Abstract
The critical role of Aurora kinase in cell cycle progression and its deregulation in cancer has garnered significant interest. As such, numerous Aurora targeted inhibitors have been developed to date, almost all of which target the ATP cleft at the active site. These current inhibitors display polypharmacology; that is, they target multiple kinases, and some are being actively pursued as therapeutics. Currently, there are no general approaches for targeting Aurora at sites remote from the active site, which in the long term may provide new insights regarding the inhibition of Aurora as well as other protein kinases, and provide pharmacological tools for dissecting Aurora kinase biology. Toward this long term goal, we have recently developed a bivalent selection strategy that allows for the identification of cyclic peptides that target the surface of PKA, while the active site is blocked by an ATP-competitive compound. Herein, we extend this approach to Aurora kinase (Aurora A), which required significant optimization of selection conditions to eliminate background peptides that target the streptavidin matrix upon which the kinases are immobilized. Using our optimized selection conditions, we have successfully selected several cyclic peptide ligands against Aurora A. Two of these inhibitors demonstrated IC50 values of 10 μM and were further interrogated. The CTRPWWLC peptide was shown to display a noncompetitive mode of inhibition suggesting that alternate sites on Aurora beyond the ATP and peptide substrate binding site may be potentially targeted.
The Aurora family of serine/threonine kinases, which consist of Aurora A, B, and C, play a central role in coordinating cytoskeletal and chromosomal events during mitosis.1 Specifically, Aurora A localizes to the spindle poles and is involved in centrosome maturation and separation, initiation of mitosis, spindle assembly, and cytokinesis.2, 3 On the other hand, Aurora B, an important element of the chromosomal passenger complex,4 functions at the kinetochore to regulate proper alignment of the chromosomes on the mitotic spindle.5, 6 Aurora C, although not as extensively studied, is believed to be complementary in function to Aurora B.7 Both Aurora A and Aurora B are regarded as oncogenes, showing transformative potential when overexpressed in vitro and have been shown to be aberrantly expressed and amplified in several cancers.8–11 As such, both kinases have been extensively targeted for potential cancer therapeutics.8
In general, the development of truly selective protein kinase inhibitors has proven to be extremely challenging, as the structure of the kinase catalytic domain and particularly the ATP-binding region are highly conserved among the greater than 500 members of the human kinome,12 while numerous enzymes also utilize ATP as a substrate. The favored methods of generating kinase inhibitors, namely screening small molecule libraries against the catalytic domain of a chosen kinase, generally result in compounds that bind in the ATP-binding site (so-called Type I inhibitors) and are usually poorly selective across the kinome.13 More recently, a few compounds have been discovered that exploit non-conserved regions of the ATP-binding site, such as a hydrophobic pocket blocked in many kinases by a bulky “gatekeeper” residue or a pocket present in the inactive, or “DFG out” conformation of several kinases.14, 15 This has lead to heightened interest in developing strategies to identify kinase inhibitors that not only do not occupy the ATP-binding site but perhaps target kinases outside the core catalytic domain (true allosteric inhibitors).16
Unexplored regions of the kinase, namely anywhere but the ATP cleft, hold the potential to reveal novel sites for inhibitor development. Owing to the intricate regulation of protein kinases and their conformational flexibility, such allosteric sites may possibly exist. Recently several allosteric kinase inhibitors have been identified through novel screening methods. For example, the inclusion of regulatory domains and the use of differential screening with varying ATP concentration have identified several allosteric ligands of AKT isoforms.17, 18 However, methods for identifying allosteric ligands that target the kinase domain directly have been more elusive. A recent approach combining HTS using MS and NMR has identified MAPK inhibitors (biaryl-tetrazole class) with 11 – 16 μM Kd values for the unactive kinase and prevent activation.19 In another example, differential cytotoxicity screening against BCR-ABL positive cells was utilized and after discarding hits resembling known ATP-competitive compounds, a new class of inhibitors containing a 4,6-pyrimidine core were discovered. These new inhibitors were shown to operate in an allosteric fashion by targeting a distal myristoyl binding pocket of c-ABL.20, 21 Betzi and coworkers in another example of allosteric inhibitor screening combined fluorescent probes and protein crystallography where the probe, 8-anilino-1-naphthalene sulfonate (ANS), bound an allosteric pocket near the ATP site in CDK2 with an apparent Kd of 37 μM.22 Due to the lower affinity of most initial allosteric hits, which are typically greater than 10 μM, many allosteric ligands may be potentially missed during traditional HTS campaigns. However, the potential for selectivity for these new classes of allosteric ligands provides the impetus for redesigning current methodologies to discover such inhibitors.
