disruption that leads to losses in a-p asymmetry coincides with the time of cytoplasmic flow and posterior localization of P granules (Strome and Wood 1983; Hill and Strome 1988; Hird et al. 1996). These results suggest that a single mechanism of microfilament-mediated cytoplasmic flow could suffice to generate all a-p asymmetry after fertilization, although it is also possible that multiple microfilament-dependent functions are involved.
Later in C. elegans embryogenesis, the microfilament cytoskeleton plays a role in establishing the morphology of the hatching larva (Priess and Hirsh 1986). During the latter half of embryonic development, after completion of most embryonic cell divisions, the embryo undergoes a dramatic shape change (Sulston et al. 1983). The embryo contracts circumferentially and elongates dramatically along the a-p axis before hatching. Laser ablation and drug studies have shown that the microfilament cytoskeleton within hypodermal cells is needed for this elongation process (Priess and Hirsh 1986). After hypodermal cells enclose the embryo, their microfilament bundles become circumferentially arranged. Contractile forces generated by these bundles appear to cause elongation along the a-p axis, perpendicular to the microfilament bundles.
Because the microfilament cytoskeleton is crucial for establishing cell polarity and for morphology, understanding the regulatory mechanisms that converge on the cytoskeleton is of fundamental importance in understanding how these processes are controlled. Genetic screens for early regulators of pattern formation in C. elegans have identified some factors that likely interact with the microfilament cytoskeleton during development. For example, six par genes (for partitioning defective) have been identified that are required for some aspects of a-p polarity in the fertilized zygote (for review see Kemphues and Strome 1997). The par genes encode proteins that colocalize with actin microfilaments in the cortical regions of the cytoplasm. Moreover, the serine/threonine kinase PAR-1 has a COOH-terminal domain that binds a nonmuscle myosin II heavy chain (NMY-2) present in early embryos (Guo and Kemphues 1996). Consistent with this interaction, NMY-2 is required for proper a-p polarity and for cytokinesis in the early embryo (Guo and Kemphues 1996). Though the molecular identities of the other par genes are known, the mechanisms through which they regulate a-p polarity remain unclear.
Genetic analyses also have identified genes that may regulate the hypodermal microfilament cytoskeleton during embryonic elongation. Mutational and expression analyses of the let-502 gene, which encodes a protein similar to rho-associated kinases, have demonstrated its role in the proper elongation of the embryo (Wissmann et al. 1997). In addition, genetic interactions are seen between let-502 and mel-11, which encodes a myosin phosphatase subunit. These genes have been postulated to interact with and regulate the microfilament cytoskeleton in hypodermal cells, likely through a nonmuscle myosin regulatory light chain (nmRLC; Wissmann et al. 1997). Other genes required for embryonic elongation encode an α- and a β-catenin and a cadherin, which are required for hypodermal cell migrations and for anchoring the actin cables at hypodermal cell junctions (Costa et al. 1998).
To identify genes that regulate the microfilament cytoskeleton and cell polarity in the early embryo, we took advantage of a technique called RNA-mediated genetic interference, or RNAi (Fire et al. 1998) to systematically assay the function of embryonic transcripts. Using this method, we identified a gene, mlc-4, which appears to encode the sole regulatory light chain for nonmuscle myosin in C. elegans. nmRLCs are key regulators of myosin function and phosphorylation of nmRLC by several regulatory kinases leads to the assembly and increased ATPase activity of myosin (for review see Bresnick 1999). We show that mlc-4 is required for at least three different processes in the C. elegans embryo. Maternal expression of mlc-4 is required for cytokinesis and for some, but not all, aspects of a-p polarity. A zygotic role for mlc-4 was revealed through our isolation of a deletion that almost entirely removes the mlc-4 locus. Embryos homozygous for this deletion allele, mlc-4(or253), fail to fully elongate during embryonic morphogenesis. This elongation defect appears to result from a defect in the ability of a subset of hypodermal cells, called seam cells, to change shape during embryonic elongation. We suggest that the let-502/mel-11 regulatory pathway interacts with MLC-4 to generate contractile forces required for proper elongation of the embryo.
Materials and Methods
Early Embryo cDNA Library Construction and RNAi Screening
30 embryos younger than the 12-cell stage were devitellinized using chitinase and chymotrypsin (Edgar 1995; Shelton and Bowerman 1996) and lysed in reverse transcription buffer (Dulac and Axel 1995) with a small bore pipette. cDNA was synthesized as in Dulac and Axel, 1995. cDNA was partially digested with Tsp509 (New England Biolabs Inc.), ligated into Lambda Zap II (Stratagene), and packaged with Gigapack Gold (Stratagene). The resulting library was plated at low density and individual plaques were picked into the wells of a 96-well microtiter plate containing 200 μl of SM (100 mM NaCl, 20 mM Tris, pH 7.4, and 1 mM MgCl2). 2.5 μl of the suspended phage was transferred to PCR reactions, also in 96-well microtiter plates, and T3 and T7 primer sequences were used to amplify the insert contained in the phage plaque using PCR. Aliquots from four different PCR reactions were pooled and phenol was extracted. The insert DNA was ethanol precipitated and resuspended in a T7 transcription reaction (40 mM Tris-HCl, pH 8.0, 6 mM MgCl2, 10 mM DTT, 2 mM spermidine, 2 mM each ribonucleotide, 20 U RNase inhibitor [Promega], 50 U T7 RNA polymerase [Stratagene] incubated 2 h at 37°C).
The synthesized RNA was phenol extracted and ethanol precipitated. RNA from the transcription reactions was resuspended to a concentration of ∼1 mg/ml. RNA from pools of four inserts (see above) were combined pairwise to form pools of RNA from eight different inserts. RNA was injected into both gonad arms of four N2 hermaphrodites per pool of eight. Healthy injected animals were transferred to fresh media plates daily and their eggs were monitored for hatching. 1,024 inserts were assayed in 139 RNA pools. 24 h after injection, 13 pools resulted in sterility of the injected animals, 45 pools resulted in 100% penetrant embryonic lethality in the brood of injected animals, and 7 pools gave a partially penetrant embryonic lethality. Phenotypes of embryos from injected animals were examined under high power (630×) using a Zeiss Axioskop and were categorized as follows: defects in early cleavages (14 pools), defects in proliferation (21 pools), and defects in patterning and/or morphogenesis (17 pools). Early embryonic phenotypes were examined by cutting open injected animals and mounting the embryos on an agar pad (Sulston et al. 1983), whereas terminal defects were examined from embryos washed from the media plates.
Identification of the Gene Associated with an RNAi Phenotype
Upon identification of an RNA pool with an RNAi phenotype, the two pools of RNA from four inserts that constituted the pool of eight were injected individually into four N2 hermaphrodites. Once the pool of RNA from four inserts with the RNAi phenotype was identified, RNA was synthesized and injected from each individual PCR-amplified insert to identify the individual insert that was responsible for the RNAi phenotype. The sequence of the insert was determined by sequencing the PCR-amplified insert at the University of Oregon DNA Sequencing Facility, using an ABI 377 Prism automated fluorescent sequencer (Perkin-Elmer Corp.). The insert sequence was used in a BLAST analysis at the National Center for Biotechnology Information or at the C. elegans Genome Project at the Sanger Center to identify the corresponding gene from data released by the C. elegans Sequencing Consortium.
RNA Interference of the mlc-4 Locus
Detailed analysis of the mlc-4(RNAi) phenotype was performed by injecting double stranded RNA (dsRNA) prepared using T7 and T3 RNA polymerases (Stratagene) in separate reactions on the same template DNA. The complementary strands were mixed and injected. Although dsRNA was used, the phenotypic effect of impure single strand RNA and dsRNA preparations were indistinguishable for mlc-4 (impure preparations of single strand RNA have been shown previously to be effective in RNAi experiments [i.e., Guo and Kemphues 1996; Fire et al. 1998]). The mlc-4(RNAi) phenotype described in Results appeared fully penetrant and maximally expressive among the progeny of animals cultured 24 h at 22°C after injection into both gonad arms (In one experiment, no attempt at cytokinesis was seen in 81 out of 81 early embryos obtained from 11 healthy animals injected in both gonad arms 24 h earlier). We did not see evidence of sterility among injected animals, which differs from a deletion allele in mlc-4 (see Results). This suggests that mlc-4 function may not be completely removed by RNAi.
Analysis of Cortical Flow
Cortical flow was analyzed by time-lapse digital video microscopy. Data acquisition was accomplished using a Power Macintosh 8100 equipped with a Ludle focus drive controller and a Scion frame-grabber card. Using an algorithm written by C.A. Shelton, multiple focal planes were captured at consecutive timepoints. The captured frames were compiled into a fourth-dimensional movie using the Hardin 4D Player hypercard stack written by Jeff Hardin at the Department of Zoology at the University of Wisconsin and modified by C.A. Shelton. Individual yolk granules were followed in a time-lapse montage of the cortical surface of embryos and traced using Adobe Photoshop software. Spatial calibration was accomplished by imaging uniform 2.5-μm beads.
