Abstract
Epigenetic modifications play an essential role in the regulation of gene expression and ultimately cell fate. Methylation of cytosine at CpG dinucleotides (mCpG) is an important epigenetic mark that has been correlated with cancer when present at promoter sites of tumor suppressor genes. In order to develop a rapid methodology for the direct assessment of global levels of DNA methylation, we first interrogated the methyl-CpG binding domains (MBDs), the Kaiso family of Cys2-His2 zinc fingers and a SET- and RING-associated (SRA) domain using a split-luciferase reassembly methodology. We identified MBD1 as the most selective domain for the discrimination between mCpG and CpG sites with over 90-fold selectivity. Utilizing a bipartite strategy, we constructed a purely methylation-dependent bipartite sensor for the direct detection of global levels of DNA methylation by attaching MBD1 domains to each of the split-luciferase halves. This new sensor was validated for the direct determination of genomic DNA methylation levels in in vitro studies without any intervening chemical or enzymatic processing of DNA. Finally, we demonstrated that this bipartite sensor can be utilized for monitoring dose-dependent changes in global levels of methylation in DNA from HeLa cells challenged with 5-aza-2′-deoxycytidine, a DNA methyltransferase inhibitor.
Introduction
Methylation of genomic DNA allows for epigenetic control of genomic stability, gene expression, and repression of transposable elements.1–2 More recently, tumor-type specific hypermethylation at promoter regions of tumor suppressor genes has been observed and correlated with cancer sub-types.3–4 Furthermore, genome-wide hypomethylation has also been shown to occur in tumor cells.5–7 In mammals DNA methylation occurs almost exclusively in CpG dinucleotides, where DNA methyltransferases catalyze the transfer of a methyl group from S-adenosylmethionine to cytosine.8–9 The elucidation of the mechanism for DNA demethylation, however, remains elusive.10–12 Interestingly, a 5′-hydroxymethylcytosine modification has recently been identified and is being actively investigated.13 The recognition of genomic methylation patterns in humans is mainly orchestrated by a family of proteins known as methyl-CpG binding domains (MBDs) along with the Kaiso family of methylation-specific Cys2-His2 zinc finger domains (ZBTBs, or zinc finger and BTB domain-containing proteins) and the SET- and RING-associated (SRA) domains of the E3 ubiquitin-protein ligase UHRF1 (ubiquitin-like PHD and RING finger domain-containing protein 1).14–17 These mCpG-binding proteins are reported to bind hypermethylated sites in gene promoters, act as transcriptional repressors and recruit DNA methyltransferases.18 The importance of identifying methylation sites at both the sequence-specific and global levels has lead to numerous approaches, including bisulfite sequencing9 and immunoprecipitation-based techniques.8 Bisulfite sequencing is often limited by DNA degradation as well as incomplete conversion of 5′-methylcytosine to uracil,19 while immunoprecipitation-based techniques for global DNA methylation can be time intensive. Therefore, the development of more direct methods for the detection of methylated DNA at both the sequence-specific and global levels has generated significant interest.9, 20–25
We have previously reported several split-protein reassembly or complementation-based sequence-specific methylation sensors. These sensors comprise genetically fragmented split-signaling proteins, including the green fluorescent protein (GFP),26 β-lactamase,27 and firefly luciferase,28 where MBD2 is attached to one fragment as a methylation specific recognition domain and a sequence-specific zinc finger (ZF) is attached to the complementary fragment for targeting a defined sequence (Figure 1a). The formation of a ternary complex drives reassembly of the fragmented protein to afford a detectable output. With this in mind, we explored the use of a sensitive, methylation-specific domain for the development of a new sensor that could directly report on global levels of DNA methylation in a genomic context. We envisioned that a sensor comprising one copy of the chosen domain attached to each half of the split-luciferase enzyme could potentially read out global levels of methylation (Figure 1b). Thus we turned to a systematic exploration of the human MBD, ZBTB and UHRF families in the context of our most sensitive reporter, the split-firefly luciferase system.
Figure 1. Schematic representation of split-luciferase complementation systems.
a) A methyl-binding domain (MBD) is tethered to the N-terminal fragment of firefly luciferase (NFluc) and a Cys2-His2 zinc finger (ZF) domain is tethered to the C-terminal fragment of firefly luciferase (CFluc). Addition of the DNA target containing a symmetrically methylated CpG dinucleotide (green) and the appropriate ZF binding site (ZFBS, blue) will result in DNA-binding and luciferase reassembly. b) The firefly luciferase fragments are both tethered to identical MBDs. Addition of a DNA target containing multiple adjacent mCpG dinucleotides results in mCpG dependent DNA-binding and luciferase reassembly.
