Abstract
Mitochondrial dysfunction may contribute to the development of insulin resistance and type 2 diabetes. Nucleoside reverse transcriptase inhibitors (NRTIs), specifically stavudine, are known to alter mitochondrial function in human immunodeficiency virus (HIV)-infected individuals, but the effects of stavudine on glucose disposal and mitochondrial function in muscle have not been prospectively evaluated. In this study, we investigated short-term stavudine administration among healthy control subjects to determine effects on insulin sensitivity. A secondary aim was to determine the effects of stavudine on mitochondrial DNA (mtDNA) and function. Sixteen participants without personal or family history of diabetes were enrolled. Subjects were randomized to receive stavudine, 30 – 40 mg, twice a day, or placebo for 1 mo. Insulin sensitivity determined by glucose infusion rate during the hyperinsulinemic euglycemic clamp was significantly reduced after 1-mo exposure in the stavudine-treated subjects compared with placebo (−0.8 ± 0.5 vs. +0.7 ± 0.3 mg· kg−1 · min−1, P = 0.04, stavudine vs. placebo). In addition, muscle biopsy specimens in the stavudine-treated group showed significant reduction in mtDNA/nuclear DNA (−52%, P = 0.005), with no change in placebo-treated subjects (+8%, P = 0.9). 31P magnetic resonance spectroscopy (MRS) studies of mitochondrial function correlated with insulin sensitivity measures (r2 = 0.5, P = 0.008). These findings demonstrate that stavudine administration has potent effects on insulin sensitivity among healthy subjects. Further studies are necessary to determine whether changes in mtDNA resulting from stavudine contribute to effects on insulin sensitivity.
Keywords: human immunodeficiency virus, insulin resistance, magnetic resonance spectroscopy
Recent data suggest that mitochondrial dysfunction precedes the development of diabetes in insulin-resistant offspring of patients with type 2 diabetes mellitus (DM) (15). Resistance to insulin action at the skeletal muscle is the earliest abnormality in such patients. Evaluation of gene expression in muscle samples from individuals with type 2 DM and impaired glucose tolerance has identified alterations in several genes involved in mitochondrial oxidative phosphorylation (12, 14). For example, both peroxisomal proliferator activator receptor-γ coactivator (PGC1) and nuclear respiratory factor (NRF) are reduced in prediabetic and diabetic human muscle tissue. These data suggest that both individuals with diabetes and those in the prediabetic state without the clinical effect of hyperglycemia show diminished gene expression of elements essential for the mitochondrial respiratory chain oxidative phosphorylation.
Animal and human models of DM suggest that mitochondrial dysfunction is significantly related to the development of insulin resistance. Analysis of the muscle tissue from the insulin-resistant obese ob/ob mice compared with thin ob/+ littermates revealed a reduction in the expression of a subunit of mitochondrial ATP synthase (20). In addition, data from Kelley et al. (6) and others (1, 19) have shown a reduction in mitochondrial size, number, and activity associated with a reduction in insulin-stimulated glucose disposal. Petersen et al. (15) demonstrated a 30% decrease in mitochondrial oxidative phosphorylation and an 80% increase in intramyocellular lipid content in insulin-resistant lean adult relatives of patients with type 2 DM, suggesting that a decrease in mitochondrial function leads to decreased lipid oxidation, increased intramyocellular lipid (IMCL) accumulation, and insulin resistance.
Nucleoside reverse transcriptase inhibitors (NRTIs) have been shown to impair mitochondrial function by molecular quantification and functional assay in individuals infected with human immunodeficiency virus (HIV) (5, 9). Cote et al. (3) demonstrated a 68% reduction in mitochondrial DNA (mtDNA)/nuclear DNA levels in peripheral white blood cells from NRTI-treated patients. Several investigators have demonstrated a reduction in mtDNA copies per cell in fat biopsies from HIV-infected patients treated chronically with NRTIs (13, 22). Moreover, a recent investigation of NRTI administration in healthy non-HIV-infected patients demonstrated significant inhibitory effects on mitochondrial gene expression in fat after 2 wk of combined NRTI therapy (10). Other studies have suggested that respiratory chain activity, an index of mitochondrial function, is reduced in lipodystrophic patients receiving NRTIs (23). The effects of NRTIs, specifically zidovudine (AZT), have also been shown using 31P magnetic resonance spectroscopy (MRS) in vivo. Steady-state exercise resulted in a greater depletion in gastrocnemius muscle phosphocreatine in the AZT-treated patients compared with non-AZT-treated HIV-infected patients and healthy controls (18).
Although prior studies suggest a potent effect of NRTIs on mitochondrial function in peripheral blood mononucleated cells (PBMCs) and adipocytes, these studies have neither investigated insulin resistance and glucose disposal nor prospectively investigated the effects of NRTIs on mitochondrial function in muscle. In addition, the role of NRTIs may be confounded by HIV itself, other causes of insulin resistance, and metabolic abnormalities. For example, Reeds et al. (16) demonstrated reduced mtDNA in muscle among HIV-infected patients compared with non-HIV-infected patients, independent of treatment status, in a recently reported cross-sectional study. In contrast, administration of such agents to healthy subjects may be a useful model to prospectively determine the effects of mitochondrial dysfunction on insulin resistance, independent of confounding factors such as family history of DM, hyperglycemia, and related metabolic disorders. The current study was therefore designed to prospectively investigate the hypothesis that NRTI-induced altered mitochondrial function leads to insulin resistance, using a randomized, double-blinded, placebo-controlled design in healthy non-HIV-infected patients. Mitochondrial function was evaluated by both mtDNA quantitation in muscle biopsy and phosphocreatine recovery with the use of 31P-MRS to investigate the relationship of mitochondrial function to insulin sensitivity.