Immunocytochemistry
For immunocytochemical localization of microtubules, NMY-2, PAR-2, PAR-3, and P granules, the following embryo fixation was used: 10–15 wild-type or RNA-injected worms (24 h after injection) were cut open on a polylysinated slide. Using a coverslip and wicking action, the carcasses were flattened until embryos were visibly deformed. The slides were frozen on dry ice for 5 min, coverslips flicked off (freeze-cracked), and the slides submerged in room temperature methanol for 15 min. For P granule staining, the slides were allowed to dry in room air for 5 min. Slides were placed in PBS (140 mM NaCl, 2.7 mM KCl, 6 mM phosphate, pH 7.3) for 5 min. For immunocytochemical localization using the MH27 antibody and visualization of actin in postbean stage embryos and larvae, the following fixation was used after the freeze-crack step: 10 min in 3% paraformaldehyde, 100 mM NaPO4 buffer, pH 7.4, and 0.5 mM EDTA. Slides were blocked for at least 2 h in PBS plus 1% BSA. Primary antibody incubation was typically done overnight at 4°C.
For PAR-2 and PAR-3 staining, primary antibody incubation was done for at least 36 h at 4°C. Antibodies recognizing NMY-2, PAR-2, and PAR-3 were a gift of Ken Kemphues (Cornell University, Ithaca, NY) (Etemad-Moghadam et al. 1995; Boyd et al. 1996; Guo and Kemphues 1996). Antibodies recognizing P granules (OIC1D4) were a gift of Susan Strome (Indiana University, Bloomington, IN) (Strome and Wood 1983). After primary antibody incubation, slides were washed three times for 8 min in Tris-Tween (100 mM Tris-HCl, 1% Tween 20). Slides were incubated in rhodamine- or FITC-conjugated secondary antibody (Tago) for at least 2 h at room temperature. Slides were washed as before, incubated in PBS plus 10 ng/ml 4′6-diamidino-2-phenylindole (DAPI) for 2 min, washed in PBS for 1 min, and mounted in Slow-Fade medium (Molecular Probes Inc.). For visualization of filamentous actin (F-actin) in early embryos, embryos were prepared as above to the point of removing the coverslip. Immediately after coverslip removal, embryos were simultaneously fixed and stained by application of 200 μl of staining solution (75% methanol, 3.7% formaldehyde, and 330 nM rhodamine-conjugated phalloidin in 0.5× PBS; [Boyd et al. 1996]) for 20 min. Slides were washed, stained with DAPI, and mounted in Slow-Fade as above. Confocal microscopy was performed on a laser scanning microscope (model 310; Zeiss).
Generation of the mlc-4::GFP Expression Strain
PCR was used to generate a DNA fragment containing 1.2 kbp of sequence 5′ of the mlc-4 translational start site, the coding regions, and a Kpn I site in place of the stop codon in the final exon. This fragment was cloned into the Kpn I site of TU61, a gift of M. Chalfie (Columbia University, New York, NY) in which the lacZ fragment of pPD16.43 (Fire et al. 1990) has been replaced with green fluorescent protein (GFP; Chalfie et al. 1994). This plasmid was coinjected following the procedure of Mello and Fire (Mello et al. 1991) with a plasmid encoding rol-6(su1006) providing a dominant marker. Independent lines exhibiting identical expression patterns were obtained.
Generation of the mlc-4(or253) Deletion Allele and Genetic Manipulation
Nematode culture methods are described in Brenner 1974. The mlc-4(or253) deletion was isolated by essentially following the procedure of Jansen et al. 1997 except that primers were selected to give a final product of 1.8 kbp and the bank of mutagenized worms was generated differently: 500,000 F1 animals derived from 50,000 mutagenized P0 were distributed to 960 plates and allowed to grow for two generations. A portion of the animals was washed from each plate, DNA was extracted, and PCR analysis performed to identify plates with animals harboring a deletion allele. After several rounds of sibling selection and PCR detection, single worms were isolated harboring the deletion. The deletion allele was outcrossed to N2 Bristol wild-type worms five times and balanced over qC1. Animals from this strain produced essentially wild-type brood sizes with 26.5% (106/400) of the hatched larvae manifesting the phenotype illustrated in Fig. 7. No significant difference in hatching rate was seen between wild-type and mutant embryos. Genetic and PCR analyses of mutant and wild-type animals from such a brood indicate that the mutant phenotype is strictly correlated with a genotype homozygous for the deletion, indicating that the mlc-4(or253) allele is recessive and that the phenotype results from complete loss of mlc-4 function.
Figure 7.
Embryonic elongation phenotype of mlc-4(or253). (A) Nomarski photomicrograph of a wild-type embryo just before hatching (12 h at 22°C). (B) Photomicrograph of an mlc-4(or253) embryo of similar age as in A. Note that the mutant embryo has only elongated to the twofold stage, whereas the wild-type embryo in A has elongated to the threefold stage (stages as in Wood 1988). mlc-4(or253) embryos do not elongate further and hatch as shortened, thicker larvae. (C) Fluorescence micrograph of phalloidin staining of actin in a wild-type larva shortly after hatching. Two of four bands of bodywall muscles are shown (micrograph is the same magnification as A, but the larva is no longer folded into the eggshell). (D) Phalloidin staining of actin in an mlc-4(or253) larva. Two of four bands of bodywall muscles are shown. Although the muscles appear structurally similar to wild type, the larva is shorter and thicker than wild-type larva. (E) Fluorescence micrograph of MH27 antibody staining in a wild-type larva of similar age as A. The epitope recognized by MH27 marks the boundaries of hypodermal cells. The focal plane shows 7 of 10 seam cells present along one side of the larvae. (F) MH27 staining in an mlc-4(or253) embryo of similar age to A, B, and E. 8 of 10 seam cells along one side of the larva are shown. Note that the seam cell morphology in the mlc-4(or253) embryo lacks the thin, elongated, appearance of seam cells in the wild-type embryo. (G) MH27 staining in a bean stage embryo that harbors an mlc-4::GFP expression construct and is just beginning the elongation process. Shown are 8 of 10 seam cells. (H) Same embryo as in G, double stained with antibodies recognizing GFP. Note that the anti-GFP antibody stains the seam cells, as revealed by comparison to the MH27 staining in G. All photomicrographs are at equivalent magnification. Bar, 10 μm.
Results
Phenotypic Screening of Early Embryonic Transcripts
RNAi refers to the potent and specific ability of double-stranded exonic RNA, when microinjected into the syncytial ovary of an adult C. elegans or into syncytial Drosophila embryos, to reduce or eliminate the function of the corresponding gene (Fire et al. 1998; Kennerdell and Carthew 1998). RNAi phenocopies loss-of-function mutations (Rocheleau et al. 1997; Fire et al. 1998). In addition, RNAi can eliminate detectable protein expression (Guo and Kemphues 1996; Powers et al. 1998; Schumacher et al. 1998b; Skop and White 1998). Instead of using RNAi in a gene-directed approach, we used RNAi to systematically assay randomly selected embryonic transcripts for early embryonic functions.
We first constructed an embryonic cDNA library, using early embryos as a source of mRNA. In an initial screen of this library, 1,024 randomly chosen cDNA inserts were used as starting material for RNA microinjection. After microinjection of pooled RNAs into the syncytial ovary of adult C. elegans hermaphrodites, the injected animals and their progeny were monitored for RNAi-mediated defects during oogenesis and embryogenesis. The 1,024 inserts were assayed in 139 RNA pools and phenotypes were observed after injection of RNA representing 65 pools (see Materials and Methods). The phenotypes could be categorized in the following categories: sterility of the injected animals (13 pools), early cleavage defects among embryos of injected animals (14 pools), proliferation defects (21 pools), and morphogenesis/patterning defects (17 pools). After initial identification, pools were broken down by injecting RNA synthesized from the individual inserts composing a pool. Corresponding genes were identified by sequence analysis of the individual inserts that produced an RNAi defect. Genes identified included a number that have already been described, including the protein kinase air-1 (early cleavage class; Schumacher et al. 1998a), the p34 cdc2 homologue ncc-1 (early cleavage class; Mori et al. 1994), and the cell fate regulator pos-1 (morphogenesis/patterning class; Tabara et al. 1999). In addition, genes that have no previously described embryonic role were identified (5/10 identified genes in the early cleavage class).
