Abstract
Esophageal squamous cell carcinoma (ESCC) is the sixth most frequent cause of cancer death in the world, and cigarette smoke is a key factor in esophageal carcinogenesis. To identify molecular changes during cigarette smoke-induced ESCC, we examined the methylation status of 13 gene promoters in the human immortalized, nontumorigenic esophageal epithelial cell line (Het-1A) that were exposed to mainstream (MSE) or sidestream cigarette smoke extract (SSE) for 6 months in culture. The promoter of sequence-specific single-stranded DNA-binding protein 2 (SSBP2) was methylated in the Het-1A cells exposed to MSE (MSE-Het-1A). Promoter methylation (86%, 56/70) and downregulation of SSBP2 expression were frequently detected in tumor tissues from ESCC patients. In addition, reintroduction of SSBP2 in an ESCC cell line (TE1) that does not express SSBP2 and in the MSE-Het-1A cells inhibited expression of LRP6 and Dvl3, which are mediators of the Wnt signaling pathway. SSBP2 expression markedly decreased the colony-forming ability of ESCC cell lines and significantly inhibited cell growth of the MSE-Het-1A cells. Our results indicate that cigarette smoking is a cause of SSBP2 promoter methylation and that SSBP2 harbors a tumor suppressive role in ESCC through inhibition of the Wnt signaling pathway.
Keywords: ESCC, cigarette smoking, SSBP2, promoter methylation
Esophageal squamous cel1 carcinoma (ESCC) is the eighth most common type of caner, accounting for more than 90% cases of esophageal cancer worldwide.1 Most patients with ESCC are diagnosed at an advanced stage, and metastasis to the regional lymph nodes occurs frequently.2 There is considerable epidemiological evidence suggesting that alcohol, tobacco, diets deficient in vitamins/protective antioxidants, carcinogens and thermal injuries are important in the pathogenesis of ESCC.3,4 Among all these, tobacco smoking is a key risk factor for the development of ESCC,5 and the risk increases with long-term and/or passive smoking and rises markedly for people who both smoke and drink.
The progressive transformation of a normal cell to a malignant cell is accompanied by a series of genetic/epigenetic changes, which include inactivation of tumor suppressor genes (TSGs) and activation of oncogenes.6 One of major epigenetic events involved in functional inactivation of TSG in the process of malignant transformation is loss of gene expression by aberrant DNA methylation, which has appeared as an emerging molecular marker in human cancer.7 We have identified novel TSGs inactivated by promoter methylation in ESCC; e.g., N-methyl-d-aspartic acid receptor 2B (NMDAR2B)8,9 and neurofilament heavy chain polypeptide (NEFH).8 In addition, correlations between aberrant gene methylation and smoking exposure are often observed in smoking-related human cancer; for example, hypermethylation of the APC and p16 promoters is associated with tobacco smoking in nonsmall cell lung carcinoma,10 colon cancer11 and cervical squamous cell carcinoma.12 The methylation level of NEFH in noncancerous esophageal mucosa has a significant correlation with smoking duration.13 Interestingly, promoter methylation of TSG occurs more frequently in cancers from smokers than nonsmokers,10 suggesting that a tobacco signature could emerge from distinctive patterns of gene promoter methylation.
Among the two types of cigarette smoke, “first-hand” smoke is mimicked by mainstream cigarette smoke (MS, inhaled by the smoker), and “second-hand” smoke by sidestream cigarette smoke (SS, inhaled by nonsmokers in places where smoking is allowed). It is well known that most of the cigarette smoke carcinogens are initiators and promoters of carcinogenesis in many organs such as the lung, stomach, liver and colon.13 Cigarette smoke extract (CSE) is used as a surrogate for cigarette smoke carcinogens because it contains most of the particulate chemicals identified in cigarette smoke14 and is a highly genotoxic substance.15 By usin water-soluble CSE, we recently established two resistant cell lines form a nonmalignant esophageal epithelial cell line, Het-1A, after exposure to either mainstream (MSE) or sidestream cigarette smoke extract (SSE). The chronic exposure of Het-1A cells to MSE or SSE caused alterations in cellular phenotypes, leading to acquisition of tumorigenic characteristics.16
Sequence-specific single-stranded DNA-binding protein 2 (SSBP2) at chromosome 5q13.3 is closely related with ubiquitously expressed genes, SSBP3 (1p31.3) and SSBP4 (19p13.1).17 Here, we report induction of SSBP2 promoter methylation by cigarette smoke exposure in the Het-1A cells exposed to MSE (MSE-Het-1A). Moreover, we found aberrant methylation of SSBP2 in primary ESCC and a tumor suppressive role of SSBP2 through inhibition of Wnt signaling pathway.
Material and Methods
Cel1 lines and tissues
HEK293 cells were purchased from ATCC and maintained in DMEM with 10% FBS, and ESCC cell lines were grown as described.8,9 An immortalized, nontumorigenic esophageal epithelial cell line, Het-1A, was purchased from ATCC and grown in BEGM (Lonza Group Ltd., Basel, Switzerland) as recommended. Cell passage number was counted from the first cell propagation (+1) on arrival from ATCC. Twenty pairs of ESCC and normal esophagus tissues (patient no. 1–20) were obtained from the Gastroenterology Division, Department of Medicine, University of Maryland. Fifty cases of primary ESCC genomic DNA, a normal esophageal tissue cDNA (PN) and five ESCC tissue cDNA (T3–T7) were obtained from patients who underwent surgery at the Medical Institute of Bioregulation Hospital, Kyushu University and the Saitama Cancer Center. gDNA of 10 normal esophageal epithelial tissues (NN) were extracted from formalin fixed, paraffin-embedded sections, which had been prepared from biopsy of patients without cancer at Department of Pathology, The Johns Hopkins University.
Establishment of CSE-resistant cells
The preparation of CSE and establishment of resistant cells lines were previously described.16 Control-, MSE- and SSE-Het-1A cells at the passage between +24 and +32 were examined.
Bisulfite sequencing
Bisulfite-modified genomic DNA was amplified by polymerase chain reaction (PCR) as described.8 Primer sequences are shown in Supporting Information Table S1. a11 the PCR products were gel-extracted (Qiagen, Valencia, CA) or cloned into Topo-TA plasmid (Invitrogen, Carlsbad, CA) and sequenced with an amplification primer (F1) using the ABI BigDye cycle sequencing kit (Applied Biosystems, Foster City, CA). The criteria to determine methylation in ce11 lines and tissues were described previously.18 When “methylated” CpG was found in more than 30% of total CpGs in an amplified PCR product, it was considered as “methylation-positive.” A search for CpG islands in each gene promoter was done by using the on-line accessible software, Methprimer.
Methylation-specific PCR
Bisulfite-treated DNA was amplified with methylation-specific primer sets for individual gene as described.18 Unmethylated SSBP2 was examined by the PCR with unmethylation-specific primers.