The human members of the MBD family, namely MeCP2, MBD1, MBD2, MBD3, and MBD4), are defined by a significant degree of sequence and structural similarity.29–31 Numerous studies have focused on characterizing the ability of MBD family proteins to bind mCpG sites by utilizing common biochemical techniques, including electrophoretic mobility shift assays (EMSAs)32–33, DNase footprinting assays and chromatin immunoprecipitation (ChIP),34 and in vivo immunostaining.35 However, to our knowledge there are no reports of a direct evaluation of the methylation-specific binding of this set of MBDs. Additionally, characterization of the methyl-binding abilities of the ZBTB and UHRF proteins has yielded novel domains that preferentially bind to methyl-CpG over its unmethylated counterpart.36–37 We anticipated that the information gained by directly comparing all methyl-CpG binding domains would allow us to select an appropriate domain for developing more sensitive split-protein sensors for detecting global levels of DNA methylation. Herein, we report our systematic investigation of the mCpG binding capabilities of the human MBD, ZBTB and UHRF families in the context of a split-luciferase sensor. Building on these results, we also describe the development of a general di-MBD sensor for the direct detection of global levels of DNA methylation, which can potentially be utilized for assessing the effect of 5-aza-2′-deoxycytidine, a DNA methyltransferase inhibitor, in HeLa cells.
Materials & Methods
Plasmid Construction and mRNA Production
DNA encoding the N-terminal (residues 2 – 416) and C-terminal (residues 398 – 550) firefly luciferase fragments were generated by PCR with appropriate primers, inserted into a pETDuet-1 vector (Novagen) using standard techniques, and verified by dideoxyoligonucleotide sequencing. These fragments are herein referred to as NFluc and CFluc, respectively. Fragments encoding the MBDs (MeCP2, MBD1, MBD2, MBD3, and MBD4), the ZBTBs (ZBTB4, ZBTB33, ZBTB38), the UHRFs (UHRF1 and UHRF2) and ZFs (Zif268 and E2C) were generated by PCR starting from their respective plasmids (Supporting Information, Table S1). DNA-binding domains were conjugated to their respective firefly luciferase halves through a short glycine-serine linker. The fusion proteins were generated using standard cloning techniques and subsequently verified using dideoxyoligonucleotide sequencing (Supporting Information, Table S2). For preparation of templates for in vitro transcription, PCR fragments corresponding to the desired fusion proteins were generated using a forward primer containing a T7 RNA polymerase promoter and a Kozak sequence and a reverse primer containing a 3′ hairpin loop. The purified PCR products were subsequently used as templates for in vitro transcription using the RiboMAX™ Large Scale RNA Production System - T7 (Promega) following the manufacturer’s protocol.
DNA Target Preparation
All oligonucleotide targets were obtained HPLC purified from IDT (Supporting Information, Table S3). All designed DNA targets were annealed in 1x BamHI Buffer (NEB) using the following procedure: heating to 95 °C for 7 min, cooling to 56 °C at a rate of 0.1 °C sec−1, equilibrating at 56 °C for 5 min, cooling to 25 °C at a rate of 0.1 °C sec−1 and finally equilibrating at 25 °C for 10 min using a Labnet Multi Gene II thermocycler. The pETDuet recombinant plasmid variant, pET (7667 bp), and purified HeLa genomic DNA were methylated using M. SssI CpG methyltransferase (NEB) according to the manufacturer’s protocol. The extent of protection was determined by using the methylation sensitive endonucleases BstUI, where the cleavage of the target sequence is blocked by CpG methylation and McrBC, where the cleavage of the target sequence requires either symmetrical methylation or hemimethylation.
Treated HeLa Genomic DNA Preparation
HeLa cells were cultured in T-25 flasks at 1×105 cells per flask in 90% DMEM/F12 1:1 media (Lonza) supplemented with 10% FBS (Lonza), penicillin-streptomycin (Mediatech), and ampotericin B (JR Scientific). After 24 hours, triplicate flasks were treated with 1 μM, 250 nM, or 100 nM 5-aza-2′-deoxycytidine (Sigma) or the vehicle DMSO. Compound was removed and media was replaced after 24 hours of treatment, and cells were allowed to proliferate for 3 days before collection. At harvest the cells in each flask were detached with trypsin-EDTA, and DNA was purified using the Wizard® Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions. The effect of varying concentrations of 5-aza-2′-deoxycytidine on genomic DNA methylation was determined using the methylation sensitive endonuclease McrBC.