METHODS
Study Design
The single-site placebo-controlled study was approved by the Partners (Massachusetts General Hospital) and the Massachusetts Institute of Technology (MIT) Institutional Review Boards. Written informed consent was obtained from all participants, and the investigation was conducted according to the principles of the Declaration of Helsinki. Participants were recruited by local advertisements over a 1-yr period from April 2005 to April 2006. All participants were healthy individuals, aged 20–55 yr old, without chronic medical illness, and with a body mass index (BMI) between 18 and 27 kg/m2. Participants were excluded for a history of anemia or renal or liver disease; personal or family history of DM; or hyperinsulinemia, impaired fasting glucose, glucose intolerance, or diabetes based on a 2-h oral glucose tolerance test. The use of medications known to alter glucose metabolism, significant alcohol use, pregnancy, or lack of a barrier contraceptive method were also exclusion criteria.
After signing consent, all qualified participants underwent a screening evaluation, which included a complete medical and family history, physical examination, baseline laboratory testing of complete blood count, liver and renal function, insulin, lipid panel, lactate, and HIV antibody status. In addition, baseline and 2-h post-75-g glucose load glucose values were obtained. Participants were enrolled in the baseline evaluation if they were found to have normal glucose tolerance and were without laboratory abnormalities.
Baseline evaluation included repeat fasting laboratory studies, hyperinsulinemic-euglycemic clamp to evaluate insulin sensitivity, muscle biopsy for mitochondrial analysis, 31P-MRS studies to evaluate mitochondrial function via phosphocreatine recovery rates, 1H-MRS studies to evaluate intramyocellular lipid content, and densitometry for body composition. In addition, activity questionnaires were completed. Participants were subsequently randomized to receive stavudine at the standard therapeutic dose of 30 – 40 mg twice daily, depending on baseline weight, or matching placebo for 1 mo. Weekly safety and compliance visits were completed. An end-of-study visit at 1 mo was performed, repeating the procedures from the baseline visit. Randomization was blinded to subjects and investigators.
Hyperinsulinemic Euglycemic Clamp
A primed infusion of regular insulin (40 mU × m2 × min−1) was given over 2 h. The priming dose (200 mU × m2 × min−1) was for 2 min. A variable infusion of 20% dextrose was used to maintain plasma glucose concentrations at the euglycemic value of 5 mmol/l (90 mg/dl). Arterialized blood samples collected from a retrograde IV catheter in a warmed hand vein were obtained for blood glucose determinations every 10 min during the 30 min before the insulin infusion and every 5 min during the 2-h clamp. The analysis of the glucose infusion rate, GIR (mg· kg−1 · min−1 of glucose infusion), was performed during the final hour of the clamp procedure. We did not specifically investigate hepatic insulin resistance and do not know the effects of stavudine on hepatic glucose production.
Muscle Biopsies
Muscle biopsies of the nondominant vastus lateralis were performed by a physician with extensive experience in the percutaneous needle biopsy technique (4). The procedure was performed using sterile technique and local xylocaine. Samples of ~100 mg of muscle were obtained, and aliquots were stored frozen for DNA analysis. Both biopsies were done in the same anatomic location and performed after the euglycemic hyperinsulinemic clamp had been completed.
mtDNA Analysis
Muscle tissue was immediately frozen and stored at −80°C. Whole blood was collected in CPT tubes (Becton Dickenson), and the PBMCs were washed several times at low speed (300 g) to eliminate platelets before being stored frozen until used. Total DNA was extracted using the QIAAmp DNA tissue kit with an overnight lysis step for muscle tissue or the QIAAmp DNA kit for the PBMCs. The mtDNA-encoded COX1 gene was quantified, while the accessory subunit of the polymerase-γ gene (ASPG) was amplified for the nuclear DNA quantification, as described elsewhere (3). The assay was modified to include a plasmid-based standard curve, and each gene was quantified by real-time PCR on a Roche Lightcycler 480. Quantification of mtDNA in tissue samples and peripheral blood was reported as the ratio of mtDNA to nuclear DNA.
31P-MRS Protocol
Mitochondrial function was determined using 31P-MRS to assess phosphocreatine resynthesis following exercise. 31P-MRS was performed at the Massachusetts General Hospital NMR Center using a 45-cm bore, Siemens 3.0 T magnet, with a 31P operating frequency of 49.879 MHz. 31P spectra were obtained every 2 s over a 10-min period. Maximum voluntary contraction (MVC) for the quadriceps was determined before commencement of the exercise protocol, based on the maximum weight that could be lifted. The exercise protocol consisted of 2 min of rest, followed by 3 min of repetitive bilateral quadriceps contractions, lifting a predetermined, individualized load (30 – 40% MVC), followed by 5 min of recovery. To standardize the protocol, subjects performed knee extensions at a constant frequency of 0.5 Hz (every 2 s) during the 3 min of exercise. Mitochondrial function was determined from the 31P-MRS by plotting the phosphocreatine (PCr) peak integrated area vs. time during exercise recovery. The PCr recovery curves were fit by a single exponential function to determine the recovery time constant (τPCr). Intracellular pH (pHi) was calculated by comparing the chemical shift difference between the PCr and inorganic phosphate (Pi) peaks in ppm, using the following equation: pHi = 6.85 + log[(δ − 3.56)/(5.64 − δ)]. pHi determined at the start of the recovery phase immediately postexercise did not differ between study days for patients with paired data available at baseline and end of study. For validation, the linear slope of the PCr recovery curve in the immediate postexercise period, during which the pH does not change significantly, was calculated using τPCr/3 as the time window duration. 31P-MRS spectroscopy could not be performed on all subjects, as a software upgrade required the scanner to come off-line before the study finished.