Quantitative methylation-specific PCR (TaqMan-MSP)
The level of SSBP2 methylation was analyzed as described.18 The methylation ratio (TaqMan methylation value, TaqMeth V) was defined as the quantity of fluorescence intensity derived from promoter amplification of SSBP2 divided by fluorescence intensity from β-actin amplification and multiplied by 100. This ratio was used as a measure for the relative level of methylated DNA in samples. Statistical Analysis was performed as described.18
5-Aza-dC treatment and RT-PCR
Cells were treated with 5 μM 5-aza-2′-deocycytidine (5-Aza-dC) (Sigma, St. Louis, MO) every 24 hr for 3 days, and RNA was extracted using Trizo1 (Invitrogen, Carlsbad, CA) and reverse transcribed as described.18
Real-time RT-PCR
cDNA from a patient without cancer (NN) and two ESCC patients (T1 and T2) were purchased from BioChain Institute, Inc. (Hayward, CA). One microliter of each cDNA was used for real-time RT-PCR using QuantiFast SYBR Green PCR Kit (Promega, Valencia, CA) as described.18 For Cyclin D1, MMP-7 and TCF1, TaqMan predesigned primers and probes (Applied Biosystems, Foster City, CA) were mixed with 2X Master Mix and proceeded for PCR.
Western blot analyses
Whole cell lysates extracted in RIPA buffer were separated on 4–12% gradient SDS-PAGE, and western blotting was performed as described.18 Anti-SSBP2 antibody was purchased from Abcam (Cambridge, MA), and anti-β-actin antibody was purchased from Sigma (St. Louis, MO). The other antibodies were purchased from Cell Signaling Technology (Danvers, MA).
Immunohistochemistry
An esophagus cancer test tissue array with self-matching normal adjacent tissue (ES241) was purchased from Biomax Inc. (Rockville, MD). After deparaffinization and rehydration, tissue slides were incubated with anti-SSBP2 rabbit polyclonal antibody (1:250 dilutions) at 4°C overnight, and further procedure was followed as described.18
Cel1 growth assay
Cells were plated on a 24-well plate at a density of 1–2 × 104 cells/well and incubated overnight at 37°C. Cells were transfected with plasmids or siRNA or treated with sulindac sulfone (Sulindac, Calbiochem, San Diego, CA) and human recombinant Wnt3a (R&D systems, Minneapolis, MN) for indicated time to inhibit and activate Wnt pathway, respectively, and the tetrazolium-based cell viability (MTT) assay was performed. The results were expressed as absorbance at 570 nm or a percentage of absorbance in samples compared to control (100%).
Colony focus assay
SSBP2 plasmid (pSSBP2) was kindly provided by Dr. Stephen J. Brandt (Vanderbilt University Medical Center, Nashville, TN), and colony focus assays were performed in ESCC cell lines transfected with pSSBP2 in the presence of G418 for 2 weeks as described.8
Luciferase reporter assay
The control- and MSE-Het-1A cells in 24-well plates (5 × 104/well) were transfected with pSSBP2 or control plasmids (0.5 μg/well) and incubated for 24 hr. Then, cells were cotransfected with TOPflash (200 ng/well) and Renilla (10 ng/well) or FOPflash (200 ng/well) and Renilla plasmids and incubated for further 48 hr. TOPflash and FOPflash have wild-type (TOP) or mutated (FOP) binding sites for the β-catenin-TCF4/Lef complex, respectively. β-catenin/TCF/Lef transcriptional reporter activity was performed using TOP-flash/FOPflash TCF Reporter Kit (Upstate Biotechnology, Lake Placid, NY).
Results
To identify cigarette smoking-related gene methylation in esophageal cancer, we investigated the methylation status of 13 gene promoters (OPCML, HOP/OB1, PGP9.5, NEFH, NMDAR2B, DLC, B4GALT1, Trypsinogen-4, DCC, APC, SSBP2, FHIT and p16) in Het-1A cells exposed to MSE (MSE-Het-1A) or SSE (SSE-Het-1A) for 6 months. Although aberrant methylation of these genes in ESCC has been reported (Table 1), little is known about whether cigarette smoking is a cause of gene methylation. Genomic DNA was extracted for bisulfite conversion from the MSE- and SSE-Het-1A cells at passage +24 at which point cells displayed cellular resistance to MSE and SSE, respectively, and increased tumor-promoting activity in vitro.16 Genomic DNA from parental Het-1A cells at the same passage was also examined in parallel (control-Het-1A). We performed bisulfite sequencing for OPCML, HOP/OB1, PGP9.5, NEFH, DLC and NMDAR2B and methylation-specific PCR (MSP) for B4GALT1, Trypsinogen-4, DCC, APC, FHIT, P16 and SSBP2.
Table 1.
Methylation profile in Het-1A cells
| Methylation1 (%) | References | C | M | S | Method | |
|---|---|---|---|---|---|---|
| OPCML | 21/32 (66) | 19 | U | U | U | BS |
| HOP/OB1 | 10/20 (55) | 20 | U | U | U | BS |
| PGP9.5 | 21/50 (42) | 21 | M | M | M | BS |
| NEFH | 12/20 (60) | 8 | M | M | M | BS |
| NMDAR2B | 19/20 (95) | 8 | U | U2 | U2 | BS |
| DLC | 48/94 (51) | 22 | U | U2 | U2 | BS |
| B4GALT1 | 12/20 (60) | 23 | M | M | M | MSP |
| Trypsinogen-4 | 25/49 (51) | 24 | M | M | M | MSP |
| DCC | 52/70 (74) | 25 | U | U | U | MSP |
| APC | 16/32 (50) | 26 | U | U | U | MSP |
| SSBP2 | 15/18 (83) | 23 | U | M | U | MSP |
| FHIT | 49/95 (51) | 27 | U | U | U | MSP |
| p16 | 53/95 (56) | 27 | U | U | U | MSP |
C, control-Het-1A cells; M, MSE-Het-1A cells; S, SSE-Het-1A cells; M, methylation; U, unmethylation; BS, bisulfite sequencing; MSP, methylation-specific PCR.
Methylation reported in ESCC.
Three or four of new methylated CpGs were found in the gene promoter.
PGP9.5 was methylated in the Het-1A cells regardless of CSE exposure (Supporting Information Fig. S1A), whereas OPCML1 (data not shown) and HOP/OB1 (Supporting Information Fig. S1B) were not methylated before or after CSE exposure (Table 1). None of the CpG sites in the promoters of NMDAR2B and DLC were methylated in the control-Het-1A cells, whereas three or four newly methylated CpGs were found in the MSE- and SSE-Het-1A cells (data not shown). In the NEFH promoter, 12 of 33 CpG dinucleotides were methylated in the control-Het-1A cells (partial methylation), but MSE or SSE exposure induced de novo methylation in 12 or 13 more CpG dinucleotides (Supporting Information Fig. S1C). By MSP analysis, methylation of B4GALT1 and Trypsinogen-4 was observed in the control-Het-1A cells and remained methylated in CSE-exposed cells (Fig. 1a). Promoter methylation of DCC, APC, FHIT and P16 (Fig. 1a) was not observed in the control-Het-1A cells and did not changed after CSE exposure. For SSBP2, methylation of the promoter was clearly observed in the MSE-Het-1A cells (Fig. 1a, M-SP) with weak intensity of any unmethylated alleles (Fig. 1a, UnM-SP) but not in the control-Het-1A cells and SSE-Het-1A cells. Therefore, among the genes examined, CpG dinucleotides in the NMDAR2B, DLC, NEFH and SSBP2 promoters seemed to be most susceptible to CSE exposure.
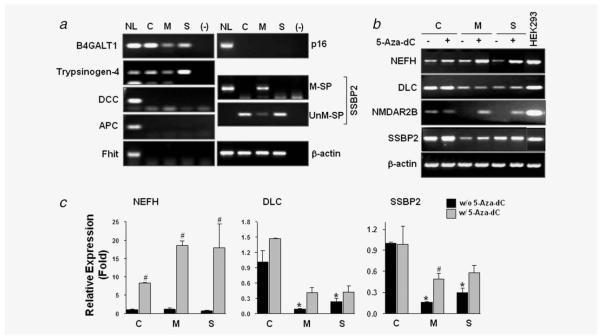
Figure 1.