Identification of a Myosin Light Chain Gene, mlc-4, Required at an Early Step during Embryonic Cytokinesis
Injected RNA from three cDNA inserts identified in the screen resulted in cytokinesis and polarity defects during the first cell cycle of C. elegans embryogenesis. All three inserts represent a single gene, identified as C56G7.1 in data from the C. elegans genome sequencing consortium (Consortium, 1998; GenBank accession number 3877857), predicted to encode an nmRLC. This gene, which we name mlc-4, codes for a protein 74% identical to the nmRLC protein encoded by the Drosophila spaghetti-squash gene (Karess et al. 1991) and is more similar to nmRLCs than the other two identified myosin regulatory light chains in the C. elegans genome, mlc-1 and mlc-2, both of which are only 47% identical to Spaghetti-squash (Rushforth et al. 1998). Sequence searches of the C. elegans genome and expressed sequence tag data revealed no additional C. elegans sequences with the potential to encode a myosin regulatory light chain (the mlc-3 gene encodes an essential myosin light chainlike protein). Because dsRNA derived from the mlc-1 and mlc-2 genes resulted in no early embryonic phenotype when injected either singly or together (data not shown), we believe that the phenotypic effects described here are specific to the mlc-4 gene and that mlc-4 may provide all nmRLC function in C. elegans. In addition, although RNAi may not completely eliminate all mlc-4 function, we found the phenotypes described here to be consistent and fully penetrant starting 24 h after injection of dsRNA corresponding to the mlc-4 locus.
Reducing mlc-4 function by RNAi resulted in a cytokinesis defect at both meiosis and mitosis (Fig. 1; see Materials and Methods). Shortly after fertilization in wild-type embryos, products of meiosis are extruded from the embryo in structures known as polar bodies. In mlc-4(RNAi) embryos, polar body extrusion after meiosis was initiated, but invariably failed, resulting in the retention of extra chromosomes that often form a nuclear structure (Fig. 1). During the first mitotic cell cycle, a mitotic spindle formed but no cytokinetic furrow was seen, resulting in a multinucleate, single cell embryo after the completion of mitosis (Fig. 1). Additional mitoses continued without cytokinesis (data not shown).
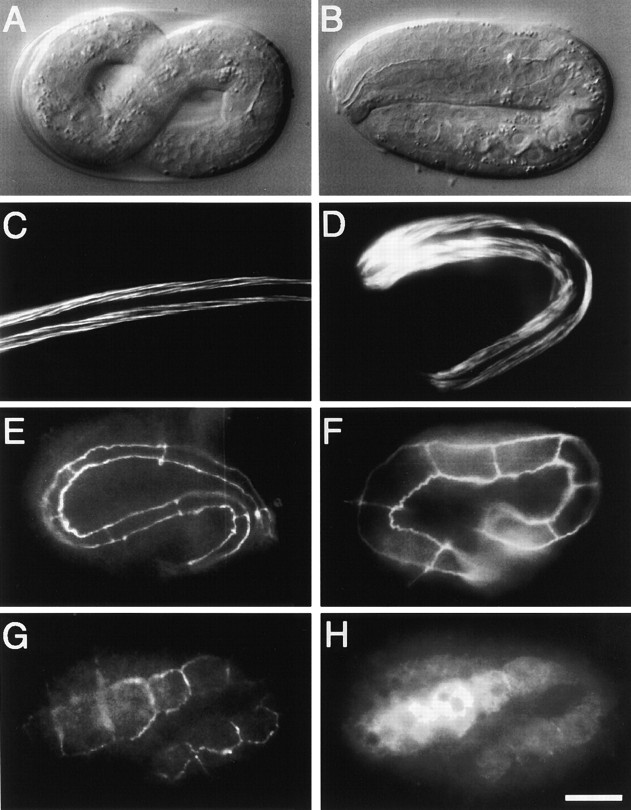
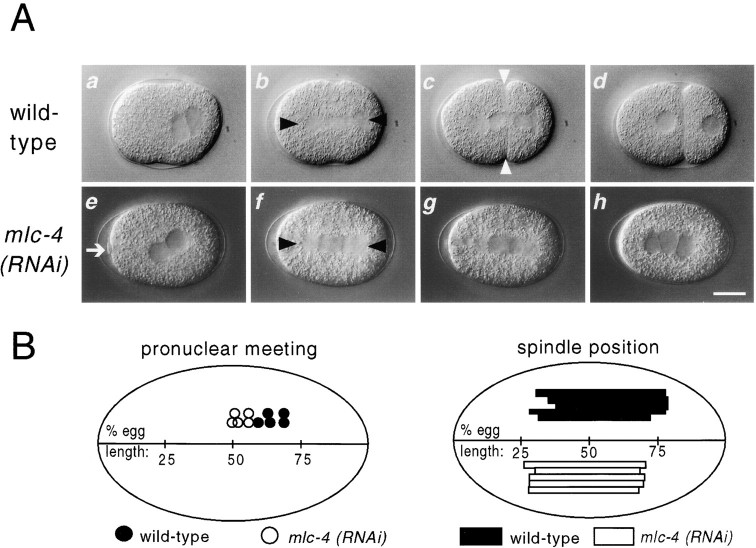
Figure 1.
RNAi phenotype of mlc-4. In this and later figures, embryos are oriented with anterior pole to the left and dorsal pole up. All photomicrographs are reproduced at equal magnification. (A) Defects during the first embryonic mitosis in mlc-4(RNAi) embryos. Black triangles mark the two poles of the mitotic spindle in each embryo; white triangles indicate the cytokinetic furrow in each embryo. (a–d) First mitosis in a wild-type embryo. Note the posterior displacement of pronuclear meeting (a), the mitotic spindle (b), and the resulting formation of two unequally sized cells (d). (e–h) First mitosis in an mlc-4(RNAi) mutant embryo. Pronuclei meet in the center of the embryo (e), polar body extrusion fails (e–g), a symmetrically placed spindle is formed (f), and no attempt at a cytokinetic furrow is seen (the same defects were seen in all mlc-4(RNAi) embryos examined from pronuclear migration through the first mitotic cell cycle, n > 10). An abortive attempt to form a polar body is seen in the first frame. This polar body regresses during the first mitosis and results in the formation of a supernumerary nuclear structure, as seen in the last frame. (B) The position of pronuclear meeting and first mitotic spindle position in five different wild-type and mlc-4(RNAi) embryos are illustrated. Compared with wild type, the positions of pronuclear congression and of the first mitotic spindle is symmetric in mlc-4(RNAi) embryos. Bar, 10 μm.
To investigate how mlc-4 inactivation affects the microfilament cytoskeleton, we examined the distribution of myosin and F-actin in mlc-4(RNAi) embryos. NMY-2 is a maternally expressed nonmuscle myosin II, identified previously as a regulator of embryonic polarity and as being required for embryonic cytokinesis (Guo and Kemphues 1996). In wild-type embryos, NMY-2 is cortically distributed throughout all early embryonic cells (Fig. 2 A). In mlc-4(RNAi) embryos, NMY-2 was still cortically localized but was dramatically enriched at sites where cytokinetic furrows were expected to form (Fig. 2 A). For example, NMY-2 was localized to a small ring at the apparent attempted site of polar body extrusion in mlc-4(RNAi) embryos. In addition, after the first mitotic cycle, a strong ring of cortical NMY-2 staining was seen in the portion of the cell cortex overlying the midzone of the mitotic spindle. During subsequent attempts at cytokinesis, bands of NMY-2 staining again accumulated at presumptive sites of cleavage furrow formation. Despite the strong accumulation of NMY-2 at sites of the cytokinetic furrow, little, if any, contractile force appeared to be generated as little or no ingression of the membrane was observed in mlc-4 (RNAi) embryos.
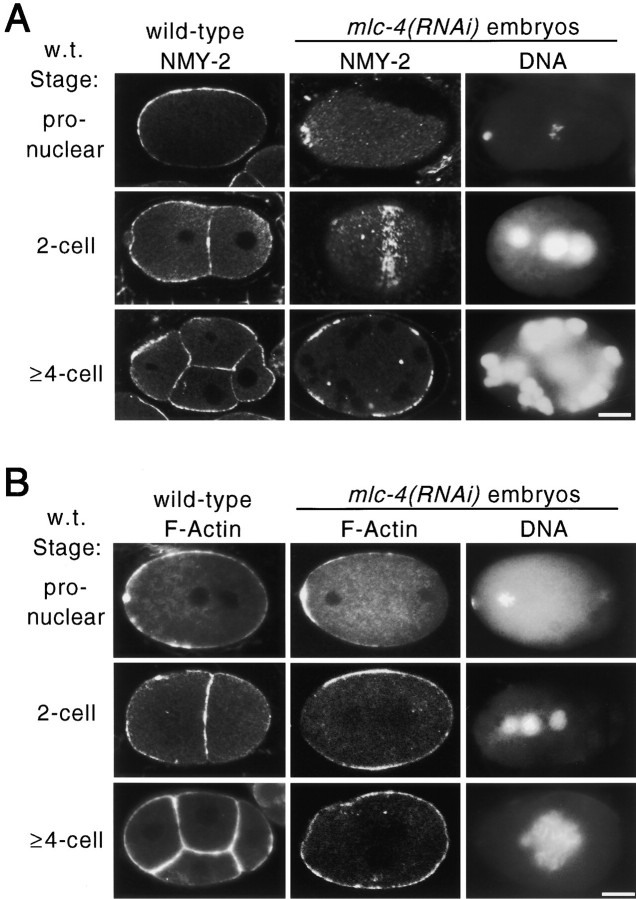
Figure 2.