Dual X-Ray Absorptiometry
Whole body DEXA was performed using a Hologic QDR-4500 densitometer to determine total body and regional fat and lean body mass.
1H-MRS and MRI Scanning
MRS of calf muscles was performed for determination of lipid concentrations in skeletal muscle at the baseline and 1-mo visits. Scans were performed using a 1.5 Tesla system (Signa LX, version 12.0; GE Medical Systems, Milwaukee, WI). 1H-MRS of tibialis anterior and soleus muscles was performed after 8-h overnight fasting. Subjects were positioned feet first in the magnet bore, and the right calf was placed in a standard radiofrequency transmit and signal receive quadrature extremity coil. A triplane gradient echo localizer pulse sequence with echo time (TE) of 1.6 ms and repetition time (TR) of 54.0 ms and axial T1-weighted images (TR, 700 ms; TE, 14 ms; slice thickness, 4 mm; interslice gap, 1 mm) of the calf was performed for voxel placement. The first axial slice was always placed at the level of proximal fibular tip. Single-voxel MRS data were acquired using point-resolved spatially localized spectroscopy (PRESS) pulse sequence with TE of 25 ms, TR of 3,000 ms, 32 acquisitions, and 8 number of excitations (NEX). In all cases, a 3.4-ml voxel was placed on the largest cross-sectional area of the muscle, avoiding visible interstitial tissue, fat, or vessels. To ensure consistent positioning in follow-up examination, the axial slice used for voxel placement (counted from proximal fibular tip) was annotated in a logbook and screen-captured with voxel overlays and x-y coordinates. Fitting of all 1H-MRS data was performed using LCModel software (version 6.0-2, Stephen Provencher; Oakville, ON, Canada) running on a Linux (Red Hat, Raleigh, NC) workstation. Metabolite quantification was performed using eddy current correction and water scaling. The signal corresponding to IMCL (1.3 ppm) methylene protons was automatically scaled to unsuppressed water peak (W) (4.7 ppm), with values being expressed in institutional units (IU). Data for IMCL and extramyocellular lipid (EMCL) are reported for the tibialis anterior {[T(IMCL/W)], [T(EMCL/W)]} and the soleus {[S(IMCL/W)], [S(EMCL/W)]} muscles.
Axial T1-weighted single slices of the abdomen (at the level of L4 pedicle) and midthigh (half-distance between acetabulum and medial femoral condyle) were obtained using a water-suppressed fast spin-echo pulse sequence (TR 350 ms; TE 17.8 ms; echo train of 4; 5122 matrix; 1 NEX; 10-mm slice thickness; 40-cm field of view). The areas of visceral, abdominal subcutaneous, and midthigh subcutaneous fat were determined using graphic analysis software and expressed in square centimeters (Accuview, version 3.130; AccuImage Diagnostics, San Francisco, CA).
Assays
Assays were performed at the MIT General Clinical Research Center (GCRC) core laboratory facility. Serum insulin was measured using radioimmunoassay (RIA; Diagnostic Products, Los Angeles, CA). Intra-assay and interassay coefficients of variation (CVs) range from 3.1 to 9.3% and from 4.9 to 10.0%, respectively. TNF-α was measured by enzyme immunoassay (R&D Systems, Minneapolis, MN) with intra-assay and interassay CVs of ±6.7 and ±13.4%, respectively, and a sensitivity of 1.2 pg/ml. Adiponectin was measured using an RIA (Linco Research, St. Charles, MO). Intra-assay and interassay CVs range from 1.78 to 6.21% and from 6.90 to 9.25%, respectively. Serum leptin levels were determined using an RIA kit (Linco Research; intra-assay CV, 3.4%; interassay CV, 3.0 – 6.2%; sensitivity, 0.5 ng/ml). Serum glucose, lipids, and chemistries were measured using standard methodologies.
Statistical Analysis
Statistical analyses were performed using JMP SAS-based software. Student t-tests and ANOVA were utilized to detect differences between placebo and stavudine-treated participants, with a two-sided P value of <0.05 considered to indicate statistical significance. Paired t-tests were performed for within-group changes. Data are expressed as means ± SE unless otherwise indicated. For the GIRs, a single outlier who improved by >3 mg· kg−1 · min−1 was excluded from the analysis.
RESULTS
Subject Characteristics
Sixteen participants completed the baseline evaluation, and fifteen completed the 1-mo study period. Ten women and six men, with a mean age of 31.3 ± 2.4 yr and a mean BMI of 22.9 ± 0.5 kg/m2, were enrolled (Table 1). Two participants were African American, one was Asian, and thirteen were Caucasian. Participants had normal lipid profiles, glucose and insulin values, liver function, and renal function at baseline. None of the subjects had hypertension. Hyperinsulinemic-euglycemic clamp procedures were performed during the baseline evaluation and demonstrated normal insulin sensitivity with a mean GIR during the final hour of the clamp procedure of 8.0 ± 0.7 mg· kg−1 · min−1. Insulin sensitivity, body composition, and mitochondrial content and function did not differ between the groups at baseline.
Table 1.
Baseline demographics of participants
| Placebo (n = 9) | Stavudine (n = 7) | P Value | |
|---|---|---|---|
| Age, yr | 31±4 | 32±3 | 0.81 |
| Sex | |||
| Male | 3 (33%) | 3 (43%) | 0.70 |
| Female | 6 (67%) | 4 (57%) | |
| Race | |||
| Caucasian | 6 (67%) | 6 (86%) | 0.47 |
| African American | 1 (11%) | 1 (14%) | |
| Hispanic | 1 (11%) | 0 (0%) | |
| Asian | 1 (11%) | 0 (0%) | |
| BMI, kg/m2 | 22.7±0.8 | 23.0±0.5 | 0.77 |
Age and body mass index (BMI) values are presented as means ± SE. %Group in parentheses.