Unlike most small molecule inhibitors, peptides are potentially amenable to targeting the peptide binding site or kinase surface as opposed to binding the ATP-cleft, and hence have the potential advantage of probing less conserved regions. An exciting application of these surface-targeting ligands has been in the generation of selective bivalent inhibitors, which covalently combine surface binding peptide moieties with small molecules that are known to target the ATP-binding site. This combined targeting has been successfully employed against protein kinases to produce inhibitors of enhanced potency and selectivity compared to their starting fragments.23–26
Recently, we have developed an approach to generate bivalent inhibitors utilizing phage displayed peptide libraries, and successfully demonstrated its feasibility in creating a new class of potent and selective inhibitors of a model kinase, cAMP-dependent protein kinase A (PKA).27, 28 In our approach, the ATP-binding site is occupied with a pan-inhibitor, staurosporine, and a phage displayed peptide library is directed to the kinase surface via the non-covalent assembly of two coiled-coils conjugated to each moiety, allowing for their simultaneous binding (Figure 1). After several rounds of in vitro selection, the two ligands (staurosporine and a selected peptide) are covalently linked to generate a potential bivalent inhibitor with greater binding affinity and perhaps an enhanced selectivity profile, due to the targeting of the kinase surface. The initial application of this approach to PKA produced bivalent ligands that are >90-fold more potent than the starting staurosporine derivative alone.27 Importantly, kinetic analysis of the cyclic peptide demonstrated it to be a noncompetitive inhibitor.28 In our efforts to test the generality of this approach and potentially discover noncompetitive inhibitors against therapeutically relevant kinases, we chose to target the most extensively studied kinase of the Aurora family1, Aurora Kinase A (Aurora A).
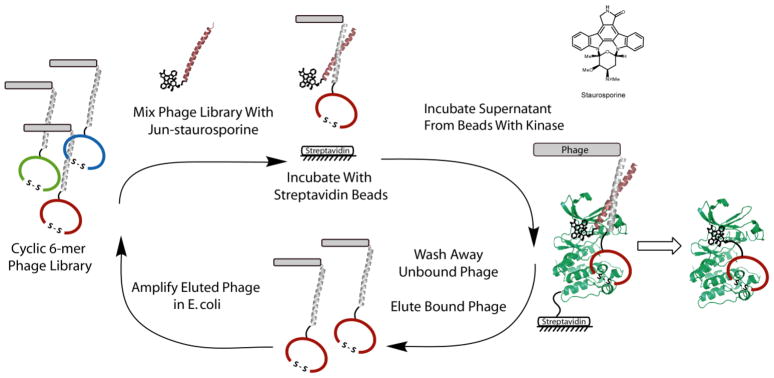
Figure 1. The Bivalent Selection Strategy Applied to Aurora A (AUR2).
Left The selection process is shown where the initial phage library (fos-cyclic peptides) is mixed with jun-staurosporine, incubated with “blank” streptavidin beads and then unbound phage/jun-staurosporine assemblies are transferred to AUR2 kinase streptavidin beads. Unbound phage are washed away after a 30-minute incubation period and the bound phage are eluted with low pH and amplified in E. coli. After this iterative process, selected peptides are identified by DNA sequencing and subsequently synthesized and tested by kinase assays. Right The general approach is outlined where the phage library and ATP-competitive compound are bound simultaneously via the non-covalent association of the leucine zipper pair, fos and jun. After an in vitro selection, peptides can be conjugated to the ATP-competitive compound, staurosporine, to generate a bivalent ligand.
Our bivalent phage display approach shown in Figure 1 was applied to Aurora A as described previously for PKA;27, 28 however, challenges arose concerning high background binding phage (streptavidin binding) and low potencies of selected sequences for Aurora A. These problems were overcome by appropriate changes in selection conditions. The final selection protocol resulted in the discovery of two peptides with low micromolar IC50 values for Aurora A, which to our knowledge are among the most potent peptides identified to date for Aurora A. One of these peptides was further interrogated by kinetic analysis and showed a noncompetitive mode of inhibition.