Methylation status of genes in Het-1A cells exposed to MSE or SSE. (a) MSP was performed for analysis of gene promoters of B4GALT1, Trypsinogen-4, DCC, APC, FHIT, P16 and SSBP2. PCR was performed with methylation-specific primers, and PCR products were run on 4% agarose gels prestained with ethidium bromide. In vitro methylated, bisulfite-treated human normal lymphocyte DNA (NL) was used as a positive control for MSP, and distilled water was used as a negative PCR control (—). β-actin was used to confirm integrity of bisulfite-treated DNA. PCR products were gel purified and sequenced to verify true amplification and CpG methylation of the genes. C, control-Het-1A cells; M, MSE-Het-1A cells; S, SSE-Het-1A cells. For SSBP2, MSP was performed with methylation-specific (M-SP) and unmethylation-specific primers (UnM-SP). (b) Expression of NEFH, DLC, NMDAR2B and SSBP2 was examined by RT-PCR analysis, and gene reactivation was examined after the 5-Aza-dC treatment (5 μM). Total RNA was extracted from cells at the passage +28. PCR products were visualized under UV light. β-actin was used as a loading control. C, control-Het-1A cells; M, MSE-Het-1A cells; S, SSE-Het-1A cells. Because of its low expression level, touch-down PCR was performed for the NMDAR2B as described.8 (c) Real-time RT-PCR was performed, and relative expression (fold) was calculated by comparing the ratios of mRNA expression of genes to an internal control gene, β-actin. Experiments were done in duplicate, and independent experiments were performed twice. Values indicate means ± SD. *p < 0.05 compared to the control-Het-1A cells without W/o) 5-Aza-dC treatment; #p < 0.05 compared to each cell line without 5-Aza-dC treatment (t-test). No significance in NEFH expression was found between control and MSE- or SSE-exposed cells.
To investigate whether CpG methylation induced by CSE exposure was associated with decreased gene expression, RT-PCR analysis was performed. NEFH expression was found to be slightly decreased in the SSE-exposed cells compared to the control (Fig. 1b). The demethylating agent 5-Aza-dC increased the NEFH level in the control cells and further enhanced it in the MSE- and SSE-Het-1A cells. Real-time RT-PCR analysis revealed about an 18-fold increase in NEFH expression in the MSE- and SSE-Het-1A cells and an 8-fold increase in the control cells (Fig. 1c), suggesting that increased CpG methylation sites further suppressed gene expression. DLC expression level was decreased by chronic exposure of MSE or SSE but not increased by 5-Aza-dC treatment, indicating that 5 μM 5-Aza-dC treatment for 72 hr might not be sufficient to reactivate this gene or that the downregulation of DLC in the MSE- and SSE-exposed cells is not caused by promoter methylation. NMDAR2B was expressed in the Het-1A cells but its basal expression was much lower than that in HEK293 cells (Fig. 1b). NMDAR2B was silenced in the MSE- and SSE-exposed cells and reactivated by 5-Aza-dC treatment. In addition, the mRNA level of SSBP2 decreased in the MSE- and SSE-Het-1A cells compared to the control, but an increase in expression by 5-Aza-dC was only observed in the MSE-Het-1A cells, correlating with its promoter methylation. A significant decrease of DLC and SSBP2 expression in the MSE- and SSE-Het-1A cells was also confirmed by the real-time RT-PCR analysis. These results indicate that the SSBP2 methylation participates, at least in part, in the downregulation of gene transcription under MSE exposure. No methylation (Fig. 1a) or reactivation of SSBP2 by 5-Aza-dC in the SSE-Het-1A cells indicates that decreased SSBP2 expression by SSE exposure is not related to promoter methylation. Therefore, among the genes examined, SSBP2 was selected for further study due to its specific promoter methylation and downregulation in the MSE-Het-1A cells.
We previously observed SSBP2 methylation in 15 of 18 ESCC (83%) but not in any of the 10 (0%) normal esophageal tissues from noncancer patients (0%).28 Because only the frequency of SSBP2 methylation was reported in our previous study, SSBP2 methylation was further investigated in ESCC cell lines and a larger number of ESCC samples. SSBP2 methylation was not observed in the nontumorigenic cell lines, HEK293 and Het-1A, but was observed in 8 of 11 ESCC ce11 lines (TE1, TE2, KYSE70, KYSE140, KYSE150, KYSE200, KYSE410 and KYSE520) by MSP with methylation-specific primers (M-SP) (Fig. 2a). However, an unmethylated allele for SSBP2 (UnM-SP) was always observed in all cell lines tested.
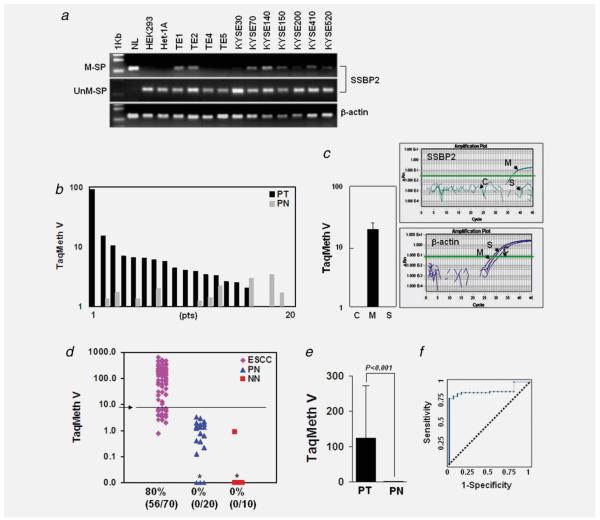
Figure 2.
Methylation of SSBP2 in ESCC cel1 lines and primary tissues. (a) Methylation status of SSBP2 was examined in HEK293, Het-1A and ESCC cell lines. In vitro methylated, bisulfite-treated human normal lymphocyte DNA (NL) and Het-1A cells were used as a positive and negative control, respectively, for MSP with methylation-specific primers (M-SP). β-actin was used to confirm the integrity of bisulfite-treated DNA. (b) Quantitative level of SSBP2 methylation in paired normal and tumor tissues prepared from 20 individual patients (Pts) (Wilcoxon matched-pairs signed-ranks test). Only three patients harbored a higher methylation level in PN than in PT. (c) Methylation level of SSBP2 in Het-1A cells exposed to MSE or SSE. Amplification curves of β-actin show adequate amplification of β-actin for all samples tested. (d) A scatter plot of methylation values of SSBP2 gene in tissues. No cases of normal esophageal tissue from ESCC patients (PN) and from patients without cancer (NN) displayed TaqMeth V over the optimal cut off (3.912) indicated as an Arrow. *Samples with a ratio equal to zero could not be plotted correctly on a log scale, so are presented here as 0.01. All assays were performed in duplicate or triplicate, and experiments were repeated twice. Data showed reproducible and concordant results in triplicate. TaqMeth V is described in Material and Methods section. (e) The overall TaqMeth V detected in primary ESCC was significantly higher than that in corresponding normal tissues (Wilcoxon unmatched-pairs signed-ranks test). (f) ROC curve analysis of TaqMeth V of SSBP2. The area under ROC (AUROC, 0.895 ± 0.033) conveys the accuracy in distinguishing PN from ESCC in terms of its sensitivity and specificity (p < 0.001). The optimal cut off was calculated from this ROC analysis. Solid line, SSBP2; dashed line, no discrimination. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
To compare SSBP2 methylation in normal and tumor tissues, TaqMan-MSP analysis was performed in 20 pairs of ESCC (paired tumor, PT) and matched normal esophageal tissues (paired normal, PN) with a probe targeted to the CpG island of SSBP2. We compared the methylation level (TaqMan methylation value, TaqMeth V) in individual patients. Higher values of SSBP2 methylation in PT than in PN were observed in 15 of 20 ESCC patients, indicating that the majority of the tumor tissues harbor higher levels of SSBP2 methylation than matched normal esophageal tissues (p = 0.0015) (Fig. 2b). When the methylation level of SSBP2 was compared in the Het-1A cells with or without CSE exposure, amplification of methylated SSBP2 DNA was not observed in the contro1- and SSE-Het-1A cells, but typical real-time-MSP curves were generated in the MSE-Het-1A cells (Fig. 2c, right). The TaqMeth V of SSBP2 in the MSE-Het-1A cells was 19.25 ± 5.47 (mean ± SD) (Fig. 2c, left).