Response to Stavudine
Insulin sensitivity
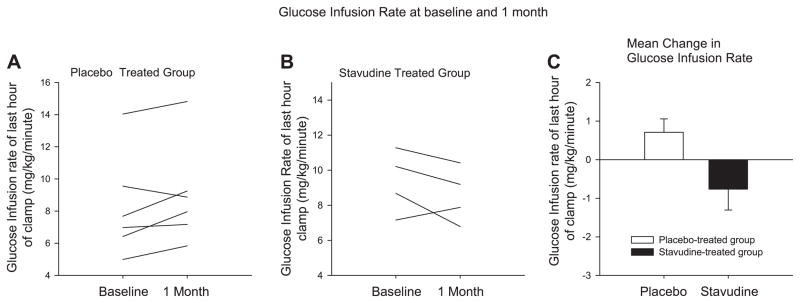
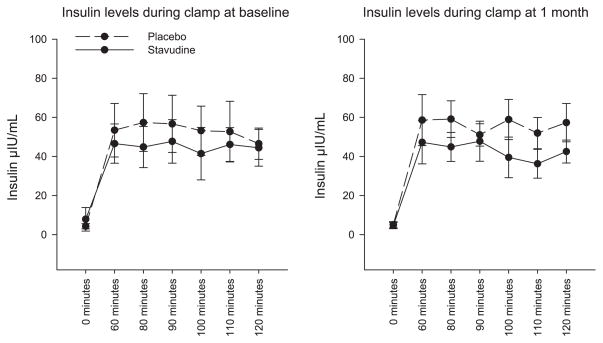
Insulin sensitivity was assessed by hyper-insulinemic-euglycemic clamp. The majority of stavudine-treated participants had a reduction in insulin sensitivity at the 1-mo time point. The change in GIR was significantly different between the groups over this time period (−0.8 ± 0.5 vs. +0.7 ± 0.4 mg· kg−1 · min−1, P = 0.04, stavudine vs. placebo, respectively; Fig. 1). When correcting for lean body mass, similar results were obtained (−1.0 ± 0.7 vs. 1.0 ± 0.6 mg· kg lean−1 · min−1, stavudine vs. placebo, P = 0.056). Individual data in all patients, pre- and poststavudine vs. placebo, are shown (Fig. 1). Corresponding mean insulin levels during the clamp procedure are shown. There was no difference in insulin levels between groups at baseline and the end of the study by MMANOVA (Fig. 2). Normal insulin levels are defined as <15 μIU/ml, based on the 90th percentile of available Framingham data (personal communication, Dr. James Meigs, Massachusetts General Hospital, Boston, MA). Corresponding values for M/I (glucose disposal adjusted for insulin) were − 0.03 ± 0.03 mg· kg−1 · min−1 · insulin−1 for stavudine and +0.01 ± 0.01 mg· kg−1 · min−1 · insulin−1 for placebo.
Fig. 1.
Change in glucose infusion rate (GIR; determined from the euglycemic hyperinsulinemic clamp) from baseline to the end of the study, shown for each participant in the placebo (A) and the stavudine-treated groups (B). C: mean changes in GIRs during euglycemic hyperinsulinemic clamp for each group (P = 0.04, stavudine vs. placebo). Results are means ± SE.
Fig. 2.
Comparison of insulin levels in stavudine-treated and placebo groups during clamp at baseline and 1 mo. P values for the difference between groups were P = 0.4 at baseline and P = 0.3 at 1 mo, as determined by MMANOVA. Error bars represent SD.
Mitochondrial quantification
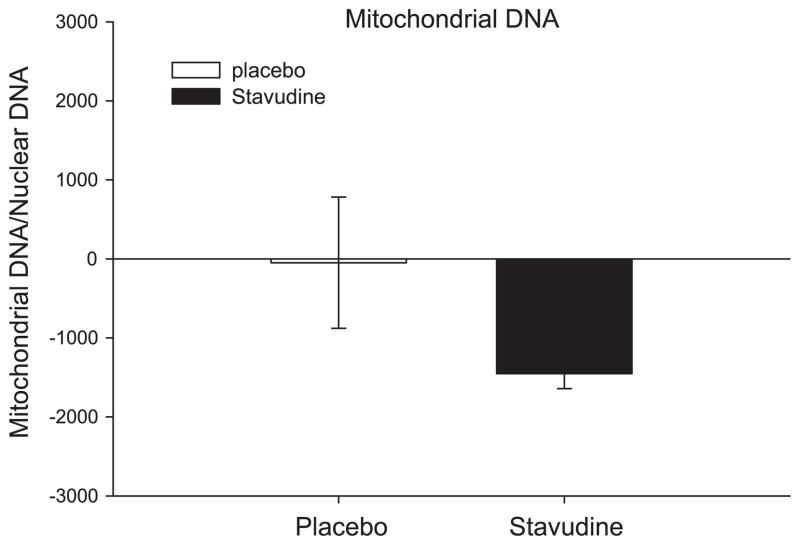
All the stavudine-treated participants showed a reduction in mtDNA content in muscle biopsies. The stavudine-treated participants showed a 52% reduction in mtDNA/nuclear DNA after the 1-mo study period, while the placebo-treated participants showed a mean 8% increase. The change within the stavudine-treated group was statistically significant (P = 0.005), whereas this was not the case among the placebo-treated patients (P = 0.9) (Fig. 3). The change in mitochondrial quantification in muscle biopsy tissues correlated with the change found in blood PBMCs (r2 = 0.6, P = 0.03), although the comparisons of mitochondrial changes in the PBMCs did not reach statistical significance. The change in mtDNA was not significantly related to change in GIR (r = 0.47, P = 0.24).
Fig. 3.