RESULTS AND DISCUSSION
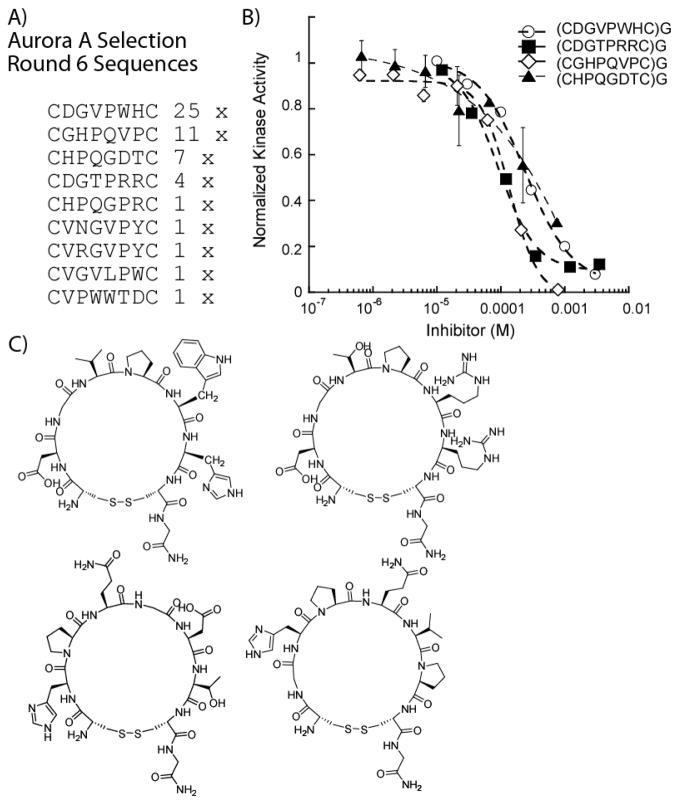
Phage display, essentially as described previously,27 was carried out against biotinylated Aurora A immobilized on streptavidin modified magnetic beads. After six rounds of selection, convergent sequences were found and the four most prevalent peptides were synthesized via solid phase peptide synthesis and characterized through kinase inhibition assays (Figure 2). Of the selected peptides, a motif consisting of the tri-amino acid HPQ was found in several clones, which has been previously shown to target streptavidin.29 However, since several sequences did not contain known streptavidin binding motifs, all four peptides were synthesized to characterize their Aurora A inhibitory potential. Each of the selected peptides was found to inhibit Aurora A at relatively high micromolar concentrations (91–243 μM), alluding to a potential lack of kinase specificity (Figure 2b). The presence of the previously known HPQ motif suggested that the isolated peptides may preferentially target the streptavidin beads over the immobilized kinase, even after a pre-incubation step using fresh streptavidin beads without immobilized kinase. Since the selected peptides inhibited weakly or were potentially background sequences that bind either the beads or streptavidin (supplementary data), we decided to further optimize selection conditions to favor more potent peptides. Furthermore, we also performed a background selection on the streptavidin beads alone (without the addition of jun-staurosporine) to understand the full breadth of streptavidin and magnetic bead binding sequences, HPQ-containing or otherwise, that are generated from our library
Figure 2. Selection Results and Analysis by Kinase Assays of Aurora A Selected Cyclic Peptides.
(A) Initial selection results yielded clear consensus peptides. (B) The four most prevalent selected cyclic peptides were assayed against 0.5 nM Aurora A and displayed very high micromolar IC50 values: 242 μM for (CDGVPWHC)G, 91 μM for (CDGTPRRC)G, 137 μM for (CHPQGDTC)G and 234 μM for (CGHPQVPC)G. (C) Structures of the selected peptides.
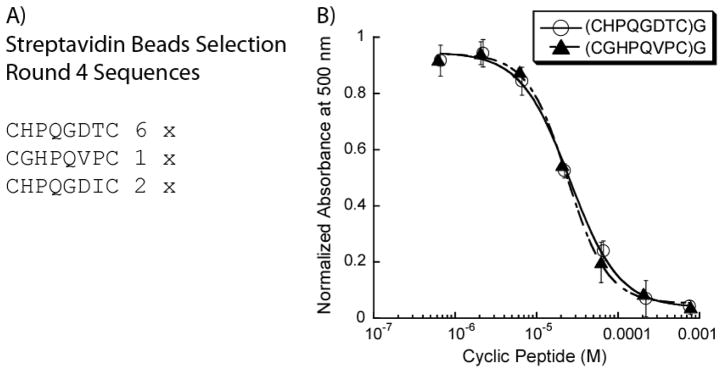
The results of the streptavidin magnetic beads background selection (4 rounds) are shown in Figure 3. The two most prevalent peptides (from 24 sequences) are (CHPQGDTC)G and (CGHPQVPC)G, which were amongst the most prominent HPQ-containing sequences during the first Aurora A selection. To a first approximation, this confirmed our hypothesis that the phage display conditions needed to be further optimized to eliminate streptavidin binding peptides. In order to adequately optimize the negative selection process for complete removal of the streptavidin binding HPQ sequences, we also determined the affinity of (CHPQGDTC)G and (CGHPQVPC)G for streptavidin. It has been established through X-ray crystallography that HPQ-containing peptides bind in the biotin binding pocket of the streptavidin monomer,30, 31 and thus our background peptides would be amenable to the HABA dye competitive displacement assay. 32, 33 Through our HABA assays, we were able to obtain IC50 values of 25.6 μM and 24 μM for (CHPQGDTC)G and (CGHPQVPC)G (figure 3), respectively, while the determination of the streptavidin-HABA dissociation constant (supplementary data) allowed the calculation of the respective dissociation constants (Kd = 20.7 μM and 19.4 μM).