The SSBP2 methylation level was also compared between 70 ESCC and 20 normal corresponding tissues from ESCC patients (PN). Ten normal esophageal epithelial tissues from noncancer patients (NN) were also included to compare methylation levels between cancer and noncancer patients. The distribution of methylation values in each group of samples is shown in Figure 2d. In 70 ESCC samples, methylation values (TaqMeth V) in tumor tissue ranged from 0 to 631.23 (median value 65.36) and in PN from 0 to 3.40 (median value 1.00). The overall TaqMeth V detected in primary ESCC (124.67 ± 148.53, mean ± SD) was significantly higher than that in PN (1.09. ± 1.01, mean ± SD) (p < 0.001) (Fig. 2e). Testing methylation of SSBP2 resulted in a highly discriminative receiver-operator characteristic (ROC) curve profile, clearly distinguishing ESCC from PN (Fig. 2f). The optimal cut off (value, 3.912) was calculated from the ROC analysis to maximize sensitivity and specificity. Neither NN nor PN samples exhibited a value over the cutoff, yielding 100% specificity, whereas 86% (56/70) of primary ESCC displayed SSBP2 methylation (p < 0.001, ESCC vs. PN, Fisher’s exact test). No correlations were found between SSBP2 methylation and clinical features of ESCC patients. Unfortunately, a smoking history was not available in these ESCC samples.
To investigate the transcriptional level of SSBP2, RT-PCR was performed using primers specific for SSBP2. Expression of SSBP2 was clearly observed in the TE4, TE5, and KYSE30 cells where SSBP2 was not methylated, and a faint or undetectable level of SSBP2 was observed in TE1, TE2 and most KYSE cells; only two cel1 lines among the KYSE series, KYSE70 and KYSE140, expressed SSBP2 (Fig. 3a, left). Because SSBP2 methylation was observed in KYSE70 and KYSE140 cells, it seems that promoter methylation is not a major contributor to SSBP2 expression in these cell lines. SSBP2 mRNA expression was reactivated by treatment with 5-Aza-dC in KYSE410 and KYSE520 cells but not in KYSE30 cells (Fig. 3b, right), indicating regulation of SSBP2 expression, at least in part, by methylation of its promoter. SSBP2 expression was also examined by RT-PCR in cDNA prepared from patients with or without cancer. Compared to the mRNA level in two normal esophageal tissues (NN and PN), downregulation of SSBP2 was observed in al1 ESCC patients examined (T1–T7) (Fig. 3b, left). Real-time RT-PCR analysis revealed five times lower expression of SSBP2 in ESCC compared to NN (Fig. 3b, right) (t-test, p < 0.05).
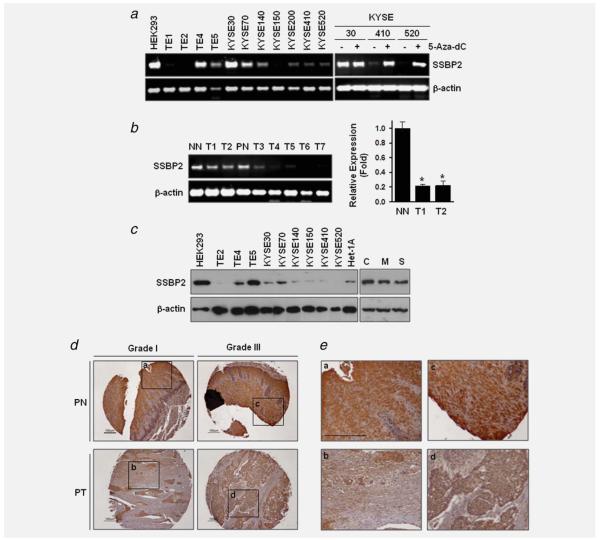
Figure 3.
mRNA expression SSBP2 in ESCC cell lines and primary tissues. (a) Expression of SSBP2 in cell lines was examined by RT-PCR analysis (left). Downregulation of the SSBP2 transcript was reactivated by treatment of the demethylating agent, 5 μM 5-Aza-dC (right). (b) Left, SSBP2 expression was examined by RT-PCR in cDNA prepared from normal and tumor tissue from ESCC patients (PN and T1–T7) and from a patient without cancer (NN). β-actin was used as a loading control. PCR products were visualized under UV light. Right, Real-time RT-PCR was performed in cDNA from patients with ESCC (T1 and T2) and without cancer (NN). Relative expression (fold) was calculated by comparing the ratios of mRNA expression of SSBP2 to an internal control gene, β-actin. Experiments were done in duplicate, and independent experiments were performed twice. Values indicate means ± SD. *p < 0.05 compared to NN (t-test). (c) Total cell lysates were extracted run on 4–12% polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were then blotted for the expression of SSBP2. Left, Lysates from HEK293, Het-1A and ESCC cell lines. Right, cell lysates from Het-1A cells at the passage +31. β-actin was used as a loading control. C, control-Het-1A cells; M, MSE-Het-1A cells; S, SSE-Het-1A cells. (d) Immunohistochemical analysis was performed in an ESCC tissue array (PT) with cancer adjacent normal esophageal tissues (PN). A strong expression of SSBP2 was detected in all PN. Tumor grades are indicated. (e) Selected representative portions in tissues are shown at high magnification (a-d). Scale bar, 100 μm.
Western blot analysis revealed that SSPB2 protein expression correlated with the mRNA levels (Fig. 3c, left). Decreased SSBP2 protein was also observed in the MSE- and SSE-Het-1A cells compared to the contro1, consistent with the RT-PCR results (Fig. 3c, right). To examine expression of SSBP2 protein in primary tissues, immunohistochemical staining was performed in a tissue microarray including normal esophageal (PN) and cancer tissue sections (PT) prepared from six individual ESCC patients. Moderate or strong expression of SSBP2 was detected in all adjacent normal appearing esophageal tissues in patients with cancer, whereas downregulation of SSBP2 in PT was detected in five of six ESCC (Fig. 3d and 3e) (Supporting Information Table S3). A similar level of SSBP2 between PN and PT was observed only in one patient. These results suggest a specific decrease of SSBP2 mRNA and protein in ESCC development.