Change in mitochondrial DNA/nuclear DNA in muscle tissue. P = 0.005 for change in stavudine group, and P = 0.9 for change in placebo group. Results are means ± SE. P = 0.15 for the change between groups.
31P-MRS studies
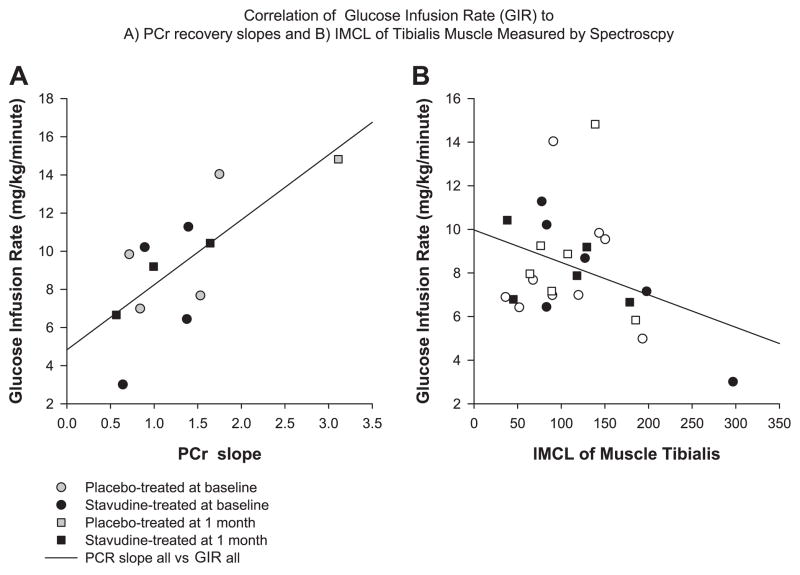
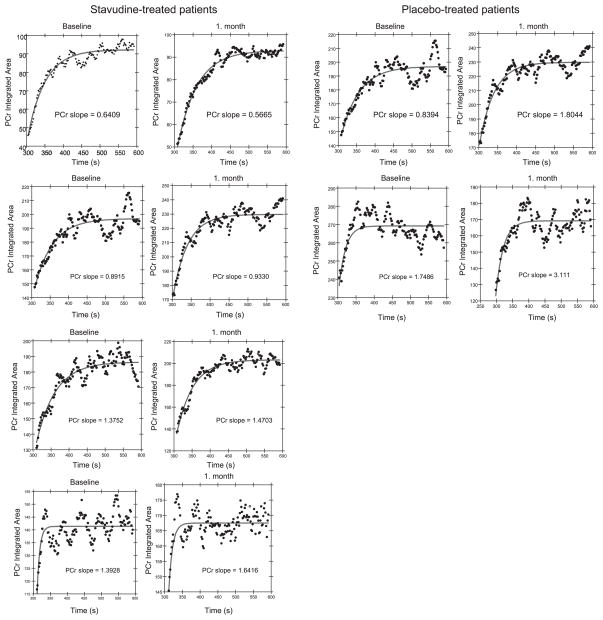
Data from spectroscopy studies were analyzed for PCr recovery. Fifteen spectroscopy studies were performed on nine participants. Six participants had completed studies at both the baseline and 1-mo visits. The PCr recovery period showed a significant negative correlation to the measurement of insulin sensitivity by hyperinsulinemic-euglycemic clamp during the final hour of the procedure (GIR = mg· kg−1 · min−1; time, 60 –120 min) when all observations, baseline and 1 mo, were evaluated (r2 = 0.50, P = 0.01). The negative correlation indicates that participants with a more rapid recovery, and therefore better mitochondrial function, were more insulin sensitive. Similarly, a reanalysis for confirmation, using a linear fit of the initial PCr recovery period, as previously described (7), showed a significant positive correlation (r2 = 0.52, P = 0.008), representing a more rapid recovery for more sensitive individuals (Fig. 4). Six participants had analysis of PCr recovery rate by initial slope analysis at baseline and 1 mo. Subjects in the placebo groups showed an increase in the slope recovery, whereas this was not seen among patients receiving stavudine, and the difference between the two groups was significant (0.1 ± 0.1 vs. 1.2 ± 0.2 s−1, P = 0.002, stavudine vs. placebo, respectively). The PCr recovery curves on all subjects for whom paired data are available are shown (Fig. 5).
Fig. 4.
A: correlation of phosphocreatine (PCr) recovery slope by 31P magnetic resonance spectroscopy (MRS) and GIR by hyperinsulinemic euglycemic clamp (r2 = 0.52, P = 0.008). B: correlation of intramyocellular lipid (IMCL) of tibialis muscle and GIR by hyperinsulinemic euglycemic clamp (r2 = 0.35, P = 0.08).
Fig. 5.
PCr recovery curves for each participant at baseline and at 1 mo. The PCr recovery slope for each patient is indicated.
1H-MRS studies
Alterations in the IMCL content of the soleus and tibialis muscles were evaluated by proton-based MRS. The association between IMCL and GIR tended to be significant (r = 0.35, P = 0.08), such that higher IMCL tended to be associated with lower insulin sensitivity in a combined analysis of all subjects (Fig. 4). However, the changes in IMCL were not significantly different in the stavudine and placebo-treated groups (Table 2). Similarly, EMCL did not change between the two groups.
Table 2.