Figure 3. Characterization of Selected Cyclic Peptides From the Streptavidin Coated Magnetic Beads Biopanning.
(A) The background selection against streptavidin coated magnetic beads reached consensus by the fourth round, with the HPQ motif was clearly present in the majority of selected peptides. (B) Two selected peptides were evaluated in a HABA-competition binding assay for streptavidin and the IC50’s were found to be 25.6 μM ((CHPQGDTC)G) and 24 μM ((CGHPQVPC)G).
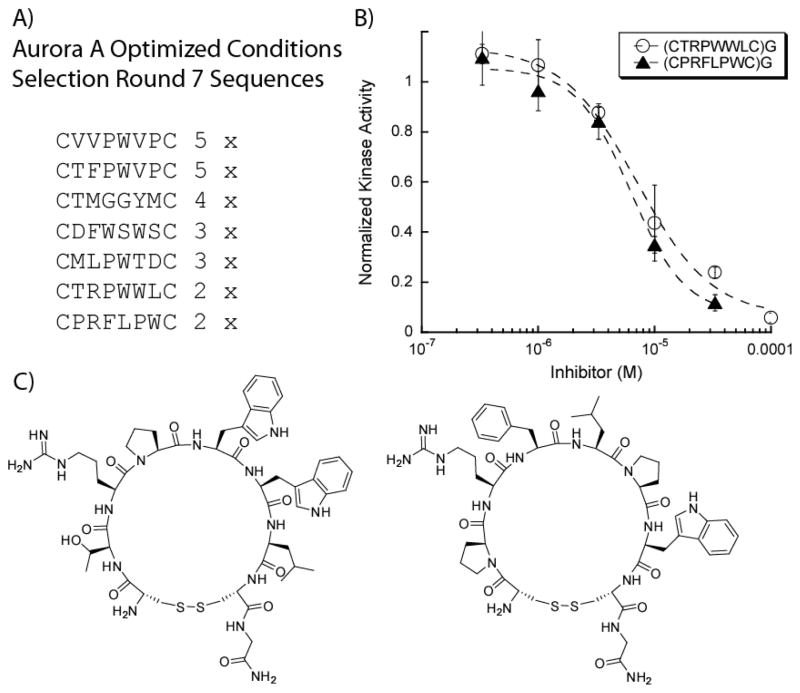
As the background peptides clearly have significant affinity for streptavidin compared to Aurora A, we sought to increase stringency in order to favor Aurora A binding peptides. Therefore, our objectives were two-fold: (1) prevent enrichment of streptavidin binding sequences through a more rigorous negative selection protocol and (2) test harsher conditions to potentially increase the potency of our target binders against Aurora A. The harsher wash conditions involved increasing the amount of detergent, Tween 20, and BSA to further diminish nonspecific background binding. We evaluated the effects of these modifications (0.1% Tween 20, 0.2% BSA) through a selection against Aurora A and a control selection against streptavidin. After several rounds of selection against the both Aurora A and streptavidin alone, no clear motifs emerged in either case (Table 1). This suggested that these conditions are too stringent as even the streptavidin binding HPQ motif was not observed. We next increased the number of washes and include free streptavidin (10 μM) so that the HPQ peptide should readily be removed during washes with free streptavidin while not effecting Aurora A binding peptides. Under these new set of conditions, an Aurora A selection was performed resulting in convergent sequences that may potentially target Aurora A with no appearance of the HPQ motif indicative of streptavidin binding (Figure 4A).
Table 1.
Results From Selections under New Selection Conditions
| AURORA A Kinase Round 3 | Streptavidin Beads Round 3 |
|---|---|
| CFNTSSQC | CQLPYRQC |
| CQELTQRC | CEVRQRWC |
| CFTKVCSC | CTSSLQQC |
| CGNRHYQC | CPIQQFRC |
| CEKRPRRC | CQLLGRVC |
| CVDNRKGC | CQWFHSIC |
Figure 4. Aurora A Assays of Selected Peptides Show Low Micromolar Inhibition Constants.
(A) The optimized selection conditions yielded several convergent sequences with no dominating sequence. (B) Two peptides from the selection have potent IC50’s of 10.5 μM for (CTRPWWLC)G and 6.2 μM for (CPRFLPWC)G at 0.5 nM AUR2. (C) Two selected peptides that were synthesized and evaluated in AUR2 kinase assays.