Split-Luciferase Reassembly using MBD/ZBTB/UHRF Fusion Proteins
Duplicate 25 μL translations were carried out in Flexi® Rabbit Reticulocyte Lysate (Promega) according to the manufacturer’s protocol using 1 pmol of MBD/ZBTB/UHRF-NFluc and CFluc-Zif268 mRNA, 10 μM ZnCl2, 0.5 μL of RNasin® Plus (Promega), and either 10 nM C10Z(m) or C10Z(u) target DNA. To investigate MBD1 sensitivity for hemimethylation, duplicate 25 μL translations were carried out in Flexi® Rabbit Reticulocyte Lysate (Promega) according to the manufacturer’s protocol using 1 pmol of MBD1-NFluc and CFluc-Zif268 mRNA, 10 μM ZnCl2, 0.5 μL of RNasin® Plus (Promega), and 10 nM C2Z(m), C2Z(tm), C2Z(bm) or C2Z(u) target DNA. To determine the detection limit, duplicate translations were carried out in Flexi® Rabbit Reticulocyte Lysate using 1 pmol MBD1-NFluc and CFluc-E2C mRNA, 10 μM ZnCl2, 0.5 μL of RNasin® Plus and 100, 75, 50, 25, 10, or 5 pM of the DNA target C2E(m). In all cases, translations were incubated at 30 °C for 90 minutes and assayed by adding 80 μL of Steady-Glo® Luciferase Assay System (Promega) to 20 μL of translated lysate. Light emission was monitored 1 minute after Steady-Glo addition using a Turner 20/20n luminometer with a 3 second delay and a 10 second integration time.
35S-Methionine SDS-PAGE
For verification of protein synthesis, 25 μL translations were carried out in Flexi® Rabbit Reticulocyte Lysate (Promega) according to the manufacturer’s protocol using 1 pmol of MBD-NFluc, 0.5 μL of RNasin® Plus (Promega), 1 μL 35S-Methionine (1175 Ci/mmol) and allowed to incubate for 90 minutes at 30 °C. The samples were applied to an SDS-PAGE gel, and the dried gel was exposed overnight to a Storage Phosphor Screen (Molecular Dynamics) and scanned using a Typhoon 9410 Variable Mode Imager (GE Healthcare).
Split-Luciferase Reassembly of the diMBD1-Fluc Sensor
In all experiments pertaining to the diMBD1-Fluc sensor, 23.75 μL translations were carried out in Flexi® Rabbit Reticulocyte Lysate according to the manufacturer’s protocol using 1 pmol of MBD1-NFluc and CFluc-MBD1 mRNA, and 0.5 μL of RNasin® Plus and allowed to incubate for 90 minutes at 30 °C. For the limit of detection using synthetic oligonucleotides, 0.5 pmol of RNA was used instead of the typical 1 pmol. Target DNA (synthetic oligonucleotides, plasmids, or genomic DNA) was added post-translation and incubated for 60 minutes at 4 °C. For the initial test using synthetic oligonucleotides, the DNA targets C6C(m) and C21C(m) were added to a final concentration of 10 nM. For the limit of detection, the DNA target C6C(m) was added to a final concentration of 20, 10, 8, 6, 4, 2, or 1 nM. For the initial test using methylated plasmid DNA, the DNA targets pET(m) or pET(u) were added to a final concentration of 16 pM. For the limit of detection, the methylated plasmid DNA target pET(m) was added to a final concentration of 5, 2.5, 1.0, 0.50, or 0.25 pM. For the initial test using HeLa genomic DNA, 500 ng of the native DNA target HeLa(n) was added to the in vitro translated fusion proteins. For the 5-aza-2′-deoxycytidine HeLa genomic DNA, 350 ng of genomic DNA isolated from HeLa cells that had been treated with either vehicle or 100 nM, 250 nM, or 1.0 μM 5-aza-2′-deoxycytidine was added to the in vitro translated fusion proteins. In the case of the HeLa(m) and HeLa(n) DNA titrations, 30, 25, 20, 15, or 10 ng of the DNA target HeLa(m) or 30, 25, or 20 ng of the DNA target HeLa(n) was added to the in vitro translated fusion proteins. All samples were assayed by the addition of 80 μL of Steady-Glo® Luciferase Assay System to 20 μL of translated lysate. Light emission was monitored 1 minute after Steady-Glo addition using a Turner 20/20n luminometer with a 3 second delay and a 10 second integration time. All experiments were carried out in duplicate, with the exception of HeLa genomic DNA from cells treated with 5-aza-2′-deoxycytidine, which was carried out in triplicate.