To investigate the possible tumor suppressive activity of SSBP2, we transfected SSBP2 expression plasmids (pSSBP2) into TE1 and TE2 cells where basal levels of SSBP2 were barely detectable and then performed a colony focus assay. Contro1 TE1 and TE2 cells displayed strong colony-forming ability (Fig. 4a and Supporting Information Fig. S2B), whereas reintroduced SSBP2 markedly decreased the colony formation in both cell lines.
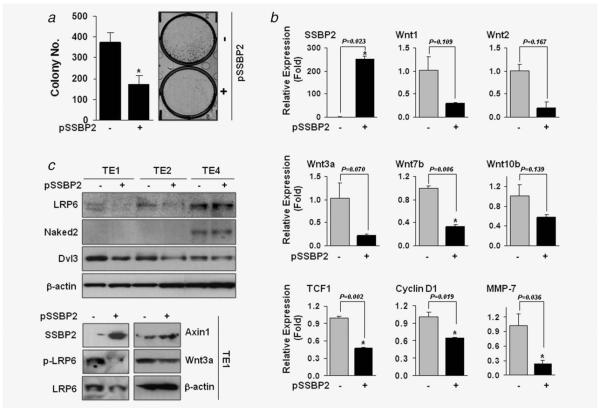
Figure 4.
Tumor suppressive activity of SSBP2. (a) pSSBP2 (+) or empty vector control (−) was transfected into TE1 cells, and the colony focus assay was performed after incubation of cells in the presence of G418 (1 mg/ml) for 2 weeks. Colonies were fixed and stained with 0.4% crystal violet solution (MeOH/Acetic acid, 3:1) and counted. *p < 0.05 compared to control (t-test). (b) Real-time RT-PCR was performed in TE1 cells transfected with pSSBP2 or control, and relative expression (fold) was calculated by comparing the ratios of mRNA expression of genes to an internal control gene, β-actin. Experiments were done in duplicate, and independent experiments were performed twice. Values indicate means ± SD. *p < 0.05 compared to the control (t-test). (c) The expression of LRP6, Naked and Dvl3 was examined in TE1, TE2 and TE4 cells with or without transfection of pSSBP2 (upper). The expression of p-LRP, Axin1 and Wnt3a was examined in TE1 cells with or without transfection of pSSBP2 (lower).
Wnt members are critical mediators of cigarette smoke-induced lung cancer,29,30 and SSBP2-overexpressing PC3 cells demonstrated downregulation of Wnt signaling.31 To see whether SSBP2 can regulate Wnt expression in ESCC, we performed real-time RT-PCR analysis in TE1 cells after transfection of pSSBP2. SSBP2 expression significantly decreased Wnt7b level with a trend toward a decrease of Wnt1, Wnt2, Wnt3a and Wnt10b (Fig. 4b). In addition, the protein level of LRP6 was decreased by SSBP2 in TE1 and TE2 cells but not in TE4 cells (Fig. 4c, upper). The expression of Naked2 was not affected by SSBP2 overexpression. Dvl3 expression was decreased by SSBP2 in TE1 and TE2 and slightly in TE4. In TE1 cells, LRP6 phosphorylation and Wnt3a levels were decreased, but Axin1 expression was increased by SSBP2 (Fig. 4c, lower).
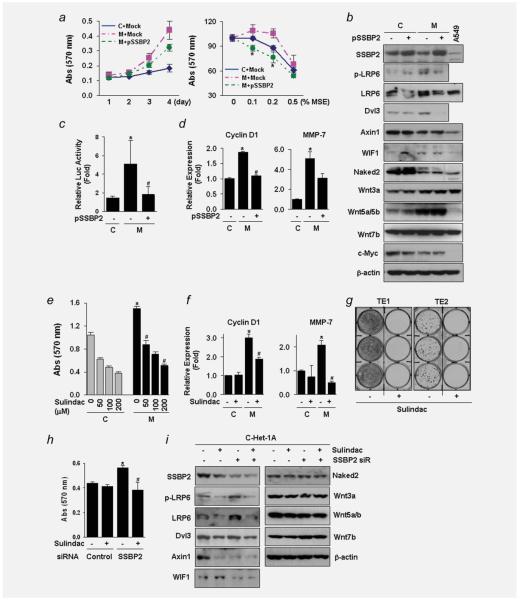
Cell growth (Fig. 5a, left) and cellular resistance to MSE treatment (Fig. 5a, right) were significantly decreased by SSBP2 overexpression in MSE-Het-1A cells, indicating that SSBP2 likely suppresses cell growth stimulated by chronic MSE exposure. Protein expression of LRP6, Dvl3, Wnt3a and Wnt5a/b increased in the MSE-Het-1A cells compared to that in the control-Het-1A cells, whereas the level of Axin1 and Naked3 decreased in the MSE-Het-1A cells (Fig. 5b). In addition, SSBP2 overexpression in the MSE-Het-1A cells resulted in decreased LRP6 and Dvl3 expression, without changing the levels of p-LRP, Axin1, Naked2, Wnt3a and Wnt5a/b.
Figure 5.
Suppression of the Wnt/β-catenin signaling by SSBP2. (a) Cell growth (left) and cellular sensitivity to MSE treatment (right) were measured by the MTT assay after incubation of cells (passage +31) transfected with pSSBP2 or empty vector control (Mock) for indicated time. MSE was treated for 48 hr. The results were expressed as absorbance at 570 nm. Values indicate means ± SD. *p < 0.05 compared to the Mock transfected-MSE-Het-1A cells (t-test). C, control-Het-1A cells; M, MSE-Het-1A cells; S, SSE-Het-1A cells. Expression of E-cadherin, an epithelial marker, was not changed, but N-cadherin level, a mesenchymal marker, was downregulated by SSBP2 expression in the MSE-Het-1A cells (data not shown). (b) Expression of proteins involved in the Wnt signaling was examined by western blotting in the control- and MSE-Het-1A cells (passage +31) with or without transfection of pSSBP2. Total cell lysates (30 μg/lane) were extracted run on 4–12% polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were then blotted for the indicated Wnt proteins. β-actin was used as a loading control. A protein lysate from the A549 lung cancer cell line (15 μg/lane) was used to compare expression level of each protein with that in Het-1A cells. No difference in p-LRP6 and Wnt7b expression was observed between the control- and MSE-Het-1A cells. (c) Luciferase assay after transfection of TOPflash or FOPflash reporter constructs into the control- and MSE-exposed cells (passage +29). After pSSBP2 was transfected for 24 hr, reporter constructs were transfected and incubated for further 24 hr. Renilla luciferase plasmids were cotransfected with TOPflash or FOPflash as an internal control. Each reporter activity was normalized to Renilla, and relative luciferase activity (fold) was calculated by dividing TOPflash activity by FOPflash activity. Experiments were done in four replicates and repeated twice. Values indicate means ± SD. *p < 0.05 compared to the control-Het-1A cells; #p < 0.05 compared to the empty vector-transfected MSE-Het-1A cells (t-test). (d) mRNA levels of Cyclin D1 (left) and MMP-7 (right) determined by real time-RT-PCR. (e) Treatment with Sulindac (100 μM) for 48 hr inhibited cell growth in a dose-dependent manner in both the control- and MSE-exposed cells (passage +28) and decreased Cyclin D1 and MMP-7 expression in the MSE-Het-1A cells (f). No difference was observed in TCF1 expression (data not shown). Experiments were done in four replicates and repeated twice. Values indicate means ± SD. *p < 0.05 compared to the control-Het-1A cells in the absence of Sulindac (t-test); #p < 0.05 compared to MSE-Het-1A cells in the absence of Sulindac. C, control-Het-1A cells; M, MSE-Het-1A cells. (g) TE1 and TE2 cells were treated with Sulindac (100 μM) three times a week for 2 weeks, and colony focus assays were performed. Colonies were fixed and stained with 0.4% crystal violet solution (MeOH/Acetic acid, 3:1) and taken pictures. (h) A siRNA pool targeting SSBP2 and a nontargeting control were transfected into control-Het-1A cells (C-Het-1A at the passage, +29). After 4 hr of transfection, Sulindac (100 μM) was added and incubated for a further 48 hr. Cellular growth was assessed by the MTT assay. Experiments were done in four replicates and repeated twice. Values indicate means ± SD. *p < 0.05 compared to cells transfected with control siRNA in the absence of Sulindac (t-test); #p < 0.05 compared to cells transfected with SSBP2 siRNA in the absence of Sulindac. (i) Expression of proteins involved in the Wnt signaling was examined by Western blotting in Het-1A cells (passage +32) after Sulindac treatment following transfection with control or SSBP2 siRNA. Wnt inhibitory factor-1 (WIF1) is a secreted antagonist of the Wnt pathway. β-actin was used as a loading control. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
To investigate β-catenin-TCF/Lef-dependent transcription after long-term CSE exposure, the luciferase reporters, TOP-flash and FOPflash, which have either wild-type (TOP) or mutated (FOP) binding sites for the β-catenin-TCF4/Lef complex, were transfected into the control- and MSE-Het-1A cells. We found a 6-fold increase of TOP activity in the MSE-Het-1A cells compared to the contro1, which was decreased by SSBP2 overexpression (Fig. 5c). SSBP2 also reduced the mRNA level of Cyclin D1 and MMP-7 in the MSE-Het-1A cells (Fig. 5d), suggesting that SSBP2 suppresses Wnt/β-catenin signaling activated by chronic MSE exposure.