From the selection results, an apparent preference for proline containing peptides was found, often with a tryptophan residue adjacent to it in a PW motif. Interestingly, the selection results did not correlate with the initial Aurora A selection, as neither (CDGVPWHC)G or (CDGTPRRC)G appeared under the newer stringent wash conditions. Two of the most prevalent peptides, (CTRPWWLC)G and (CPRFLPWC)G, were synthesized, purified, and assayed against Aurora A. The two peptides were found to be the most potent peptide inhibitors of Aurora A evaluated thus far with our selection strategy, having IC50 values of 7 and 6 μM, respectively (0.5 nM Aurora A).
Since the selected peptides themselves are low micromolar inhibitors of similar potency despite the significantly different sequences, we next sought to probe the mode of inhibition and selectivity. The selected peptide, (CTRPWWLC)G, was tested at three different concentrations of Kemptide and showed no appreciable change in the IC50 (supplementary material), which potentially suggested a noncompetitive mode of inhibition as we have previously observed when targeting PKA.28 Further evaluations of the mode of inhibition of the selected peptide by kinetic analysis, (CTRPWWLC)G also suggests noncompetitive inhibition with respect to peptide substrate (Kemptide, LRRASLG) as shown in Figure 5 (see supplementary material for fits to competitive, noncompetitive, mixed, and uncompetitive).
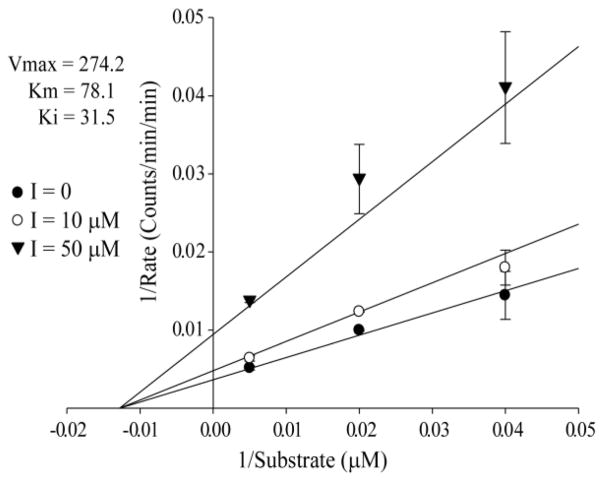
Figure 5. Aurora A Kinetic Assays of Selected Peptide, (CTRPWWLC)G, Best Fit to Noncompetitive Inhibition.
Kinetic experiments varying Kemptide concentration and inhibitor ((CTRPWWLC)G) concentrations are shown in a double reciprocal plot fit to noncompetitive inhibition. The determined Km for Kemptide of 78 μM is in agreement with literature values. The Ki of 31 μM is in agreement with the determined IC50 for Aurora A Kinase.
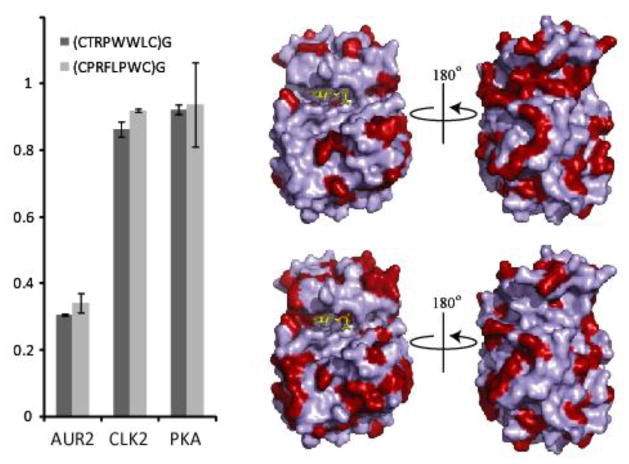
With the peptides in hand and the observation that the peptides are significantly hydrophobic with a single Arg in one case and a Thr and Arg is the other case, we asked whether the observed inhibition was selective for Aurora A or whether these peptides were potentially non-specific protein kinase binders. In order to test this, both peptides were assayed against the AGC kinase family (PKA), the CMGC family (CLK2) and the target (Aurora A, Other family) at 10 μM. Gratifyingly, as Figure 6 shows, the Aurora A selected peptides inhibit their intended kinase at 10 μM peptide concentration with negligible inhibition of either PKA or CLK2. Thus these studies demonstrate that phage display methods can be applied to discover cyclic peptide inhibitors of therapeutically relevant kinases.
Figure 6. Assessment of Kinase Selectivity of Aurora A Selected Peptides.