Results & Discussion
Direct Comparison of MBD, ZBTB and UHRF Family Members
We initially sought to establish a split-protein reassembly system that is suited for use as a platform to compare the relative binding of all members of the MBD, ZBTB, and UHRF families of domains to symmetrically methylated target oligonucleotides. Our previously established, first-generation Fluc-based methylation sensor had the C-terminal Fluc fragment (CFluc) attached to the 3-finger ZF Zif268, which binds its target 5′-GCG-TGG-GCG-3′ with a reported Kd of 6 nM,38 and the N-terminal Fluc fragment (NFluc) attached to the methyl-CpG binding domain of MBD2.28 As such, we investigated the MBD, ZBTB and UHRF families using an identical platform, where NFluc (residues 2 – 416) was conjugated to the domain to be interrogated creating MBD/ZBTB/UHRF-NFluc, while CFluc (residues 398 – 550) was left conjugated to Zif268 yielding CFluc-Zif268 (Figure 1a). An oligonucleotide, C10Z(m), which contained a symmetrically methylated CpG site followed by a 10 base pair separation and the Zif268 binding site, was designed as a target for directing reassembly. The unmethylated counterpart is defined as C10Z(u). Cell-free translations were initiated by the addition of in vitro transcribed mRNA corresponding to MBD/ZBTB/UHRF-NFluc and CFluc-Zif268 fusions in the presence of either methylated or unmethylated DNA targets.
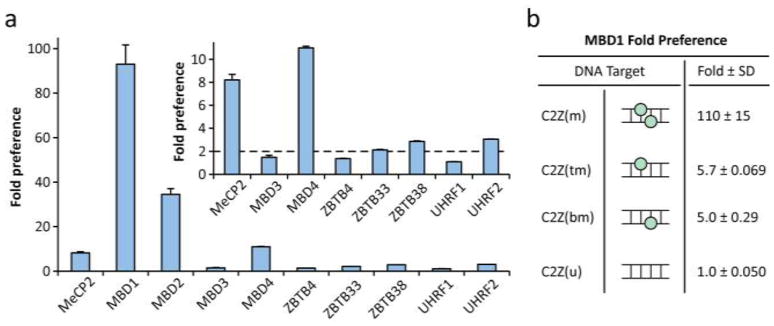
We found that four (MeCP2, MBD1, MBD2, and MBD4) of the five previously tested MBDs produced at least 2-fold luminescent signal over background in the presence of the mCpG-containing target DNA (Figure 2a). Of these four MBDs there was considerable variation in overall intensity, ranging over an order of magnitude from MeCP2 to MBD1. The most sensitive MBD was, by far, MBD1, which showed a 93-fold preference for the mCpG dinucleotide over the unmethylated counterpart. This sensitivity decreased in the following order: MBD2 (35-fold), MBD4 (11-fold), and MeCP2 (8-fold). Despite this wide spread in mCpG-binding selectivity, the tested MBD-NFluc fusions showed comparable levels of protein expression, suggesting that the results reflected the true mCpG-binding preferences (Supporting Information, Figure S1). Although the ZBTB domains reportedly bind methylated CpG dinucleotides, they require multiple adjacent mCpG sites or specific DNA sequences for high affinity binding.36 It is therefore not surprising that the ZBTB domains showed very little preference for a single mCpG dinucleotide in the context of a sequence unrelated to the preferred binding site. However, both ZBTB33 (Kaiso) and ZBTB38 showed at least a 2-fold preference for a single mCpG over the unmethylated counterpart. Lastly, the UHRF domains showed very little preference for symmetrically methylated DNA, as they reportedly bind hemimethylated DNA with a slightly higher affinity.37 However, the binding affinities of these domains for either hemimethylated or symmetrically methylated CpG dinucleotides are significantly higher than the protein concentrations used in this experiment, potentially affecting the results. Regardless, the previously studied UHRF1 showed virtually no preference for symmetrically methylated CpG, while the homologous UHRF2 showed about 3-fold preference.
Figure 2. Interrogation of MBD, ZBTB and UHRF binding affinities.
a) Initial test of MBD, ZBTB and UHRF family members’ preferences for the symmetrically methylated C10Z(m) DNA over the unmethylated counterpart C10Z(u) using the split-luciferase sensor system incorporating the 3-finger ZF Zif268 as a DNA anchor. Inset shows enlarged subset of the same data for clarity. Error bars represent +1 standard deviation. b) Fold preference of MBD1 binding to DNA using the split-luciferase sensor system incorporating Zif268 with symmetrically methylated C2Z(m) and hemimethylated (on sense strand C2Z(tm) or antisense strand C2Z(bm)) DNA as compared to the unmethylated counterpart C2Z(u).