Distribution of F-actin and nonmuscle myosin in wild-type and mlc-4(RNAi) embryos. Images were obtained by confocal microscopy (see Materials and Methods). (A) Distribution of NMY-2, a nonmuscle myosin II required for cytokinesis and polarity in the early embryo (Guo and Kemphues 1996). (first column) NMY-2 distribution in wild-type pronuclear, two-cell, and four-cell embryos. NMY-2 is uniformly localized to the cortex of early embryonic cells. (second column) NMY-2 distribution in mlc-4(RNAi) embryos. NMY-2 accumulates at the presumptive sites of cleavage furrow formation. At the pronuclear stage, a ring of NMY-2 localization is seen around the location of the polar body DNA (see third column for DNA localization; this localization was seen in 3/3 additional embryos scored at this stage). At the equivalent of the two-cell stage (middle), a surface focal plane image of the embryo shows a band of NMY-2 localization that extends around the circumference of the embryo (7/7 additional embryos at this stage showed similar localization). In later stages, shown in the bottom, NMY-2 is localized to bands that appear as patches in a confocal section. (third column) DNA localization in the same embryos as in column 2, as revealed by staining with DAPI. In the middle, note the extra, smaller, nuclear structure formed from the polar body chromosomes. (B) Distribution of F-actin revealed by rhodamine-conjugated phalloidin staining. (first column) F-actin distribution in wild-type pronuclear, two-cell, and four-cell embryos. F-actin appears uniformly distributed in the cortex of the embryonic cells. (second column) F-actin distribution in mlc-4(RNAi) embryos equivalent in age to pronuclear, two-cell, and four-cell embryos. As in wild type, F-actin is enriched at the cortex of the embryo. (third column) DNA localization in the same embryos as in column 2, as revealed by DAPI staining. Despite the localization of NMY-2 and F-actin to presumed regions of the cytokinetic furrow, no membrane ingression is seen in mlc-4(RNAi) embryos during mitotic cell cycles. Bars, 10 μm.
The distribution of F-actin in mlc-4(RNAi) embryos was largely unaffected (Fig. 2 B). F-actin is uniformly distributed at cortical surfaces in early wild-type embryos with a slight enrichment at cytokinetic furrows. Although much less dramatically than NMY-2, F-actin was somewhat enriched at presumptive sites of cytokinetic furrow formation in mlc-4(RNAi) embryos as compared with wild-type embryos. Thus, the ability of the microfilament cytoskeleton to organize appears unaffected by reducing mlc-4 function. In fact, F-actin and especially myosin become enriched at expected sites of cleavage furrow ingression. Because no furrows were formed in mlc-4(RNAi) embryos, however, we infer that the primary defect is a functional defect in force generation rather than a defect in the organization or assembly of a contractile ring.
Polarized Cytoplasmic Flow Directed at the Sperm Pronucleus–Centrosomal Complex Requires mlc-4 and nmy-2 Function
Because chemical inhibitors of actin polymerization block the polarized cytoplasmic flow directed by the sperm pronucleus–centrosomal complex (see introduction), we asked if mlc-4 and nmy-2 are required for cytoplasmic flow. The timing, speed, and direction of cytoplasmic flow have been defined in wild-type embryos by using multifocal plane, time-lapse video microscopy to follow the movements of yolk granules in pronuclear stage embryos (Hird and White 1993). The polarized cytoplasmic flow has two components: an internal movement of cytoplasm towards the sperm pronucleus and associated centrosomes and a corresponding movement of cytoplasm in the opposite direction near the cortical surface of the embryo. The internal flow is directed toward the paternal pronucleus wherever it is located, suggesting that factors associated with the sperm pronucleus or associated centrosomes trigger and direct the flow (Hird and White 1993; Goldstein and Hird 1996). Cytoplasmic flow is mainly confined to the posterior regions of a one-cell stage embryo, beginning several minutes before maternal pronuclear migration and lasting until just before pronuclear congression. In wild-type embryos, we consistently observed directional cortical flow of yolk granules at 4.5 ± 0.6 μm/min in the posterior of one-cell stage embryos during this time (Fig. 3). Reducing the function of either mlc-4 or nmy-2 by RNAi completely abolished the detectable directed movements of cortical yolk droplets (Fig. 3). We followed yolk granules in mlc-4(RNAi) and nmy-2(RNAi) mutant embryos from before pronuclear migration to late telophase of the first mitosis without noting any directed cortical flow or internal flow. These results suggest that, in addition to their role in cytokinesis, mlc-4 and nmy-2 are required for the cytoplasmic rearrangements that occur shortly after fertilization.
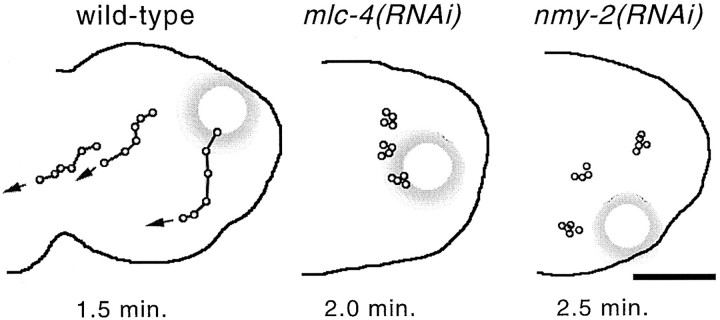
Figure 3.
Cortical movement of yolk granules in wild-type, mlc-4 (RNAi), and nmy-2(RNAi) mutant embryos. Individual yolk droplets were followed by time-lapse digital photomicroscopy (see Materials and Methods) and were traced onto an outline of the respective embryos. Each dot along a line represents the position of the same yolk droplet at consecutive time intervals; the total elapsed time is shown below each embryo. The large shadowed circles represent the position of the paternal pronucleus in each embryo. (left) Cortical flow in a wild-type embryo. Note the cortical movement away from the paternal pronucleus. Analysis of 20 droplets in four different embryos yields an average speed of cortical yolk droplets at 4.5 ± 0.6 μm/min. (middle and right) Cortical yolk droplet movement in mlc-4(RNAi) and nmy-2(RNAi) mutant embryos, respectively. In these and three other embryos for each mutant, no directed yolk droplet movement was detected (20 granules for each mutant). Bar, 10 μm.