The two selected peptides, (CCTRPWWLC)G and (CPRFLPWC)G, were separately assayed against the AGC family kinase PKA and the CMGC family kinase CLK2 at a fixed peptide concentration of 10 μM. Both peptides inhibit Aurora A while not inhibiting CLK2 and PKA. Shown to the right are surface representations of Aurora A with light blue residues representing regions that are homologous to PKA (top) and CLK2 (bottom). The red regions represent differences between each respective kinase, and are therefore possible regions for peptide binding sites on Aurora A. (PDB code: 2DWB)
In summary, the application of our bivalent selection strategy to other kinases beyond PKA such as Aurora A has necessitated significant adaptation and optimization to isolate kinase selective peptides while avoiding background peptides. The improved protocol eliminates HPQ containing streptavidin binding sequences, which were fully characterized, while yielding more potent peptide inhibitors of Aurora A. The peptides identified in the course of this study are more potent than that discovered by a recent helix-loop-helix displayed phage display approach against Aurora A,34 where the best peptide appended to the helices, GRRVVVSFAWD, showed 35% inhibition at a concentration of 100 μM.34 The present work also suggests that the Aurora A inhibiting peptides discovered by this approach may have the potential to exhibit a noncompetitive mode of inhibition with respect to the peptide substrate, which was also the case in our previous study with PKA. We can speculate that the bivalent phage display approach prevents peptide binding at the substrate site perhaps by steric occlusion. Alternatively, pre-existing protein binding sites on the protein kinases are more amenable for binding peptides selected during phage-display. The selected peptides may possibly bind inactive conformations of the kinases and thereby inhibit kinase activity. Future studies will test whether appropriate bivalent analogs provide higher affinity and selectivity as well as aim to identify the binding site on Aurora A for the newly discovered peptides. The peptides identified by our phage display approach in the long term may provide a means for identifying new sites on protein kinases that are amenable for targeting with small molecules with new mechanisms of inhibition and aid in providing selective pharmacological tools for studying Aurora A biology.
METHODS
Phage Display Panning
Biotinylation of 5 μg Aurora A (Millipore) was performed using 20 eq Sulfo-NHS-LC-LC-biotin (Pierce) with 100 μM ATP in 300 μl final reaction volume in a dialysis cassette (Pierce) at 4°C for 90 minutes. After dialysis, the biotinylated kinase was diluted (1.6 ml final volume), aliquoted and stored at −80°C until use. The extent of biotinylation was monitored by kinase assay after immobilization of one aliquot on 5 μl of M-280 Streptavidin Beads (Invitrogen) according to manufacturer’s protocol. For the first round of selection, 1.1 × 109 phage were mixed with jun-staurosporine (2 μM final concentration) and incubated on ice with 5 μl of M-280 Streptavidin Beads for 30 minutes. This solution was transferred to another 5 μl of M-280 Streptavidin Beads and incubated at room temperature for 30 minutes. After washing with PBS-T (0.05% Tween 20), the bound phage were eluted with 0.2 M glycine (pH 2) and 11 μM staurosporine for 12 minutes and neutralized with 20 μl Tris buffer (pH 11). After amplification of sequences in E. coli., samples were analyzed by DNA sequencing.
Peptide Synthesis
All peptides were synthesized as described previously.27, 28 Using standard Fmoc protection strategies in solid phase peptide synthesis, all peptides were synthesized on Rink Amide resin (substitution level = 0.4 mmol/g). Coupling conditions consisted of 3 equivalents of the appropriate Fmoc protected amino acid, 3 eq PyBOP, and 6 eq DIEA in DMF for an hour. Cleavage from the resin (94% TFA, 2.5% EDT, 2.5% water, and 1% TIPS) was carried out for 2 hours, then the peptides were precipitated in chilled ether and isolated by centrifugation. Peptide oxidation was accomplished by dissolving the peptides in 20% DMSO in PBS, pH = 7.4, and incubating at 37 °C for 36 hours and was monitored with Ellman’s Reagent. Compounds were purified by HPLC and fractions containing the peptides (confirmed by mass spectrometry) were pooled and lyophilized.
Kinase Assays
Aurora A kinase assays were performed in triplicate. In a 25 μl final volume, [γ-32P]ATP (100 μM) initiated the reaction with 0.5 nM Aurora A (Millipore) and Kemptide (LRRASLG, 200 μM) in Aurora A Assay Buffer (20 mM MOPS, 10 mM MgCl, 1 mM EDTA, pH 7) with 0.01% BSA and 2.5% DMSO. After 1 h, 20 μl of the reaction mixture was spotted on P81 phosphocellulose paper. The samples were washed three times in 500 ml (0.85% phosphoric acid) and once in 500 ml (acetone) for 3 minutes each. The amount of 32P labeling of the peptide substrate was quantified using a Beckman LS 6000IC liquid scintillation counter and data were normalized to reactions containing no inhibitors, which were run in triplicate. The selectivity assays were run in duplicate, and were performed as similarly described for Aurora A except the length of time each kinase incubated with ATP was 40 minutes instead of one hour. Kinase concentrations and substrate identities/concentrations for the selectivity assay are as follows: 0.5 nM Aurora A and Kemptide (LRRASLG, 200 μM), 2 nM CLK2 (Invitrogen) and 2.5 μg substrate (AKRRRLSSLRA), and 0.52 nM PKA (Promega) and 30 μM Kemptide.