With the large preference for symmetrically methylated CpG dinucleotides achieved using the MBD1-based sensor, we thought it would also show a moderate discrimination between hemimethylated and unmethylated dinucleotides. It has been suggested that hemimethylated CpG dinucleotides are intermediates in the processes responsible for the variable degrees of hypomethylation and hypermethylation that appear during carcinogenesis.39 Here, similar oligonucleotides with either CpG methylated on both strands [C2Z(m)], or the sense [C2Z(tm)] and antisense strands [C2Z(bm)] only with a 2 base pair spacer followed by the Zif268 binding site were used. This spacing was chosen given our previously demonstrated preference of MBD domains for this distance between the mCpG and the ZF binding site.26–27 Once again, the MBD1-based sensor showed a large preference (110-fold) for the methylated target (Figure 2b). Additionally, MBD1 discriminates between the fully methylated and hemimethylated targets by about 20-fold, while retaining a moderate 5-fold preference for the hemimethylated CpG dinucleotides. Lastly, high affinity binding of the MBD1-based sensor was confirmed using the 6-finger ZF E2C, which binds to its target sequence 5′-GGG-GCC-GGA-GCC-GCA-GTG-3′ with a reported Kd of 500 pM.40 This MBD1-based sensor was found to have a detection limit of 500 amol or 10 pg target DNA C2E(m), which was detectable over the average background signal plus three standard deviations (99% confidence levels) (Supporting Information, Figure S1). These results confirm that the MBD1-based sensor can be potentially utilized for reporting primarily upon symmetrically methylated targets.
Determination of Global Methylation in Plasmid DNA
We next turned our attention to designing sensors that can directly report on global changes in genomic methylation, which have been observed in both disease progression and aging.41–42 A global methylation sensor also has the potential for providing a convenient means for the interrogation of the activity of DNA methyltransferases as well as putative demethylating enzymes. Moreover, global methylation sensors may allow for a new method for determining the activity of small molecules that perturb methylation levels, for example by inhibition of methyltransferases. Having identified MBD1 as an appropriate mCpG recognition domain that provides high fidelity in discriminating between methylated versus unmethylated targets, we chose to utilize MBD1 as the detection domain for evaluation of global DNA methylation. Toward this end, MBD1 was tethered to each half of the fragmented firefly luciferase, creating MBD1-NFluc and CFluc-MBD1, which we collectively refer to as diMBD1-Fluc (Figure 1b). In contrast to our previous sequence-specific methylation sensors that localize to specific sites in the genome, this new sensor should potentially report upon the average methylation density of the DNA being analyzed.
As an initial test, we translated the split-sensor in either the presence or absence of designed targets containing symmetrically methylated CpGs separated by a 6 base pair (bp) [C6C(m)] or 21 bp [C21C(m)] spacer (Figure 3a). The 6 bp separation was expected to provide adjacent binding sites for the MBDs, allowing for reassembly of the diMBD1-Fluc system based on previous studies,26–27 while the 21 bp separation was engineered to test the limit for feasible reassembly between the luciferase halves, where the linkers (33 amino acids can potentially span ~93 Å) should theoretically be long enough to span two turns of DNA (~70 Å linearly).43 The C6C(m) target generated considerable signal over background, while the 21 bp spacer also yielded a modest signal increase over background, demonstrating the feasibility of split-luciferase reassembly despite the large separation between adjacent mCpG sites. The previously reported split-GFP and split-lactamase sensors are significantly more sensitive to DNA length as compared to split-luciferase, which may be a consequence of the inherent flexibility at the N- and C-termini of firefly luciferase, where a large degree of disorder has been observed at both termini in the crystal structure.44 Finally, the diMBD1-Fluc split-sensor was reassembled in the presence of decreasing amounts of the C6C(m) target, revealing a limit of detection of 200 fmol or 2.6 ng, which was detectable over the average background signal plus three standard deviations (Figure 3b).
Figure 3. Detection of CpG methylation with diMBD1-Fluc methylation sensor.

a) Initial experiment utilizing the diMBD1-Fluc bipartite sensor for the detection of multiple methylated CpGs using the doubly methylated targets CpG-6-CpG(m) and CpG-21-CpG(m) or in the absence of DNA. b) Relative luminescence in the presence of decreasing concentrations of CpG-6-CpG(m), showing a linear trend across the tested concentrations. c) Detection of global methylation of a fully methylated pETDuet plasmid, pET(m), generated by treatment with M. SssI CpG methyltransfrerase compared to unmethylated pET, pET(u), or no DNA target. Inset shows the nearest neighbor distance separation between all 530 predicted mCpGs in pET(m), with 81% of predicted mCpG sites within 6 bp and 90% of predicted mCpG sites within 21 bp. d) Relative luminescence of the diMBD1-Fluc sensor in the presence of decreasing concentrations of pET(m), which shows a linear trend across the tested concentrations. Error bars represent +1 standard deviation for all bar graphs, and ± 1 standard deviation for all scatter plots.