Metabolic parameters
| Placebo (n = 9)
|
Stavudine (n = 7)
|
P Value for Comparison of Change Between Groups by ANOVA | |||
|---|---|---|---|---|---|
| Baseline | Change From Baseline | Baseline | Change From Baseline | ||
| Lipids | |||||
| HDL, mg/dl | 52±2 | 0±2 | 58±7 | 0±3 | 0.94 |
| LDL, mg/dl | 90±7 | −3±3 | 84±8 | 4±3 | 0.13 |
| Total cholesterol, mg/dl | 160±8 | −2±4 | 159±11 | 3±6 | 0.45 |
| Triglycerides, mg/dl | 69±14 | 5±9 | 45±3 | 2±6 | 0.77 |
| FFA, mEq/l | 0.57±0.06 | 0.06±0.09 | 0.64±0.10 | 0.08±0.11 | 0.88 |
| Body composition and nutritional data | |||||
| BMI, kg/m2 | 22.7±0.8 | 0.1±0.1 | 23.0±0.5 | 0.0±0.2 | 0.50 |
| WHR | 0.83±0.01 | −0.01±0.01 | 0.87±0.02 | −0.01±0.01 | 0.91 |
| Total fat, kg | 14.6±1.9 | −0.1±0.2 | 14.9±2.3 | 0.0±0.2 | 0.76 |
| Total lean, kg | 49.8±2.8 | −0.3±0.3 | 50.7±3.9 | −0.7±0.2 | 0.46 |
| VAT abdomen, cm2 | 29.8±5.5 | 0.5±1.7 | 36.8±5.1 | 2±2.7 | 0.65 |
| SAT abdomen, cm2 | 141.1±26.3 | 3.7±2.8 | 152.6±31.4 | 6.2±3.6 | 0.60 |
| SAT thigh, cm2 | 60.8±10.8 | 0.9±1.3 | 57.0±10.5 | 0.8±1.2 | 0.94 |
| %Body fat | 21.7±2.3 | 0.1±0.2 | 22.1±3.3 | 0.3±0.2 | 0.50 |
| Leptin, ng/ml | 5.2±1.3 | −0.9±0.7 | 6.5±2.6 | −0.3±1.1 | 0.64 |
| Tibialis (IMCL/W), IU | 105±17 | −2±12 | 131±33 | −38±23 | 0.18 |
| Soleus (IMCL/W), IU | 347±53 | −29±57 | 409±71 | −94±48 | 0.39 |
| Tibialis (EMCL/W), IU | 221±57 | −7±18 | 199±47 | −2±15 | 0.83 |
| Soleus (EMCL/W), IU | 584±82 | 107±85 | 726±120 | 115±80 | 0.95 |
| Inflammatory markers | |||||
| Adiponectin, μg/ml | 11.7±0.5 | 0.0±0.4 | 10.5±1.4 | 0.7±0.6 | 0.38 |
| TNF-α, pg/ml | 1.26±0.10 | 0.05±0.09 | 1.32±0.11 | 0.08±0.05 | 0.78 |
| Safety measurements | |||||
| Lactate, mmol/l | 0.9±0.1 | −0.2±0.2 | 0.8±0.1 | 0.1±0.1 | 0.15 |
| RBC count, million cells/μl | 4.2±0.1 | 0.1±0.1 | 4.4±0.3 | 0.0±0.1 | 0.67 |
| Hemoglobin, g/dl | 12.8±0.4 | 0.1±0.3 | 13.1±0.8 | 0.2±0.3 | 0.77 |
| Hematocrit, % | 37.4±1.1 | 0.6±0.6 | 38.9±2.4 | 0.1±0.7 | 0.60 |
| Creatinine, mg/dl | 0.8±0.1 | 0.0±0.0 | 0.7±0.1 | 0.0±0.0 | 0.93 |
| AST, U/l | 19±1 | 1±1 | 20±2 | 5±2 | 0.19 |
Values are means ± SE. FFA, free fatty acids; WHR, waist-to-hip ratio; VAT, visceral adipose tissue; SAT, subcutaneous adipose tissue; IMCL, intramyocellular lipid; W, unsupressed water peak; EMCL, extramyocellular lipid; RBC, red blood cell; AST, aspartate aminotransferase.
Body composition
As anticipated, because of the short duration of the study, there were no changes in body composition by densitometry, including total fat mass, total lean mass, and percent body fat mass, nor were there changes in visceral adipose tissue (VAT), abdominal subcutaneous adipose tissue (SAT), or midthigh SAT. In addition, anthropomorphic measurements, such as waist-to-hip ratio, were unchanged in both groups. Thus the changes noted in insulin sensitivity and mitochondrial quantification were not the result of changes in weight or fat distribution (Table 2).
Biochemical indexes
No significant changes between the groups were seen in lipid levels, free fatty acids, adiponectin, TNF-α, or leptin (Table 2).
Safety
No medication-related adverse events were reported during the 1-mo study. Participants had weekly safety visits and midstudy laboratory analysis. One participant required a dose reduction, to once daily, for complaints of dizziness and headache. However, the participant was randomized to placebo group. There were no significant between-group differences in levels of lactate, lipid parameters, blood pressure, renal or liver function, red blood cell count, hematocrit, or hemoglobin (Table 2).
DISCUSSION
Data from this prospective, randomized study demonstrate that short-term administration of stavudine, an NRTI, reduces insulin sensitivity in healthy subjects. In addition, stavudine results in a significant 52% reduction in muscle mtDNA within the treated subjects. Petersen et al. (15) demonstrated a decrease in mitochondrial function of 30% in the insulin-resistant offspring of individuals with type 2 DM, suggesting that mitochondrial dysfunction might contribute to the insulin resistance. The current investigation expands this area of inquiry by prospectively assessing the effects of a drug previously shown to result in mitochondrial depletion in peripheral white blood cells and in fat (3, 10, 13, 22). To our knowledge, this is the first prospective report of the effects of stavudine on insulin sensitivity and mtDNA in muscle. Importantly, we show this effect after a brief administration in healthy volunteers with no confounding by HIV, diabetes, family history of diabetes, or other disease states. These data support the hypothesis that altered mitochondrial function in muscle may be an important factor in the development of insulin resistance.