Kinetic Assays
Aurora A kinetic assays were performed in duplicate with no inhibitor, 10 and 50 μM inhibitor ((CTRPWWLC)G). In a 75 μl final volume, [γ-32P]ATP (100 μM) initiated the reaction with 0.5 nM Aurora A and Kemptide (LRRASLG at 25, 50 and 200 μM) in Aurora A Assay Buffer (20 mM MOPS, 10 mM MgCl, pH 7) with 0.01% BSA and 2.5% DMSO. At 10 minute intervals, 10 μl of the reaction mixture was spotted on P81 phosphocellulose paper. The samples were washed three times in 500 ml (0.85% phosphoric acid) and once in 500 ml (acetone) for 3 minutes each. The amount of 32P labeling of the peptide substrate was quantified using a Beckman LS 6000IC liquid scintillation counter.
HABA-Competition Assays
Characterization of the streptavidin background peptides were performed essentially as previously described 33 and were run either in duplicate or triplicate. Briefly, in a final reaction volume of 120 μL, an equimolar complex of streptavidin and HABA in PBS buffer (final concentration = 25 μM) was allowed to incubate with a variable concentration of (CHPQGDTC)G or (CGHPQVPC)G for 1 hour. After this time, the absorbance at λ = 500 nm was monitored with a Beckman DU 520 UV/Vis spectrophotometer. The absorbance of 25 μM HABA at λ = 500 nm was subtracted from the raw absorbance values, and all points were subsequently normalized to the HABA/streptavidin complex without peptide and fit to the Hill equation (eqn. 1).
| (1) |
Where LR is the fraction of ligand bound, LB is the concentration of bound ligand, LF is the concentration of unbound ligand, L is the total concentration of ligand and nH is the Hill coefficient.
| (2) |
From the Hill equation the IC50 is determined which can then be used to calculate the dissociation constant Kd according to equation 2. Where KL2 is the dissociation constant of the selected peptide and streptavidin complex, KL1 is the dissociation constant of the HABA-streptavidin complex and L1 is the HABA concentration.
Supplementary Material
Acknowledgments
We thank members of the Ghosh laboratory for helpful discussions and the NIH (R21CA141974) and the NSF (CHE-0548264) for partial support.
Footnotes
Supplementary information containing the determination of HABA-streptavidin dissociation constant, complete results from the selections, peptides mass spectrometric data and HPLC traces are available online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Bio. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 2.Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H. Aurora-a kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278:51786–51795. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- 3.Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang DW, Nitta M, Hatakeyama K, Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 4.Vader G, Medema RH, Lens SMA. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006;173:833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Yan XM, Cao LH, Li Q, Wu YH, Zhang HX, Saiyin H, Liu XH, Zhang XQ, Shi QH, Yu L. Aurora C is directly associated with Survivin and required for cytokinesis. Genes Cells. 2005;10:617–626. doi: 10.1111/j.1365-2443.2005.00863.x. [DOI] [PubMed] [Google Scholar]
- 8.Moore AS, Blagg J, Linardopoulos S, Pearson ADJ. Aurora kinase inhibitors: novel small molecules with promising activity in acute myeloid and Philadelphia-positive leukemias. Leukemia. 2010;24:671–678. doi: 10.1038/leu.2010.15. [DOI] [PubMed] [Google Scholar]
- 9.Zhou HY, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 10.Bischoff JR, Anderson L, Zhu YF, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CSM, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. Embo J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 12.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1916. 1933–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 13.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, Mclauchlan H, Klevernic I, Arthur JSC, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (ST1571, Imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 16.Cox KJ, Shomin CD, Ghosh I. Tinkering outside the kinase ATP box: allosteric (type IV) and bivalent (type V) inhibitors of protein kinases. Future Med Chem. 2011;3:29–43. doi: 10.4155/fmc.10.272. [DOI] [PubMed] [Google Scholar]