a-p Polarity Is Lost in mlc-4(RNAi) One-cell Embryos
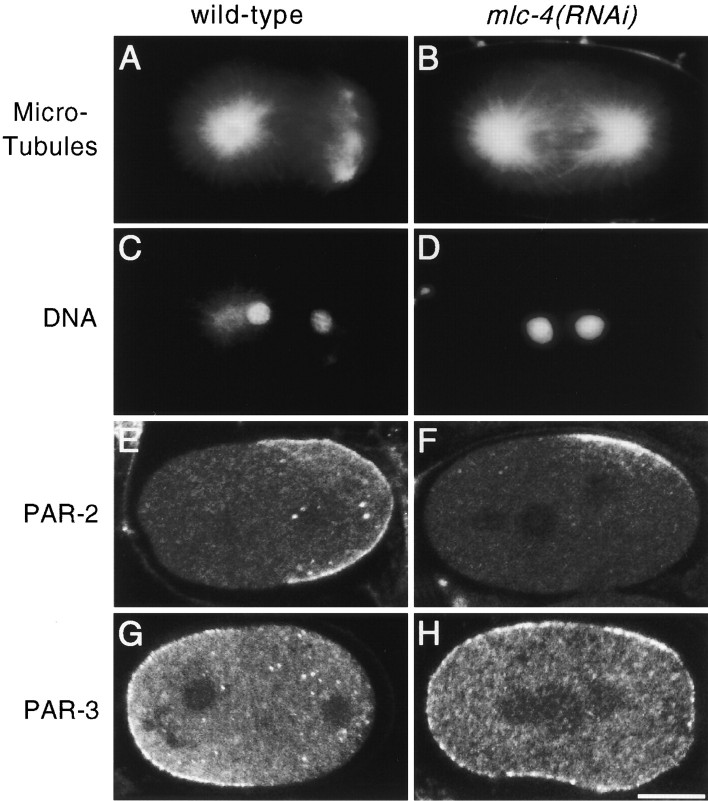
To determine if the loss of cytoplasmic flow after reducing mlc-4 or nmy-2 function correlated with a partial or complete loss of a-p asymmetry, we examined mlc-4(RNAi) embryos for asymmetries that become apparent shortly after fertilization in one-cell stage wild-type embryos. Normally, the maternal pronucleus migrates to meet the paternal pronucleus in the posterior region of the zygote (Fig. 1 A, frame a; Albertson 1984). In mlc-4(RNAi) embryos, the pronuclei instead met near the center of the embryo, with the paternal pronucleus migrating roughly the same distance as the maternal pronucleus (Fig. 1 A, frame e, and Fig. 1 B). Shortly after pronuclear congression in wild-type embryos, the first mitotic spindle forms and becomes displaced posteriorly, resulting in the production of the smaller P1 and larger AB daughters (Sulston et al. 1983). In mlc-4(RNAi) embryos, the spindle rotated normally but was positioned symmetrically along the long axis during the first attempt at cell division (Fig. 1 A, frame f, and Fig. 1 B). In addition to asymmetry in position, the first mitotic spindle in wild-type embryos exhibits a characteristic morphology during telophase of the first mitotic cell cycle: the posterior spindle pole appears flattened and disc-shaped, whereas the anterior spindle pole is spherical (Keating and White 1998; Fig. 1 A, frame c, and Fig. 4 A). However, in mlc-4(RNAi) embryos examined by Nomarski optics and by staining with antibodies to tubulin, both poles of the first mitotic spindle always appeared identical and similar to the wild-type anterior spindle pole (Fig. 1 A, frame g, and Fig. 4 B). We conclude that mlc-4 is required for the a-p asymmetries associated with pronuclear meeting and position of the first mitotic spindle as well as a-p differences in the morphology of the first mitotic spindle.
Figure 4.
First mitotic spindle morphology and PAR protein distribution in wild-type and mlc-4(RNAi) embryos. (A–D) Spindle morphology in wild-type and mlc-4(RNAi) embryos. (A and B) Spindle morphology at telophase of the first mitosis as visualized by immunolocalization with antibodies recognizing tubulin. A shows the morphology of wild-type spindle poles. The centrosome at the posterior pole splits and migrates well ahead of the anterior centrosome, giving a barlike morphology to the posterior spindle pole. B shows an mlc-4(RNAi) mutant embryo at the equivalent stage of the cell cycle. The morphology of both the anterior and posterior spindle poles resemble the anterior spindle pole in a wild-type embryo. (C and D) DAPI staining of the same embryos in A and B. Both embryos are in telophase of the first mitosis. (E–H) Distribution of PAR-2 and PAR-3 proteins in wild-type and mlc-4(RNAi) embryos. Images were obtained by confocal microscopy (see Materials and Methods). (E and F) Distribution of PAR-2. In the wild-type embryo (E), PAR-2 is localized to the posterior cortical hemisphere of the embryo. In the mlc-4(RNAi) mutant embryo (F), cortical PAR-2 is restricted to a small, laterally displaced, patch. (G and H) Distribution of PAR-3. PAR-3 is enriched in the cortex at the anterior of a wild-type embryo (G), but is uniformly distributed to the cortex in the mlc-4(RNAi) mutant embryo (H). Similar distributions of PAR proteins were seen in all mlc-4(RNAi) embryos at these early stages (5/5 for par-2, 7/7 for par-3). Bar, 10 μm.
To examine at the molecular level the extent to which a-p asymmetries are abnormal after loss of polarized cytoplasmic flow, we used indirect immunofluorescence to visualize two proteins with polarized distributions at the one-cell stage, called PAR-2 and PAR-3 (Fig. 4, E–H). In addition to a-p differences in protein localization, the par-2 and par-3 genes are required for some aspects of a-p polarity in a one-cell embryo (for review see Kemphues and Strome 1997). PAR-2 is localized to the cortical cytoplasm in the posterior half of a one-cell embryo, whereas PAR-3 is localized to the cortical cytoplasm in the anterior half (Fig. 4E and Fig. G). We found that PAR-3 was uniformly distributed around the cortex of mlc-4(RNAi) embryos (Fig. 4 H). Thus, the a-p polarity that restricts the asymmetric distribution of PAR-3 appears absent in mlc-4(RNAi) embryos. The polarized distribution of PAR-2 was less severely perturbed in mlc-4(RNAi) embryos than PAR-3. Instead of occupying the cortex throughout the posterior part of the zygote, PAR-2 was consistently localized to a small cortical patch, often laterally displaced, in the posterior half of mlc-4(RNAi) embryos (Fig. 4 F). Like the symmetrical appearance of the first mitotic spindle, the uniform localization of PAR-3 and the improper localization of PAR-2 in mlc-4(RNAi) embryos indicates that a-p polarity has been disrupted in mlc-4(RNAi) embryos.
P Granule Localization Can Still Occur in mlc-4(RNAi) and nmy-2(RNAi) Embryos
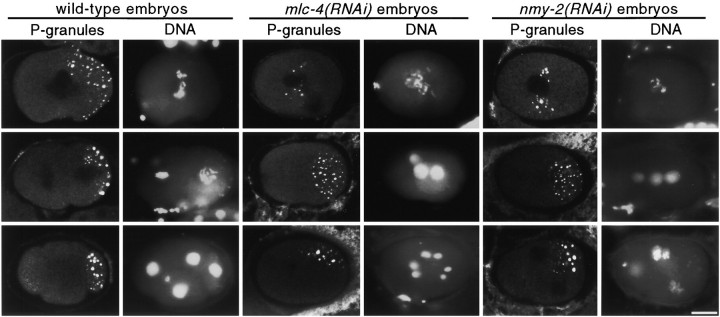
As a final test of a-p polarity defects in mlc-4(RNAi) embryos, we examined the distribution of the germline P granules. P granules in one-cell stage embryos move in an apparently identical direction and speed as cytoplasmic flow (Hird et al. 1996); both cytoplasmic flow and P granule localization are sensitive to cytochalasin D treatment (Hill and Strome 1988; Hird et al. 1996). Because cytoplasmic flow is not detectable in mlc-4(RNAi) and nmy-2(RNAi) embryos (Fig. 3), and because these RNAi embryos exhibit other losses of a-p asymmetry (Fig. 1 and Fig. 4; Guo and Kemphues 1996), we expected to detect a loss of P granule localization to the posterior cortex at the time when cytoplasmic flow occurs. Indeed, in 5/5 mlc-4(RNAi) and 4/4 nmy-2(RNAi) pronuclear stage embryos, P granules were detected in the middle of the embryo (Fig. 5). Wild-type embryos show posterior localization of P granules at this time (Fig. 5). Surprisingly, we found that P granules became localized to the posterior in 21/21 mlc-4(RNAi) embryos and 16/16 nmy-2(RNAi) embryos examined after the first mitotic cycle (Fig. 5). We conclude that polarized cytoplasmic flow is not absolutely required for P granule localization, but it is required for localization of P granules in pronuclear stage embryos.
Figure 5.