To address global methylation in the context of a larger DNA target, a derivative of the recombinant bacterial plasmid pETDuet-1, pET, was chosen for initial evaluation (Figure 3c). Since our strategy requires that two MBD1 domains bind to adjacent mCpG sites, we calculated the proximity of nearest-neighbor sites in the plasmid using our CGContentAnalyzer code (Supporting Information, Figure S2). The spacing between any two directly adjacent CpGs is 20 bp or less for 81% of the sites in the pET DNA target, and 30 bp or less for 90% of the sites (Figure 3c, inset). The plasmid was fully methylated using M. SssI CpG methyltransferase, to create pET(m). Methylation was judged to be complete as verified by digestion using the methylation-sensitive endonucleases BstUI and McrBC (Supporting Information, Figure S3). We then determined the ability of our MBD1-based split-sensor to generate differential signal in the presence of the methylated, pET(m), as compared to the unmethylated, pET(u), plasmid. We observed that the diMBD1-Fluc sensor accurately distinguishes between the plasmids as a function of methylation status, with a limit of detection of 25 amol or 125 pg of plasmid DNA being detectable above the average background signal plus three standard deviations (99% confidence level) (Figure 3d). These results establish that the diMBD1-Fluc sensor can clearly distinguish methylated oligonucleotides as well as differentiate between methylated and unmethylated plasmid DNA.
Evaluation of Global Methylation of HeLa Genomic DNA
Having established that the diMBD1-Fluc sensor can report on global methylation, we turned to investigate whether our sensor would be suitable for the semi-quantitative evaluation of the global methylation levels of HeLa genomic DNA. HeLa DNA has been reported to be methylated at ~50–60% of all CpG sites in the genome.45–46 As a first test, we purified genomic DNA from unsynchronized HeLa cells and exposed it to our MBD1 split-sensors, resulting in a 26-fold signal over background in the presence of 500 ng HeLa genomic DNA, HeLa(n) (Figure 4a).
Figure 4. Investigation of the global methylation frequency of HeLa genomic DNA.
a) Detection of global methylation of HeLa genomic DNA, HeLa(n), as compared to no DNA target with the diMBD1-Fluc sensor. b) Relative luminescence from the diMBD1-Fluc sensor in the presence of decreasing concentrations of the HeLa(n) and fully methylated, HeLa(m), generated by treatment with M. SssI CpG methyltransfrerase. A standard curve generated with HeLa(m) upon which HeLa(n) values at known DNA concentrations of 200, 250, and 300 pg/mL have been interpolated based on luminescence of diMBD1-Fluc. The analysis of this data shows that the overall genomic HeLa(n) DNA methylation frequency is ~ 50% by this method. c) The dose dependent effect of 5-aza-2′-deoxycytidine, a DNA methyl transferase inhibitor, on genomic DNA methylation in HeLa cells detected by the diMBD1-Fluc sensor. Error bars represent +1 standard deviation for all bar graphs, and ± 1 standard deviation for all scatter plots.
We next sought to establish a semi-quantitative approach for determining the percent of methylated CpG sites in genomic DNA. To accomplish this for natively methylated HeLa genomic DNA, HeLa(n), we first generated a standard curve using known concentrations of fully methylated HeLa DNA, HeLa(m), prepared by treatment with the M. SssI CpG methyltransferase. The following analysis makes the assumption that the total number of sensor accessible methylated CpGs is equal in both samples. By interpolating the signal from native HeLa DNA in comparison to the standard curve generated from the fully methylated HeLa genomic DNA (Figure 4b), we determined that the methylation frequency of native HeLa DNA is ~50% of all possible CpG sites in the genome. The value of ~50% determined by this approach is on the lower spectrum of previously determined global methylation levels of HeLa DNA.45–46 It is possible that CpG islands in genomic DNA from cancer cells are over-methylated in comparison to other CpG sites and that these sites are sterically less accessible to our sensor, leading to an underestimation.45 This semi-quantitative approach based on internal methylated standards may be applied when utilizing the diMBD1-Fluc sensor as a simple semi-quantitative approach to address changes in global methylation in different cell types.