Sulindac sulfone (Sulindac), a Wnt pathway-specific inhibitor,30 decreased the cell growth in a dose-dependent fashion in both the control- and MSE-exposed cells (Fig. 5e) and downregulated the level of Cyclin D1 and MMP-7 in the MSE-Het-1A cells (Fig. 5f). Sulindac almost completely abrogated the colony-forming ability of TE1 and TE2 cells (Fig. 5g), whereas treatment with human recombinant Wnt3a (hrWnt3a) resulted in increased cell growth in the control-Het-1A cells (Supporting Information Fig. S4A), as we11 as colony numbers and colony size in TE2 cells (Supporting Information Fig. S4B). We then transfected a siRNA pool targeting SSBP2 and a nontargeting control into HEK293 and Het-1A cells and examined cel1 growth. A significant decrease of cell growth by Sulindac was observed in the HEK293 (Supporting Information Fig. S5A) and Het-1A cells (Fig. 5h) with SSBP2 knockdown. The expression of LRP6, p-LRP6 and Wnt7b proteins was increased by SSBP2 knock-down in the Het-1A cells, but Dvl3, Naked2 and Wnt3a were not changed (Fig. 5i). The levels of Axin1, WIF1 and Wnt5a/b were decreased by SSBP2 gene knock-down. Sulindac inhibited the increased LRP6 and p-LRP6 by MSE exposure and baseline expression of Dvl3, Axin1 and WIF1, but had no effect on Naked2 expression. The expression of proteins in Wnt signaling is summarized in Supporting Information Table S2. These results indicate that SSBP2 may play a role in Wnt signaling pathway in esophageal carcinogenesis and may affect LRP6 expression and its phosphorylation.
Discussion
Exposure to cigarette smoke is associated with high risk in human cancers, and cigarette smoking increases the frequency of observed genetic23 and epigenetic alterations such as DNA methylation in these cancers.32 Sensitivity of CpG dinucleotides to CSE-induced methylation seems to clearly be gene specific as evidenced by our data here on 13 gene promoters. Furthermore, the consequences of this methylation varies in each gene; although new methylation was found in dozens of CpG dinucleotides, NEFH expression in the MSE- and SSE-Het-1A cells was not changed after 6 months of CSE exposure. A higher density of CpG dinucleotides may need to be methylated for gene silencing, or the expression of NEFH may be regulated by other promoter regions or additional epigenetic mechanisms. In contrast, NMDAR2B was completely silenced even though only a few CpGs were methylated in the CSE-exposed cells, indicating that methylation in these CpGs might be sufficient to induce gene silencing or that CpG concordant methylation in other unexamined regions participated in downregulation of NMDAR2B expression. In addition, SSBP2 was partially methylated, and its expression was not silenced in MSE-exposed cells, perhaps indicating that longer exposure and/or selection to MSE might be needed for complete methylation and gene silencing.
Our data also imply that mechanisms other than promoter methylation are involved in gene transcription in CSE-exposed cells. Our mRNA expression array revealed that APC level in the MSE- and SSE-exposed cells was 8- and 4-fold lower, respectively, compared to control cells (data not shown). DCC expression decreased 4- and 9-fold in the MSE- and SSE-exposed cells (data not shown); however, methylation of APC and DCC was not observed in our MSP analysis, suggesting that DNA methylation is not a major mechanism for CSE-induced downregulation of these genes. In addition, little difference was observed in mRNA expression of NEFH between control and MSE- or SSE-exposed cells, but a lower level of NEFH protein was observed in the MSE- and SSE-exposed cells compared to control (data not shown), indicating that CSE exposure regulates NEFH expression in translational rather than transcriptional level.
SSBP2 may be part of a stem cell leukemia factor, a multi-protein complex required for erythroid differentiation,33,34 and a JAK2 fusion partner in a patient with pre-B-cel1 acute lymphocytic leukemia.35 Nonrandom interstitial deletion, translocations or paracentric inversions involving 5q11 to 5q13 have been frequently reported in refractory myelodysplasia, human acute myelogenous leukemia (AML)36 and solid tumors.37-39 SSBP2 is a tumor suppressor in AML by inducing cell growth arrest at G1 phase, which is accompanied by loss of c-Myc expression.40 We recently reported aberrant methylation of SSBP2 in prostate cancer and its tumor suppressive function in human prostate cancer cell lines.31 In a microarray analysis, genes associated with proapoptosis and cel1 adhesion inhibition were upregulated in the SSBP2-overexpressing PC3 cells, whereas genes related to antiapoptosis, cell adhesion promotion and Wnt signaling were downregulated.31 Our results here further support a role for SSBP2 as a tumor suppressor in human cancer.