- 17.Barnett SF, Defeo-Jones D, Fu S, Hancock PJ, Haskell KM, Jones RE, Kahana JA, Kral AM, Leander K, Lee LL, Malinowski J, McAvoy EM, Nahas DD, Robinson RG, Huber HE. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartnett JC, Barnett SF, Bilodeau MT, Defeo-Jones D, Hartman GD, Huber HE, Jones RE, Kral AM, Robinson RG, Wu ZC. Optimization of 2,3,5-trisubstituted pyridine derivatives as potent allosteric Akt1 and Akt2 inhibitors. Bioorg Med Chem Lett. 2008;18:2194–2197. doi: 10.1016/j.bmcl.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 19.Comess KM, Sun CH, Abad-Zapatero C, Goedken ER, Gum RJ, Borhani DW, Argiriadi M, Groebe DR, Jia Y, Clampit JE, Haasch DL, Smith HT, Wang SY, Song DY, Coen ML, Cloutier TE, Tang H, Cheng XH, Quinn C, Liu B, Xin ZL, Liu G, Fry EH, Stoll V, Ng TI, Banach D, Marcotte D, Burns DJ, Calderwood DJ, Hajduk PJ. Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases. Acs Chem Biol. 2011;6:234–244. doi: 10.1021/cb1002619. [DOI] [PubMed] [Google Scholar]
- 20.Adrian FJ, Ding Q, Sim TB, Velentza A, Sloan C, Liu Y, Zhang GB, Hur W, Ding S, Manley P, Mestan J, Fabbro D, Gray NS. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JM, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun FX, Guo GR, Ding Q, Okram B, Choi Y, Wojciechowski A, Deng XM, Liu GX, Fendrich G, Strauss A, Vajpai N, Grzesiek S, Tuntland T, Liu Y, Bursulaya B, Azam M, Manley PW, Engen JR, Daley GQ, Warmuth M, Gray NS. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–U116. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schonbrunn E, Betzi S, Alam R, Martin M, Lubbers DJ, Han HJ, Jakkaraj SR, Georg GI. Discovery of a Potential Allosteric Ligand Binding Site in CDK2. Acs Chem Biol. 2011;6:492–501. doi: 10.1021/cb100410m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Mechanism-based design of a protein kinase inhibitor. Nat Struct Biol. 2001;8:37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]
- 24.Schneider TL, Mathew RS, Rice KP, Tamaki K, Wood JL, Schepartz A. Increasing the kinase specificity of K252a by protein surface recognition. Org Lett. 2005;7:1695–1698. doi: 10.1021/ol050179o. [DOI] [PubMed] [Google Scholar]
- 25.Hill ZB, Perera BGK, Maly DJ. A Chemical Genetic Method for Generating Bivalent Inhibitors of Protein Kinases. J Am Chem Soc. 2009;131:6686–6688. doi: 10.1021/ja900871y. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, Kumar S, Lawrence DS. Stepwise combinatorial evolution of Akt bisubstrate inhibitors. Chembiochem. 2008;9:507–509. doi: 10.1002/cbic.200700583. [DOI] [PubMed] [Google Scholar]
- 27.Meyer SC, Shomin CD, Gaj T, Ghosh I. Tethering small molecules to a phage display library: Discovery of a selective bivalent inhibitor of protein kinase A. J Am Chem Soc. 2007;129:13812–13814. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- 28.Shomin CD, Meyer SC, Ghosh I. Staurosporine tethered peptide ligands that target cAMP-dependent protein kinase (PKA): Optimization and selectivity profiling. Bioorgan Med Chem. 2009;17:6196–6202. doi: 10.1016/j.bmc.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devlin JJ, Panganiban LC, Devlin PE. Random Peptide Libraries - a Source of Specific Protein-Binding Molecules. Science. 1990;249:404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- 30.Katz BA. Binding to Protein Targets of Peptidic Leads Discovered by Phage Display - Crystal-Structures of Streptavidin-Bound Linear and Cyclic Peptide Ligands Containing the HPQ Sequence. Biochemistry-Us. 1995;34:15421–15429. doi: 10.1021/bi00047a005. [DOI] [PubMed] [Google Scholar]
- 31.Giebel LB, Cass RT, Milligan DL, Young DC, Arze R, Johnson CR. Screening of Cyclic Peptide Phage Libraries Identifies Ligands That Bind Streptavidin with High Affinities. Biochemistry-Us. 1995;34:15430–15435. doi: 10.1021/bi00047a006. [DOI] [PubMed] [Google Scholar]
- 32.Green NM. Avidin and Streptavidin. Methods in Enzymology. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 33.Meyer SC, Gaj T, Ghosh I. Highly selective cyclic peptide ligands for NeutrAvidin and avidin identified by phage display. Chem Biol Drug Des. 2006;68:3–10. doi: 10.1111/j.1747-0285.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 34.Fujiwara D, Ye ZM, Gouda M, Yokota K, Tsumuraya T, Fujii I. Selection of inhibitory peptides for Aurora-A kinase from a phage-displayed library of helix-loop-helix peptides. Bioorg Med Chem Lett. 2010;20:1776–1778. doi: 10.1016/j.bmcl.2010.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.