Finally, we sought to utilize the diMBD1-Fluc methylation sensor for determining changes in the global levels of genomic methylation in HeLa cells. Thus, low-confluency HeLa cells were treated with varying concentrations of the DNA methyltransferase 1 (DNMT1) inhibitor 5-aza-2′-deoxycytidine. Following three days of replication, total genomic DNA was isolated from cells exposed to each drug concentration. The changes in methylation levels of both the vehicle- and 5-aza-2′-deoxycytidine-treated HeLa DNA were futher supported by digestion using the methylation-sensitive endonuclease McrBC, which selectively cleaves symmetrically methylated or hemimethylated DNA over non methylated DNA (Supporting Information, Figure S4). The degree of degradation of genomic DNA from vehicle-treated cells was visibly greater than that of 5-aza-2′-deoxycytidine-treated cells. The diMBD1-Fluc sensor was subsequently used to assess the clinically relevant dose-dependent decrease in methylation upon addition of 5-aza-2′-deoxycytidine, where we observed a significant decrease in luminescent signal as a function of increasing inhibitor concentration, which agrees with reported literature data (Figure 4c).47 The ability to utilize the diMBD1-Fluc sensor for measuring a dose-dependent decrease in methylation levels mediated by 5-aza-2′-deoxycytidine treatment demonstrates that our new approach can be utilized for investigating reagents or environmental conditions that induce hypomethylation or hypermethylation.
Conclusion
The elucidation of binding specificities of both naturally occurring and designed zinc finger domains has paved the way for site-specific localization of nucleases,48 methylases,49 and integrases50 to targeted DNA sequences. The utility of DNA-targeting domains that recognize DNA modifications, such as DNA methylation or hydroxymethylation,13 may provide methodologies complementary to those realized with ZFs. Herein, we have systematically compared the methylation specificity for a large number of known human domains in the context of a split-luciferase biosensor. We identified MeCP2, MBD1, MBD2, and MBD4 as domains that provide selective binding to DNA containing mCpG sites, while MBD3, ZBTB4, ZBTB33, ZBTB38, UHRF1, UHRF2 did not demonstrate significant mCpG binding. Within the context of this assay platform, we found that MBD1 has a greater than 100-fold preference for mCpG sites over CpG sites as well as a 20-fold preference over identical sites that are hemimethylated. Having established the specificity of these domains, we designed a new split-luciferase sensor for interrogating global levels of DNA methylation based on MBD1, where each fragment of split-Fluc is attached to MBD1. This biosensor was shown to provide a simple and rapid method for determining the global methylation frequency in native HeLa genomic DNA preparations. Moreover, the new sensor was shown to be useful for monitoring changes in HeLa DNA methylation levels in response to treatment with a DNA methyltransferase inhibitor. Thus, we anticipate that the rapid, sensitive, and homogeneous assay systems described herein may ultimately allow for the detection of methylation changes in both specific sequences as well as in global levels of DNA methylation.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Ghosh lab for helpful discussions, and Dr. Pierre-Antoine Defossez for the gift of PAD1258. A.H.B. thanks the Beckman Foundation and. J.L.F. thanks the Philanthropic Education Organization, Chapter BB for support. This research was partially supported by the NSF (CHE-0548264) and NIH (R21CA143661 and R01GM077403). Molecular structures were rendered using PyMol; DeLano, W. L. (www.pymol.org).
Footnotes
Supporting Information Available: Methods and details regarding plasmids containing the utilized proteins, sequences of short oligonucleotide targets, methylation of both pET(u) and HeLa(n) DNA, investigation of variable methylation levels of 5-aza-2′deoxycytidine-treated HeLa genomic DNA, and the CGContentAnalyzer code.