Aberrant reactivation of embryonic signaling pathways in adult cells such as Wnt can provide a driving force for tumor growth.41 Overexpression and reactivation of the Wnt pathway result in activation of β-catenin/TCF-dependent transcription, which has been implicated in ESCC development.23, 42-44 Wnt members are critical mediators of cigarette smoke-induced lung cancer,29,30 and exposure to tobacco smoke condensate diminishes expression of Dickkopf-1 and upregulates Wnt5a expression in lung cancer cells.29 On stimulation by Wnt, lipoprotein receptor-related protein-6 (LRP6) is phosphorylated by kinases such as glycogen synthase kinase-3β (Gsk-3β). In the absence of Wnt stimulation, Axin negatively regulates Wnt signaling. Axin1 complexes with APC, Gsk-3β and β-catenin and promotes the Gsk-3β-mediated phosphorylation and subsequent β-catenin degradation.45,46 The Disheveled (Dvl) proteins promote β-catenin stabilization by inhibiting Gsk-3β,47 whereas Naked inhibits the canonical Wnt/β-catenin pathway by binding to Dvl. 48,49 Interestingly, reduced expression of Axin correlates with tumor progression of ESCC.44
Our results show that chronic exposure of MSE activates proteins related to Wnt signaling in esophageal epithelial cells. Interestingly, among Wnt signaling factors examined in our study, LRP6 was the only one that was increased by chronic exposure of the Het-1A cells to MSE and inhibited by pSSBP2 transfection. In addition, SSBP2 gene knock-down in the Het-1A cells increased the LRP6 expression and its phosphorylation level. These results suggest that LRP plays a role in cigarette smoke-associated esophageal carcinogenesis and that it is negatively regulated by SSBP. Moreover, negative Wnt regulators, Axin1 and Naked2, were downregulated in the MSE-Het-1A cells. Although Axin1 and Naked2 expression were not affected by increased SSBP2 expression in the MSE-exposed cells, Axin1 was decreased by SSBP2 knock-down in the control-Het-1A cells and slightly increased by SSBP2 expression in TE1 cells. These results indicate that Axin1 is regulated by SSBP2. However, Naked2 does not seem to be related to SSBP2; Naked2 expression was not affected by SSBP2 expression in ESCC cell lines or by SSBP2 knock-down in the control-Het-1A cells.
The consequences of SSBP2 manipulation at the mRNA and protein level of Wnt3a were not concordant; Wnt3a transcription was downregulated by SSBP2 overexpression in the PC3 prostate cancer cell line31 and in TE1 cells. SSBP2 knock-down in HEK293 (Supporting Information Fig. S5B) and Het-1A cells (Supporting Information Fig. S6) also increased the mRNA level of Wnt3a. However, the protein level of Wnt3a was not suppressed by SSBP2 overexpression in the MSE-Het-1A cells nor increased by SSBP2 gene knock-down in the control-Het-1A cells. The precise role of SSBP2 in Wnt3a signaling thus requires further study.
In contrast to sulindac sulfide, sulindac sulfone (Sulindac) does not block cyclooxygenase-2 activity or affect Hedgehog signaling.30 The inhibition of Wnt pathway signaling by Sulindac is through stimulation of protein kinase G (PKG) activity via inhibition of cyclic nucleotide phosphodiesterase. Activated PKG by Sulindac phosphorylates the c-terminal portion of β-catenin, thereby facilitating β-catenin degradation. In our study, LRP6 and its phosphorylation levels were decreased by Sulindac in cells treated with MSE or transfected with SSBP2 siRNA. Sulindac also decreased baseline or MSE-activated Wnt expression, indicating that Sulindac can inhibit Wnt signaling upstream of β-catenin. In addition, Sulindac significantly inhibited the cell growth of the MSE-Het-1A cells and the control-Het-1A and HEK293 cells with SSBP2 gene knock-down. Although it is not clear that Wnt pathway activation alone is sufficient to induce full malignant transformation of normal cells,30,50 our results suggest that the Wnt pathway plays, at least in part, a causal role in the proliferation and survival of CSE-transformed epithelial cells. Thus, Wnt pathway inhibitors (or even SSBP2 reintroduction) may provide a new rationale therapeutic approach for smoking-induced ESCC.
Our results indicate that cigarette smoking might be a cause of SSBP2 promoter methylation. Studying well-annotated primary ESCC samples with a detailed smoking history will add further support to this important observation. Regardless of this correlation, SSBP2 is frequently methylated and silenced in ESCC and represents a new and specific biomarker to test in this disease. In addition, the tumor suppressive function of SSBP2 is, at least in part, mediated through inhibition of Wnt signaling providing a personalized therapeutic approach for patients with SSBP2 methylated esophageal cancer.
Supplementary Material
Acknowledgements
this study was supported by National Cancer Institute (U01-CA84986) and the Flight Attendant Medical Research Institute (Myoung S. Kim) and Oncomethylome Sciences, SA. Under a licensing agreement between Oncomethylome Sciences, SA, and the Johns Hopkins University, D. Sidransky is entitled to a share of royalty received by the University upon sales of any products described in this article. D. Sidransky owns Oncomethylome Sciences, SA stock, which is subject to certain restrictions under University policy. D. Sidransky is a paid consultant to Oncomethylome Sciences, SA, and is a paid member of the company’s Scientific Advisory Board.
Grant sponsor: National Cancer Institute; Grant number: U01-CA84986; Grant sponsor: the Flight Attendant Medical Research Institute and Oncomethylome Sciences, SA
Footnotes
The Johns Hopkins University in accordance with its conflict of interest policies is managing the terms of this agreement.
Additional Supporting Information may be found in the online version of this article.
All financial or other interests with regard to the submitted manuscript that might be construed as a conflict of interest are noted in the disclosure;
References
- 1.Pisani P, Parkin DM, Bray F, Ferlay J. Int J Cancer. 1999;83:870–3. doi: 10.1002/(sici)1097-0215(19991210)83:6<870::aid-ijc35>3.0.co;2-9. Erratum: estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer, 83, 18-29 (1999) [DOI] [PubMed] [Google Scholar]
- 2.Goseki N, Koike M, Yoshida M. Histopathologic characteristics of early stage esophageal carcinoma. A comparative study with gastric carcinoma. Cancer. 1992;69:1088–93. doi: 10.1002/cncr.2820690503. [DOI] [PubMed] [Google Scholar]
- 3.Brockmann JG, St Nottberg H, Glodny B, Heinecke A, Senninger NJ. CYFRA 21-1 serum analysis in patients with esophageal cancer. Clin Cancer Res. 2000;6:4249–52. [PubMed] [Google Scholar]
- 4.Shimada Y, Watanabe G, Kawamura J, Soma T, Okabe M, Ito T, Inoue H, Kondo M, Mori Y, Tanaka E, Imamura M. Clinical significance of osteopontin in esophageal squamous cell carcinoma: comparison with common tumor markers. Oncology. 2005;68:285–92. doi: 10.1159/000086961. [DOI] [PubMed] [Google Scholar]
- 5.Stoner GD, Gupta A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis. 2001;22:1737–46. doi: 10.1093/carcin/22.11.1737. [DOI] [PubMed] [Google Scholar]
- 6.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–96. [PubMed] [Google Scholar]
- 7.Carmalt JE, Theis J, Goldstein E. Spinal cysticercosis. West J Med. 1975;123:311–3. [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MS, Yamashita K, Baek JH, Park HL, Carvalho AL, Osada M, Hoque MO, Upadhyay S, Mori M, Moon C, Sidransky D. N-methyl-D-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer Res. 2006;66:3409–18. doi: 10.1158/0008-5472.CAN-05-1608. [DOI] [PubMed] [Google Scholar]