P granule localization in wild-type, mlc-4(RNAi), and nmy-2(RNAi) mutant embryos. Three pairs of columns are presented: the left-hand column of each pair shows P granule localization, whereas the right-hand column shows DNA localization in the same embryos as revealed by DAPI staining. (top) Localization of P granules in wild-type, mlc-4(RNAi), and nmy-2(RNAi) mutant pronuclear stage embryos. Note the posterior bias in P granule localization in the wild-type embryo but not in the mlc-4(RNAi) or nmy-2(RNAi) mutant embryos. (middle) P granule localization in two-cell stage embryos. In the wild-type embryo, P granules are seen associated with the posterior most cortex in the posterior cell. In the mlc-4(RNAi) and nmy-2(RNAi) mutant embryos, P granules are localized to the posterior end of the embryos. (bottom) P granule localization in four-cell stage embryos. In the wild-type embryos, P granules are distributed to one cell of the four-cell embryo. Intriguingly, in the equivalently staged mlc-4(RNAi) and nmy-2(RNAi) mutant embryos, we note that P granules appear to have coalesced into a patch that is similar in size to that in the wild-type embryo. Bar, 10 μm.
par-2, but not par-1, Mutations Affect Polarized Cytoplasmic Flow
To test further if the posterior localization of P granules correlates with polarized cytoplasmic flow, we quantitated cytoplasmic flow rates in par-1 and in par-2 mutant embryos. In par-1 mutant embryos, P granule segregation is undetectable early and P granules are abnormally distributed equally to all cells in a four-cell stage embryo (Kemphues et al. 1988). However, in par-2 mutant embryos, P granules appear to localize to the posterior blastomere at the two-cell stage (Kemphues et al. 1988; Kemphues and Strome 1997). Previous work has suggested that polarized cytoplasmic flow may occur relatively normally in par-1 mutant embryos but is abnormal in par-2 mutants (Kirby et al. 1990). These results suggest that P granule localization and polarized cytoplasmic flows are not correlated in par mutant backgrounds. To address this issue quantitatively, we analyzed polarized cytoplasmic flow in par-1 and par-2 mutant embryos using videomicroscopy. We found that cortical flow was impaired substantially in par-2 mutant embryos, but appeared normal in par-1 mutant embryos. Cortical flow in par-1 mutant embryos occurred at a speed of 4.3 ± 0.5 μm/min, whereas cortical flow in par-2 was observed at a speed of 2.8 ± 0.6 μm/min (12 yolk granules in four different embryos for each mutant). Thus, as in mlc-4(RNAi) and nmy-2 (RNAi) embryos, we observe a lack of correlation between polarized cytoplasmic flow and posterior localization of P granules in par mutant embryos.
Embryos Homozygous for a Loss-of-function Mutation in mlc-4 Exhibit Elongation Defects and Larval Lethality
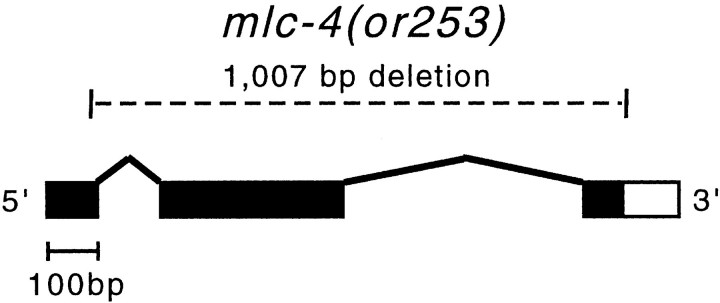
To initiate genetic analysis of mlc-4 and examine mlc-4 function at other stages of the C. elegans life cycle, we used ethyl methanesulfonate mutagenesis coupled with a PCR detection method to isolate a 1,007-bp deletion that removes most of the mlc-4 locus (Fig. 6; see Materials and Methods). This deletion allele, called mlc-4(or253), leaves only 85 bp of the first exon and can produce only the amino-terminal 28 amino acids of the predicted 172–amino acid mlc-4 protein. Comparison to the known crystal structure of a myosin regulatory light chain indicates that this amino-terminal region does not interact with the myosin heavy chain and that the truncated protein, even if stable, will be nonfunctional (Rayment et al. 1993). We found that the mutation is fully recessive and that homozygous mlc-4(or253) mutant animals hatch but die in larval stages (see Materials and Methods).
Figure 6.
Schematic of the or253 deletion allele in mlc-4. Exon/intron structure is shown as predicted by Genefinder (Consortium, 1998) and confirmed by sequence from cDNA clones; boxes represent the exon sequences and the black shading indicates the protein coding regions. The mlc-4(or253) deletion removes 1,007 bp of sequence from the 29 codon of the first exon to beyond the stop codon in the last exon.
We found that, although mlc-4(or253) mutant embryos usually hatch, they do not fully elongate at the end of embryogenesis, resulting in a morphological arrest at roughly the twofold stage (Fig. 7). Because this phenotype is superficially similar to mutations that affect bodywall muscle structure or function (Williams and Waterston 1994), we examined mutant embryos for possible muscle defects. However, we found that mutant embryos begin to twitch at similar stages of embryogenesis to wild type and they exhibit normal amounts of muscular movement from this time up to hatching (data not shown). Furthermore, no defects in bodywall muscle morphology were detected, as assayed by phalloidin staining of actin within intact animals (Fig. 7C and Fig. D). No obvious defects were seen in pharyngeal structure or function; pharyngeal pumping in mutant embryos occurred and appeared similar to wild type (data not shown). Thus, the morphological defect seen in mlc-4 (or253) mutant animals does not appear to be a consequence of defects in bodywall muscle or pharyngeal muscle function.
Hypodermal Seam Cells Show Improper Morphology in mlc-4(or253) Embryos
The hypodermis plays an important role in embryonic elongation in C. elegans. Circumferentially arranged microfilament bundles appear to be important for generating the contractile forces that squeeze the initially round postproliferation stage embryo into a normal, elongated, worm shape (Priess and Hirsh 1986; Costa et al. 1997). To investigate whether the mlc-4(or253) elongation defect was due to a defect in hypodermal function, we examined the organization of the hypodermis in wild-type and mlc-4(or253) embryos. The number and morphology of hypodermal cells can be easily assayed by staining with the antibody MH27, which recognizes an epitope associated with adheren junctions at the boundaries of hypodermal cells (Podbilewicz and White 1994). Because mlc-4(RNAi) embryos show defects in cytokinesis, we first determined whether the proper number of hypodermal cells were generated during embryogenesis in mlc-4(or253) mutants. Shortly before the bean stage of embryogenesis, hypodermal cells are organized into five longitudinal rows of cells that can be visualized by MH27 staining (Priess and Hirsh 1986; Podbilewicz and White 1994). These five rows are composed of a single dorsal row of 21 cells, two lateral rows of 11 cells each, and two ventral rows of 11 cells (Podbilewicz and White 1994). We stained embryos derived from a balanced mlc-4(or253) strain. 25% of these embryos should be homozygous for mlc-4(or253). However, we saw no difference in hypodermal cell number among these embryos. We counted 20.2 ± 0.8 dorsal cells (n = 20 embryos), 10.6 ± 0.5 lateral cells (n = 26 embryos), and 10.4 ± 0.5 ventral cells (n = 24 embryos) in the individual rows. These data, coupled with evidence that pharyngeal and bodywall muscle structure appears normal, suggest that maternal contribution of mlc-4 function is sufficient for embryonic cytokinesis and that the elongation defect is not a result of a failure to produce the proper number of hypodermal cells.
Next, we visualized the morphology of hypodermal cells late in embryogenesis, after the period in which embryos normally elongate. In wild-type embryos, a subset of lateral hypodermal cells, called seam cells, undergo a dramatic shape change during elongation. The seam cells initially are roughly oval in appearance at the apical surface, but subsequently elongate substantially in response to force generation by the microfilament cytoskeleton within the hypodermal cells (Fig. 7 E). In mlc-4(or253) homozygotes, however, we found that seam cells do not undergo this shape change as dramatically, retaining a wider appearance (Fig. 7 F). This suggests that the elongation defect seen in the mutant embryos may be due in part to a defect in the ability of seam cells to elongate to a narrow morphology.
Expression of an mlc-4::GFP Fusion Protein Encoded by a Rescuing Transgene Is Detected in Seam Cells
To examine the zygotic expression of mlc-4, we constructed a COOH-terminal translational fusion using coding sequences for GFP (see Materials and Methods). We found that this mlc-4::GFP fusion is functional, as it rescued the larval lethality of mlc-4(or253) homozygotes. However, the rescued adult animals were sterile, suggesting that the transgenic array was unable to complement the germline defects associated with a loss-of-function in mlc-4. This result is likely due to the failure of the transgene to be expressed in the germline; lack of germline expression has been noted for other transgenic arrays in C. elegans and is likely due to a germline-specific gene silencing phenomenon (Kelly and Fire 1998). The sterile phenotype is manifested by an apparent failure of oocytes to properly pinch off in the proximal arm of the gonad (data not shown). This is consistent with the cytokinetic role seen for mlc-4 in the early embryo and suggests that, in mlc-4(RNAi) embryos, mlc-4 function is not completely eliminated and some residual function remains to generate viable oocytes.
We examined MLC-4::GFP expression in the rescued lines by GFP autofluorescence and by indirect immunofluorescence using an antibody that recognizes GFP (see Materials and Methods). We did not detect expression in the adult germline or in early embryos produced by transgenic animals, consistent with a lack of germline rescue of mlc-4 loss-of-function defects. We first detected expression at the bean stage in the lateral hypodermal seam cells that are thought to play an important role in mediating embryonic elongation. This expression persisted throughout embryogenesis and into larval stages. To confirm the identity of the GFP positive cells, we fixed and double stained transgenic worms with antibodies recognizing GFP and with MH27 (Fig. 7G and Fig. H). Because the rescuing MLC-4/GFP fusion protein is expressed in seam cells and mlc-4(or253) homozygous mutant embryos show defects in seam cell morphology, we conclude that mlc-4 function is required in seam cells for proper elongation of the embryo. Postembryonic sites of mlc-4/GFP expression include strong expression in the spermathecal and uterine walls and weak expression in the gonadal sheath and intestinal muscle (data not shown). No other sites of expression were detected in embryonic, larval, or adult stages, although postembryonic dividing and/or migrating cell types were not closely examined. No expression was seen in bodywall muscles, vulval muscles, or pharyngeal muscles.