References
- 1.Meehan RR, Stancheva I. Essays Biochem. 2001;37:59–70. doi: 10.1042/bse0370059. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Huang HJS, Petrelli NJ, Zhang XL, O’dorisio MS, Held WA, Cavenee WK, Plass C. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M, Corn PG, Baylin SB, Herman JG. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 5.Feinberg AP, Vogelstein B. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 7.Ehrlich M. J Cell Biochem. 2003;88:899–910. doi: 10.1002/jcb.10464. [DOI] [PubMed] [Google Scholar]
- 8.Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, Cigudosa JC, Huang THM, Esteller M. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meissner A, Mikkelsen TS, Gu HC, Wernig M, Hanna J, Sivachenko A, Zhang XL, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Nature. 2008;454:766–U91. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ooi SKT, Bestor TH. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 12.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 13.Tahiliani M, Koh KP, Shen YH, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 15.Berger J, Bird A. Biochem Soc T. 2005;33:1537–1540. doi: 10.1042/BST0331537. [DOI] [PubMed] [Google Scholar]
- 16.Filion GJP, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. Mol Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae SH, Cheong HK, Cheong C, Kang SH, Hwang DS, Choi BS. J Biol Chem. 2003;278:45987–45993. doi: 10.1074/jbc.M306038200. [DOI] [PubMed] [Google Scholar]
- 18.Wade PA. Bioessays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- 19.Fraga ME, Esteller M. Biotechniques. 2002;33:632–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 20.Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. BMC Genomics. 2010;11 doi: 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, Mai ZB, Dai Z, Zou XY. Chem Commun. 2010;46:7781–7783. doi: 10.1039/c0cc00983k. [DOI] [PubMed] [Google Scholar]
- 22.Tainaka K, Sakaguchi R, Hayashi H, Nakano S, Liew FF, Morii T. Sensors-Basel. 2010;10:1355–1376. doi: 10.3390/s100201355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang HX, Nakata E, Hamachi I. Chembiochem. 2009;10:2560–2577. doi: 10.1002/cbic.200900249. [DOI] [PubMed] [Google Scholar]
- 24.Yamada H, Tanabe K, Nishimoto SI. Bioconjugate Chem. 2008;19:20–23. doi: 10.1021/bc7003318. [DOI] [PubMed] [Google Scholar]
- 25.Patterson A, Caprio F, Vallee-Belisle A, Moscone D, Plaxco KW, Palleschi G, Ricci F. Anal Chem. 2010;82:9109–9115. doi: 10.1021/ac1024528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stains CI, Furman JL, Segal DJ, Ghosh I. J Am Chem Soc. 2006;128:9761–9765. doi: 10.1021/ja060681j. [DOI] [PubMed] [Google Scholar]
- 27.Porter JR, Stains CI, Segal DJ, Ghosh I. Anal Chem. 2007;79:6702–6708. doi: 10.1021/ac071163+. [DOI] [PubMed] [Google Scholar]
- 28.Porter JR, Stains CI, Jester BW, Ghosh I. J Am Chem Soc. 2008;130:6488–6497. doi: 10.1021/ja7114579. [DOI] [PubMed] [Google Scholar]
- 29.Hendrich B, Bird A. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho KL, Mcnae LW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. Mol Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 31.Ohki I, Shimotake N, Fujita N, Jee JG, Ikegami T, Nakao M, Shirakawa M. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen HF, Adie K, Chaubert P, Bird AP. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klose RJ, Sarraf SA, Schmiedeberg L, Mcdermott SM, Stancheva I, Bird AP. Mol Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Saito M, Ishikawa F. J Biol Chem. 2002;277:35434–35439. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- 36.Sasai N, Nakao M, Defossez PA. Nucleic Acids Res. 2010;38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian CM, Li SD, Jakoncic J, Zeng L, Walsh MJ, Zhou MM. J Biol Chem. 2008;283:34490–34494. doi: 10.1074/jbc.C800169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavletich NP, Pabo CO. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 39.Shao CB, Lacey M, Dubeau L, Ehrlich M. Epigenetics. 2009;4:165–175. doi: 10.4161/epi.4.3.8277. [DOI] [PubMed] [Google Scholar]
- 40.Beerli RR, Segal DJ, Dreier B, Barbas CF. Proc Natl Acad Sci U S A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones PA, Baylin SB. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 42.Richardson B. Ageing Res Rev. 2003;2:245–261. doi: 10.1016/s1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 43.Ohnishi S, Kamikubo H, Onitsuka M, Kataoka M, Shortle D. J Am Chem Soc. 2006;128:16338–16344. doi: 10.1021/ja066008b. [DOI] [PubMed] [Google Scholar]
- 44.Conti E, Franks NP, Brick P. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 45.Brown SE, Fraga MF, Weaver ICG, Berdasco M, Szyf M. Epigenetics. 2007;2:54–65. doi: 10.4161/epi.2.1.3880. [DOI] [PubMed] [Google Scholar]
- 46.Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 47.Liu ZF, Liu SJ, Xie ZL, Blum W, Perrotti D, Paschka P, Kilsovic R, Byrd J, Chan KK, Marcucci G. Nucleic Acids Res. 2007;35:e31. doi: 10.1093/nar/gkl1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng XD, Noyes MB, Zhu LHJ, Lawson ND, Wolfe SA. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nomura W, Barbas CF. J Am Chem Soc. 2007;129:8676. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 50.Gordley RM, Gersbach CA, Barbas CF. Proc Natl Acad Sci U S A. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.