In this study, mtDNA decreased by 52% in those receiving stavudine, but our sample size was too small to show that changes in mtDNA were associated with insulin resistance. In addition to assessing mtDNA content in muscle, we utilized spectroscopy as a functional measure of mitochondrial activity to examine phosphocreatine recovery rates to a standardized exercise challenge (2, 17). Using 31P-MRS, we demonstrated a significant linear relationship between mitochondrial function and insulin sensitivity measured by euglycemic clamp among study participants. This technique has been used to evaluate mitochondrial function in muscle and is based on the principle that phosphocreatine recovery will be delayed in proportion to mitochondrial dysfunction. Our data utilizing spectroscopy suggested that mitochondrial dysfunction and insulin sensitivity were linked among study participants.
In contrast, our data do not suggest that mitochondrial dysfunction was associated with increased muscle fat. We studied the slow twitch oxidative soleus and the fast twitch tibialis anterior muscles by spectroscopy and found no changes in IMCL. This result does not rule out an effect on IMCL oxidation that may take longer than 4 wk to observe but does suggest that early alterations in insulin sensitivity may occur via other mechanisms. Furthermore, the sample size may have been too small to detect such changes, although trends in this direction were not observed. Tibialis IMCL tended to correlate inversely with insulin sensitivity, indicating that it is a reasonable surrogate for the measurement of muscle adiposity, with biological relevance. However, IMCL measures of vastus lateralis or biopsies from tibialis anterior and soleus might potentially provide better data that could be correlated to changes in mitochondrial function and insulin sensitivity.
The current study expands our knowledge of the effects of stavudine, a potent NRTI agent used in the treatment of HIV infection. This agent is known to result in mitochondrial toxicity, possibly due to inhibition of γ-polymerases (8, 11). More recently, a mitochondrial toxic effect secondary to intracellular depletion of uridine and metabolites has been suggested (21). The evaluation of the specific effects of stavudine has been complicated by the impact of HIV infection itself on mitochondrial function and insulin resistance. Mallon et al. (10) investigated the effects of stavudine on adipocytes in healthy individuals and found depletion in mitochondrial RNA, with upregulation of transcriptional regulators and genes involved in fatty acid oxidation, while genes involved in adipose differentiation were downregulated. In the current study, muscle tissue revealed a reduction in mtDNA/nuclear DNA after a 1-mo exposure to stavudine in healthy adults in the absence of HIV infection. These findings cannot be generalized to other NRTIs with potentially lesser effects on mitochondria.
The study was limited to a relatively short duration and small sample size but involved the use of detailed physiological techniques to provide novel information on the effects of stavudine administration on insulin sensitivity and mitochondrial function. Because of the complexity of the trial design, we could not obtain data for all participants for all endpoints; e.g., 31P-MRS scanning was not available for a few patients because of the installation of new software, and this limited the results of the study. Nonetheless, we were able to demonstrate in a prospective, randomized study significant changes in insulin sensitivity by hyperinsulinemic-euglycemic clamp in response to stavudine. Further studies are needed to investigate the degree to which the changes in muscle mtDNA, as demonstrated in this study, contribute to the rapid deterioration in insulin sensitivity seen with stavudine administration. The changes we observed were determined in healthy control subjects, with normal insulin sensitivity at baseline and no potentially confounding diseases or family history.
The current study provides prospective data demonstrating that a short course of therapy with a drug that affects mitochondrial function will result in decreased insulin sensitivity. The acquired alterations in mitochondrial number and function may contribute to worsening insulin sensitivity. Furthermore, larger studies are needed to understand the time course and mechanisms by which mitochondrial dysfunction and insulin resistance may be linked.
Acknowledgments
GRANTS
This study was funded in part by National Institutes of Health Grants R01-DK-59535 and M01-RR-01066. H. C. F. Côté holds a Michael Smith Foundation for Health Research Scholar Award [CI-SCH-050 (02-1)].
We thank the nursing and bionutrition staffs of the MIT GCRC for dedicated patient care, Jeff Breu of the MIT Core laboratory for performance of the assays needed in this protocol, and Izabelle Gadawski for the mtDNA assays.
Footnotes
DISCLOSURES
The study drug used in the current study was purchased commercially and not provided by Bristol Meyers Squibb, Inc. S. K. Grinspoon has received research grant support and honoraria from Bristol Meyers Squibb, Inc., unrelated to this project. H. C. F. Côté has pending patents related to the assessment of mtDNA.