Discussion
Using RNA-mediated interference as the basis for a general screen of embryonic phenotypes in C. elegans, we identified an nmRLC gene, mlc-4, which is required for cytokinesis, for establishment of a-p polarity, and for elongation during embryonic morphogenesis. mlc-4 appears to be the sole nmRLC gene in C. elegans and encodes a protein 74% identical to the product of the spaghetti-squash gene in Drosophila. Like spaghetti-squash, mlc-4 is required for cytokinesis. In addition, mlc-4 and a myosin heavy chain gene called nmy-2 are required for polarized cytoplasmic flow seen in one-cell embryos shortly after fertilization. Consistent with models in which polarized cytoplasmic flow plays a role in establishing a-p polarity, we detected losses in a-p asymmetry after blocking the flows by reducing maternal expression of either mlc-4 or nmy-2 with RNAi. Although delayed, we found that posterior localization of P granules can occur in the absence of cytoplasmic flow, suggesting that a second microfilament-mediated process may generate some aspects of a-p asymmetry in the early C. elegans embryo. Finally, we detect zygotic expression of an mlc-4::GFP fusion protein beginning at midembryogenesis in a subset of lateral hypodermal cells called seam cells. Phenotypic characterization of a deletion allele of mlc-4 reveals a zygotic requirement for cell shape changes of these cells associated with proper embryonic elongation.
mlc-4 Function Appears Distinct from mlc-1 or mlc-2 Functions
mlc-1, mlc-2, and mlc-4 are the three myosin regulatory light chain genes identified in the essentially complete C. elegans genomic sequence (Consortium, 1998), whereas mlc-3 encodes an essential myosin light chain. By several criteria, the genetic requirements of mlc-4 appear distinct from mlc-1 and mlc-2. Genetic analyses indicate that mlc-1 and mlc-2 have overlapping functions within bodywall and pharyngeal muscles (Rushforth et al. 1998), whereas the mlc-4(or253) deletion mutant described here has no effect on the function or morphology of bodywall and pharyngeal muscles. Consistent with these distinct functional requirements, mlc-1 and mlc-2 expression is detected only in bodywall, pharyngeal, and vulval muscles, whereas mlc-4 is expressed primarily in the gonadal sheath, uterine wall, and spermathecal wall. Finally, reducing the function of mlc-1 and mlc-2 in embryos with RNAi has no effect on early embryonic development, in contrast to the effects that we have documented in mlc-4(RNAi) embryos. In summary, mlc-4 appears to act primarily in nonmuscle cell types and may fulfill all nmRLC function in C. elegans, whereas available evidence suggests that mlc-1 and mlc-2 functions are partially redundant and limited to muscle cell types (Rushforth et al. 1998).
mlc-4 Is Required for an Early Step in Cytokinesis
Reducing the function of mlc-4 by RNAi results in a nearly complete absence of cytokinetic furrows, even though the myosin heavy chain NMY-2 becomes highly enriched at the presumptive sites of furrow assembly in mlc-4(RNAi) embryos as compared with wild-type embryos. These results indicate that mlc-4 is required for force generation but not for organization of myosin into a contractile ring. The sequence conservation of mlc-4 with nmRLC genes and the fact that nmy-2 is also required for cytokinesis (Guo and Kemphues 1996) suggests that the mlc-4 protein binds to and regulates the motor activity of the myosin heavy chains encoded by nmy-2 to affect the contractile force required for furrow ingression. A similar cytokinetic role for nmRLC is seen in Drosophila; the predicted spaghetti-squash protein is 74% identical to MLC-4 and is required for cytokinesis, apparently throughout development (Karess et al. 1991; Jordan and Karess 1997).
nmy-2/mlc-4 Are Required for Polarized Cytoplasmic Flow and the Establishment of a-p Polarity Directed by Sperm Entry
Reducing either mlc-4 or nmy-2 function with RNAi completely eliminates the polarized cytoplasmic flow that appears to be initiated by the sperm pronuclear–centrosomal complex (Hird and White 1993; Goldstein and Hird 1996), indicating that myosin function is required for this process. The role of myosin could be explained in at least two ways. First, a localized inactivation of myosin motor activity by the sperm pronuclear–centrosomal complex, resulting in a local reduction in microfilament tension, could lead to movement of the cortical microfilament network and associated cytoplasm away from the sperm pronucleus (Hird and White 1993). Alternatively, myosin could be tethered to the cortex or to the plasma membrane and, upon local activation by the sperm pronuclear–centrosomal complex, move actin filaments away from the sperm pronucleus, generating a flow of cortical material anteriorly (for review see Welch et al. 1997). In both models, a posteriorly directed internal flow would arise to replace the material that moves anteriorly along the cortex. Consistent with the hypothesis that cytoplasmic flow is required to establish a-p differences at the one-cell stage (Goldstein and Hird 1996), we see a loss in a-p polarity after reducing mlc-4/nmy-2 function and eliminating cytoplasmic flow. However, proper polarization of the a-p axis could occur independently of cytoplasmic flow while still requiring myosin function.
P Granule Distribution in the Absence of Cytoplasmic Flow Suggests a Second Role for the Microfilament Cytoskeleton in Establishing a-p Polarity
In addition to directing cytoplasmic flow, we infer a second function for microfilaments based on our analysis of P granule distribution in the absence of flow. In both mlc-4(RNAi) and nmy-2(RNAi) embryos, P granules become localized to the posterior region of the embryo during the first mitotic cycle despite the elimination of detectable cytoplasmic flow. The posterior localization of P granules in embryos lacking mlc-4 or nmy-2 function contrasts with the central positioning of P granules observed in similarly staged embryos treated with cytochalasin D to depolymerize microfilaments (Strome and Wood 1983). Thus, in the absence of nmy-2 and mlc-4 function, an additional actin-dependent process is suggested that can localize P granules to the posterior, maintaining at least one aspect of a-p polarity. One possible explanation for this unexpected result is that an actin-dependent motor activity distinct from NMY-2 and MLC-4 can segregate P granules. Alternatively, as previously proposed, a degradative activity restricted to the anterior region of the embryo coupled with a posterior P granule–anchoring function could lead to an apparent posterior localization in the absence of detectable cytoplasmic flow (Hird et al. 1996). In the latter case, anterior localization of the degradative activity would presumably depend on microfilaments but not on mlc-4/nmy-2 function. It should be noted that P granule localization is abnormal in mlc-4 or nmy-2(RNAi) embryos at the pronuclear stage, when P granules first become localized in wild-type embryos (Fig. 5). This suggests that, even if not absolutely required, cytoplasmic flow does have an early role in P granule segregation.
Our findings further suggest that the par genes may be required for two distinct functions of the microfilament cytoskeleton in the early embryo. Even though cytoplasmic flows are substantially reduced, P granules are partially localized to the posterior during the first division in par-2 mutant embryos (Kemphues et al. 1988). In contrast, eliminating par-1 function does not affect cytoplasmic flow but does result in a complete loss of P granule segregation (Kemphues and Strome 1997). Thus, par-1 and par-2 may function in distinct pathways to organize the a-p axis. Perhaps par-2 function is partially required to generate the cytoplasmic flow that localizes posterior determinants, whereas par-1 might regulate the segregation of P granules, and possibly other factors, independent of polarized cytoplasmic flow.
mlc-4 May Function in Seam Cells as Part of a Pathway to Promote Cell Shape Changes and Embryonic Elongation
mlc-4 function appears to promote the morphogenetic cell shape changes that accompany and contribute to embryonic elongation. This role is very similar to that proposed for let-502, which encodes a putative rho-associated kinase (Wissmann et al. 1997). Genetic analysis of let-502 indicates that it is required for proper embryonic elongation and let-502 expression reporter constructs, like mlc-4 reporter constructs, are expressed in seam cells at the time that elongation is occurring (Wissmann et al. 1997). In vertebrate systems, a protein similar to the let-502 kinase, rho kinase, has been shown to phosphorylate nmRLC (Kureishi et al. 1997). In addition, mutations in a C. elegans smooth muscle myosin phosphatase subunit gene called mel-11 can suppress the elongation phenotype of let-502 (Wissmann et al. 1997). These observations suggest that mlc-4 is a target of the let-502 and mel-11 regulatory pathway and that rho-like GTPases may regulate the phosphorylation of MLC-4 and, consequently, the activity of myosin and the microfilament cytoskeleton during embryonic elongation.
Although mlc-4 and let-502 function in seam cells appears essential for embryonic elongation, embryos mutant in either mlc-4 or let-502 still elongate to some extent (Fig. 7; Wissmann et al. 1997). In contrast, treatment with microfilament inhibitors blocks elongation completely (Priess and Hirsh 1986). Perdurance of maternally supplied gene product could explain partial elongation in mutant embryos. Finally, the observation that both mlc-4 and let-502 expression appears restricted to seam cells suggests that elongation in other hypodermal cells may occur via a different microfilament-dependent pathway or require lower levels of mlc-4 and let-502 expression.
mlc-4 Is Likely the Target of a Diverse Array of Regulatory Pathways
As the sole nmRLC encoded by the C. elegans genome, the mlc-4 protein is likely to be the focal point for a variety of regulatory pathways that control the microfilament cytoskeleton during the processes of cytokinesis, cell polarization, and cell shape changes during C. elegans embryogenesis. Work in mammalian cells has implicated a rho-associated kinase, citron kinase, in cytokinesis (Madaule et al. 1998). It is possible that a similar kinase could function in C. elegans to regulate mlc-4 during embryonic cytokinesis. During establishment of a-p polarity, the par genes also have essential functions. Though the relationship of the par genes with mlc-4 remains to be determined, a possible link is suggested by the observation that PAR-1 interacts with NMY-2, the likely myosin heavy chain partner for MLC-4 (Guo and Kemphues 1996). Finally, during embryonic morphogenesis, the rho-associated kinase LET-502 or the phosphatase subunit MEL-11 may interact with mlc-4. The identification of mlc-4 is an important step towards understanding how these diverse pathways regulate microfilament function in a wide range of contexts.
Acknowledgments
We are extremely grateful to Ken Kemphues and Susan Strome for supplying antibodies used in this work and for discussion of results. Thanks to Catherine Dulac (Harvard University, Cambridge, MA) for pointers on the cDNA library construction, Marty Chalfie for GFP constructs, and to Michelle Coutu (Washington University, St. Louis, MO) for supplying the MH27 antibody. Special thanks to the C. elegans Sequencing Consortium and to Yuji Kohara (National Institute of Genetics, Mishima, Japan) for providing cDNA clones. We acknowledge Jeff Hardin for writing the four-dimensional movie software and we thank Bob Barstead (Oklahoma Medical Foundation, Oklahoma City, OK) for sharing his laboratory's protocol for isolating gene deletions. Thanks to Chris Doe, Aaron Severson, Danielle Hamill, and Ann Schlesinger (all from University of Oregon, Eugene, OR) for critical comments on the manuscript.
This work supported by grants from the National Institutes of Health (R01 M58017 to B. Bowerman and F32 AM16981 to C.A. Shelton).
Footnotes
1.used in this paper: a-p, anterior-posterior; DAPI, 4′6-diamidino-2-phenylindole; dsRNA, double stranded RNA; F-actin, filamentous actin; nmRLC, nonmuscle myosin regulatory light chain; NMY-2, nonmuscle myosin II heavy chain; RNAi, RNA-mediated genetic interference
References
- Albertson D.G. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Boyd L., Guo S., Levitan D., Stinchcomb D.T., Kemphues K.J. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick A.R. Molecular mechanisms of nonmuscle myosin-II regulation. Curr. Opin. Cell Biol. 1999;11:26–33. doi: 10.1016/s0955-0674(99)80004-0. [DOI] [PubMed] [Google Scholar]
- Caenorhabditis elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 282:2012–2018. [DOI] [PubMed]
- Chalfie M., Tu Y., Euskirchen G., Ward W.W., Prasher D.C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Costa M., Draper B.W., Priess J.R. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev. Biol. 1997;184:373–384. doi: 10.1006/dbio.1997.8530. [DOI] [PubMed] [Google Scholar]
- Costa M., Raich W., Agbunag C., Leung B., Hardin J., Priess J.R. A putative catenin–cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C., Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Edgar L.G. Blastomere culture and analysis. In: Epstein H.F., Shakes D.C., editors. In Caenorhabditis elegans: modern biological analysis of an organism. Academic Press; San Diego: 1995. pp. 303–313. [Google Scholar]
- Etemad-Moghadam B., Guo S., Kemphues K.J. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Fire A., Harrison S.W., Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans . Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Goldstein B., Hird S.N. Specification of the anteroposterior axis in Caenorhabditis elegans . Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K.J. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans . Nature. 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- Hill D.P., Strome S. An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev. Biol. 1988;125:75–84. doi: 10.1016/0012-1606(88)90060-7. [DOI] [PubMed] [Google Scholar]
- Hird S.N., White J.G. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans . J. Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird S.N., Paulsen J.E., Strome S. Segregation of germ granules in living Caenorhabditis elegans embryoscell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- Jansen G., Hazendonk E., Thijssen K.L., Plasterk R.H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans . Nat. Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Jordan P., Karess R. Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila . J. Cell Biol. 1997;139:1805–1819. doi: 10.1083/jcb.139.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R.E., Chang X.J., Edwards K.A., Kulkarni S., Aguilera I., Kiehart D.P. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila . Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Keating H.H., White J.G. Centrosome dynamics in early embryos of Caenorhabditis elegans . J. Cell Sci. 1998;111:3027–3033. doi: 10.1242/jcs.111.20.3027. [DOI] [PubMed] [Google Scholar]
- Kelly W.G., Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans . Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues, K.J., and S. Strome. 1997. Fertilization and establishment of polarity in the embryo. In C. ELEGANS II, D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 335–360. [PubMed]
- Kemphues K.J., Priess J.R., Morton D.G., Cheng N.S. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kennerdell J.R., Carthew R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kirby C., Kusch M., Kemphues K. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev. Biol. 1990;142:203–215. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- Kureishi Y., Kobayashi S., Amano M., Kimura K., Kanaide H., Nakano T., Kaibuchi K., Ito M. Rho-associated kinase directly induces smooth muscle contraction through myosin light chain phosphorylation. J. Biol Chem. 1997;272:12257–12260. doi: 10.1074/jbc.272.19.12257. [DOI] [PubMed] [Google Scholar]
- Madaule P., Eda M., Watanabe N., Fujisawa K., Matsuoka T., Bito H., Ishizaki T., Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegansextrachromosomal maintenance and integration of transforming sequences. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Palmer R.E., Sternberg P.W. The identification of a Caenorhabditis elegans homolog of p34cdc2 kinase. Mol. Gen. Genet. 1994;245:781–786. doi: 10.1007/BF00297285. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B., White J.G. Cell fusions in the developing epithelial of C. elegans . Dev. Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Powers J., Bossinger O., Rose D., Strome S., Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess J.R., Hirsh D.I. Caenorhabditis elegans morphogenesisthe role of the cytoskeleton in elongation of the embryo. Dev. Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski W.R., Schmidt B.K., Smith R., Tomchick D.R., Benning M.M., Winkelmann D.A., Wesenberg G., Holden H.M. Three-dimensional structure of myosin subfragment-1a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Downs W.D., Lin R., Wittmann C., Bei Y., Cha Y.H., Ali M., Priess J.R., Mello C.C. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rushforth A.M., White C.C., Anderson P. Functions of the Caenorhabditis elegans regulatory myosin light chain genes mlc-1 and mlc-2. Genetics. 1998;150:1067–1077. doi: 10.1093/genetics/150.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J.M., Ashcroft N., Donovan P.J., Golden A. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos Development. 125 1998. 4391 4402a [DOI] [PubMed] [Google Scholar]
- Schumacher J.M., Golden A., Donovan P.J. AIR-2An Aurora/Ipl1–related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos J. Cell Biol. 143 1998. 1635 1646b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C.A., Bowerman B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 1996;122:2043–2050. doi: 10.1242/dev.122.7.2043. [DOI] [PubMed] [Google Scholar]
- Skop A.R., White J.G. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Wood W.B. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans . Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tabara H., Hill R.J., Mello C.C., Priess J.R., Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans . Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Welch M.D., Mallavarapu A., Rosenblatt J., Mitchison T.J. Actin dynamics in vivo. Curr. Opin. Cell. Biol. 1997;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- Williams B.D., Waterston R.H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A., Ingles J., McGhee J.D., Mains P.E. Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev. 1997;11:409–422. doi: 10.1101/gad.11.4.409. [DOI] [PubMed] [Google Scholar]
- Wood W.B. Embryology. In: Wood W.B., editor. In The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. pp. 215–242. [Google Scholar]