References
- 1.Antonetti DA, Reynet C, Kahn CR. Increased expression of mitochondrial-encoded genes in skeletal muscle of humans with diabetes mellitus. J Clin Invest. 1995;95:1383–1388. doi: 10.1172/JCI117790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chance B, Eleff S, Leigh JS, Jr, Sokolow D, Sapega A. Mitochondrial regulation of phosphocreatine/inorganic phosphate ratios in exercising human muscle: a gated 31P NMR study. Proc Natl Acad Sci USA. 1981;78:6714–6718. doi: 10.1073/pnas.78.11.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Côté HC, Brumme ZL, Craib KJ, Alexander CS, Wynhoven B, Ting L, Wong H, Harris M, Harrigan PR, O’Shaughnessy MV, Montaner JS. Changes in mitochondrial DNA as a marker of nucleoside toxicity in HIV-infected patients. N Engl J Med. 2002;346:811–820. doi: 10.1056/NEJMoa012035. [DOI] [PubMed] [Google Scholar]
- 4.Dietrichson P, Coakley J, Smith PE, Griffiths RD, Helliwell TR, Edwards RH. Conchotome and needle percutaneous biopsy of skeletal muscle. J Neurol Neurosurg Psychiatry. 1987;50:1461–1467. doi: 10.1136/jnnp.50.11.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gan SK, Samaras K, Thompson CH, Kraegen EW, Carr A, Cooper DA, Chisholm DJ. Altered myocellular and abdominal fat partitioning predict disturbance in insulin action in HIV protease inhibitor-related lipodystrophy. Diabetes. 2002;51:3163–3169. doi: 10.2337/diabetes.51.11.3163. [DOI] [PubMed] [Google Scholar]
- 6.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 7.Larson-Meyer DE, Newcomer BR, Hunter GR, Hetherington HP, Weinsier RL. 31P MRS measurement of mitochondrial function in skeletal muscle: reliability, force-level sensitivity and relation to whole body maximal oxygen uptake. NMR Biomed. 2000;13:14–27. doi: 10.1002/(sici)1099-1492(200002)13:1<14::aid-nbm605>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Lewis W, Kohler JJ, Hosseini SH, Haase CP, Copeland WC, Bienstock RJ, Ludaway T, McNaught J, Russ R, Stuart T, Santoianni R. Antiretroviral nucleosides, deoxynucleotide carrier and mitochondrial DNA: evidence supporting the DNA pol gamma hypothesis. AIDS. 2006;20:675–684. doi: 10.1097/01.aids.0000216367.23325.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luzi L, Perseghin G, Tambussi G, Meneghini E, Scifo P, Pagliato E, Del Maschio A, Testolin G, Lazzarin A. Intramyocellular lipid accumulation and reduced whole body lipid oxidation in HIV lipodystrophy. Am J Physiol Endocrinol Metab. 2003;284:E274–E280. doi: 10.1152/ajpendo.00391.2001. [DOI] [PubMed] [Google Scholar]
- 10.Mallon PW, Unemori P, Bowen M, Miller J, Winterbotham M, Kelleher A, Williams K, Cooper D, Carr A. Nucleoside reverse transcriptase inhibitors decrease mitochondrial and PPARgamma gene expression in adipose tissue after only 2 weeks in HIV-unifected healthy adults. 11th Conf Retrovir Opportun Infect; San Francisco. 2004. [Google Scholar]
- 11.Miro O, Lopez S, Cardellach F, Casademont J. Mitochondrial studies in HAART-related lipodystrophy: from experimental hypothesis to clinical findings. Antivir Ther. 2005;10(Suppl 2):M73–M81. [PubMed] [Google Scholar]
- 12.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 13.Nolan D, Hammond E, Martin A, Taylor L, Herrmann S, McKinnon E, Metcalf C, Latham B, Mallal S. Mitochondrial DNA depletion and morphologic changes in adipocytes associated with nucleoside reverse transcriptase inhibitor therapy. AIDS. 2003;17:1329–1338. doi: 10.1097/00002030-200306130-00007. [DOI] [PubMed] [Google Scholar]
- 14.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeds D, Cade WT, Becker A, Mohammed S, Powderly WG, de Baar MP, de Rooij E, Yarasheski KE. Muscle mitochondrial DNA and RNA copy number/cell are reduced in treatment-naive HIV-infected people, regardless of glucose intolerance. 7th Int Workshop Adverse Drug React Lipodystrophy HIV; Dublin. 2005. [Google Scholar]
- 17.Ross BD, Radda GK, Gadian DG, Rocker G, Esiri M, Falconer-Smith J. Examination of a case of suspected McArdle’s syndrome by 31P nuclear magnetic resonance. N Engl J Med. 1981;304:1338–1342. doi: 10.1056/NEJM198105283042206. [DOI] [PubMed] [Google Scholar]
- 18.Sinnwell TM, Sivakumar K, Soueidan S, Jay C, Frank JA, McLaughlin AC, Dalakas MC. Metabolic abnormalities in skeletal muscle of patients receiving zidovudine therapy observed by 31P in vivo magnetic resonance spectroscopy. J Clin Invest. 1995;96:126–131. doi: 10.1172/JCI118012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song J, Oh JY, Sung YA, Pak YK, Park KS, Lee HK. Peripheral blood mitochondrial DNA content is related to insulin sensitivity in offspring of type 2 diabetic patients. Diabetes Care. 2001;24:865–869. doi: 10.2337/diacare.24.5.865. [DOI] [PubMed] [Google Scholar]
- 20.Vicent D, Piper M, Gammeltoft S, Maratos-Flier E, Kahn CR. Alterations in skeletal muscle gene expression of ob/ob mice by mRNA differential display. Diabetes. 1998;47:1451–1458. doi: 10.2337/diabetes.47.9.1451. [DOI] [PubMed] [Google Scholar]
- 21.Walker UA, Auclair M, Lebrecht D, Kornprobst M, Capeau J, Caron M. Uridine abrogates the adverse effects of antiretroviral pyrimidine analogues on adipose cell functions. Antivir Ther. 2006;11:25–34. [PubMed] [Google Scholar]
- 22.Walker UA, Bickel M, Lutke Volksbeck SI, Ketelsen UP, Schofer H, Setzer B, Venhoff N, Rickerts V, Staszewski S. Evidence of nucleoside analogue reverse transcriptase inhibitor–associated genetic and structural defects of mitochondria in adipose tissue of HIV-infected patients. J Acquir Immune Defic Syndr. 2002;29:117–121. doi: 10.1097/00042560-200202010-00002. [DOI] [PubMed] [Google Scholar]
- 23.Zaera MG, Miro O, Pedrol E, Soler A, Picon M, Cardellach F, Casademont J, Nunes V. Mitochondrial involvement in antiretroviral therapy-related lipodystrophy. AIDS. 2001;15:1643–1651. doi: 10.1097/00002030-200109070-00006. [DOI] [PubMed] [Google Scholar]