- 9.Kim MS, Yamashita K, Chae YK, Tokumaru Y, Chang X, Zahurak M, Osada M, Park HL, Chuang A, Califano JA, Sidransky D. A promoter methylation pattern in the N-methyl-D-aspartate receptor 2B gene predicts poor prognosis in esophageal squamous cell carcinoma. Clin Cancer Res. 2007;13:6658–65. doi: 10.1158/1078-0432.CCR-07-1178. [DOI] [PubMed] [Google Scholar]
- 10.Georgiou E, Valeri R, Tzimagiorgis G, Anzel J, Krikelis D, Tsilikas C, Sarikos G, Destouni C, Dimitriadou A, Kouidou S. Aberrant p16 promoter methylation among Greek lung cancer patients and smokers: correlation with smoking. Eur J Cancer Prev. 2007;16:396–402. doi: 10.1097/01.cej.0000236260.26265.d6. [DOI] [PubMed] [Google Scholar]
- 11.Ashktorab H, Begum R, Akhgar A, Smoot DT, Elbedawi M, Daremipouran M, Zhao A, Momen B, Giardiello FM. Folate status and risk of colorectal polyps in African Americans. Dig Dis Sci. 2007;52:1462–70. doi: 10.1007/s10620-006-9236-8. [DOI] [PubMed] [Google Scholar]
- 12.Lea JS, Coleman R, Kurien A, Schorge JO, Miller DS, Minna JD, Muller CY. Aberrant p16 methylation is a biomarker for tobacco exposure in cervical squamous cell carcinogenesis. Am J Obstet Gynecol. 2004;190:674–9. doi: 10.1016/j.ajog.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 13.Oka D, Yamashita S, Tomioka T, Nakanishi Y, Kato H, Kaminishi M, Ushijima T. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412–26. doi: 10.1002/cncr.24394. [DOI] [PubMed] [Google Scholar]
- 14.Stabbert R, Voncken P, Rustemeier K, Haussmann HJ, Roemer E, Schaffernicht H, Patskan G. Toxicological evaluation of an electrically heated cigarette. Part 2: chemical composition of mainstream smoke. J Appl Toxicol. 2003;23:329–39. doi: 10.1002/jat.924. [DOI] [PubMed] [Google Scholar]
- 15.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res. 2004;567:447–74. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Kim MS, Huang Y, Lee J, Zhong X, Jiang WW, Ratovitski EA, Sidransky D. Cellular transformation by cigarette smoke extract involves alteration of glycolysis and mitochondrial function in esophageal epithelial cells. Int J Cancer. 2010;127:269–81. doi: 10.1002/ijc.25057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro P, Liang H, Liang JC, Nagarajan L. A novel, evolutionarily conserved gene family with putative sequence-specific single-stranded DNA-binding activity. Genomics. 2002;80:78–85. doi: 10.1006/geno.2002.6805. [DOI] [PubMed] [Google Scholar]
- 18.Kim MS, Louwagie J, Carvalho B, Terhaar Sive Droste JS, Park HL, Chae YK, Yamashita K, Liu J, Ostrow KL, Ling S, Guerrero-Preston R, et al. Promoter DNA methylation of oncostatin m receptor-beta as a novel diagnostic and therapeutic marker in colon cancer. PLoS One. 2009;4:e6555. doi: 10.1371/journal.pone.0006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J, Zhang Q, Jin J, Liu D, Rhim JS, Rha SY, Loyo M, Chan AT, et al. OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS One. 2008;3:e2990. doi: 10.1371/journal.pone.0002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita K, Kim MS, Park HL, Tokumaru Y, Osada M, Inoue H, Mori M, Sidransky D. HOP/OB1/NECC1 promoter DNA is frequently hypermethylated and involved in tumorigenic ability in esophageal squamous cell carcinoma. Mol Cancer Res. 2008;6:31–41. doi: 10.1158/1541-7786.MCR-07-0213. [DOI] [PubMed] [Google Scholar]
- 21.Mandelker DL, Yamashita K, Tokumaru Y, Mimori K, Howard DL, Tanaka Y, Carvalho AL, Jiang WW, Park HL, Kim MS, Osada M, Mori M, et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–68. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- 22.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, Rha SY, Tan J, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–44. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 23.Husgafvel-Pursiainen K. Genotoxicity of environmental tobacco smoke: a review. Mutat Res. 2004;567:427–45. doi: 10.1016/j.mrrev.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita K, Mimori K, Inoue H, Mori M, Sidransky D. A tumor-suppressive role for trypsin in human cancer progression. Cancer Res. 2003;63:6575–78. [PubMed] [Google Scholar]
- 25.Park HL, Kim MS, Yamashita K, Westra W, Carvalho AL, Lee J, Jiang WW, Baek JH, Liu J, Osada M, Moon CS, Califano JA, et al. DCC promoter hypermethylation in esophageal squamous cell carcinoma. Int J Cancer. 2008;122:2498–2502. doi: 10.1002/ijc.23434. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami K, Brabender J, Lord RV, Groshen S, Greenwald BD, Krasna MJ, Yin J, Fleisher AS, Abraham JM, Beer DG, Sidransky D, Huss HT, et al. Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma. J Natl Cancer Inst. 2000;92:1805–11. doi: 10.1093/jnci/92.22.1805. [DOI] [PubMed] [Google Scholar]
- 27.Guo XQ, Wang SJ, Zhang LW, Wang XL, Zhang JH, Guo W. DNA methylation and loss of protein expression in esophageal squamous cell carcinogenesis of high-risk area. J Exp Clin Cancer Res. 2007;26:587–94. [PubMed] [Google Scholar]
- 28.Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GB, Park HL, Poeta ML, Jeronimo C, Henrique R, Lendvai A, Schuuring E, Begum S, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–70. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, Caragacianu D, Schrump DS. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–8. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemjabbar-Alaoui H, Dasari V, Sidhu SS, Mengistab A, Finkbeiner W, Gallup M, Basbaum C. Wnt and Hedgehog are critical mediators of cigarette smoke-induced lung cancer. PLoS One. 2006;1:e93. doi: 10.1371/journal.pone.0000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JW, Nagpal JK, Sun W, Lee J, Kim MS, Ostrow KL, Zhou S, Jeronimo C, Henrique R, Van Criekinge W, Moon CS, Califano JA, et al. ssDNA-binding protein 2 is frequently hypermethylated and suppresses cell growth in human prostate cancer. Clin Cancer Res. 2008;14:3754–60. doi: 10.1158/1078-0432.CCR-07-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang HW, Ling GS, Wei WI, Yuen AP. Smoking and drinking can induce p15 methylation in the upper aerodigestive tract of healthy individuals and patients with head and neck squamous cell carcinoma. Cancer. 2004;101:125–32. doi: 10.1002/cncr.20323. [DOI] [PubMed] [Google Scholar]
- 33.Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, Dumenil D, Strouboulis J, Romeo PH, Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–66. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, Rodriguez P, Strouboulis J, Enver T, Vyas P, Porcher C. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides hrepressor functions in erythropoiesis. Mol Cell Biol. 2005;25:10235–50. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poitras JL, Dal Cin P, Aster JC, Deangelo DJ, Morton CC. Novel SSBP2-JAK2 fusion gene resulting from a t(5;9)(q14.1;p24.1) in pre-B acute lymphocytic leukemia. Genes Chromosomes Cancer. 2008;47:884–9. doi: 10.1002/gcc.20585. [DOI] [PubMed] [Google Scholar]
- 36.Fairman J, Wang RY, Liang H, Zhao L, Saltman D, Liang JC, Nagarajan L. Translocations and deletions of 5q13.1 in myelodysplasia and acute myelogenous leukemia: evidence for a novel critical locus. Blood. 1996;88:2259–66. [PubMed] [Google Scholar]
- 37.Achille A, Baron A, Zamboni G, Di Pace C, Orlandini S, Scarpa A. Chromosome 5 allelic losses are early events in tumours of the papilla of Vater and occur at sites similar to those of gastric cancer. Br J Cancer. 1998;78:1653–60. doi: 10.1038/bjc.1998.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atkin NB, Fox MF. 5q deletion. The sole chromosome change in a carcinoma of the bladder. Cancer Genet Cytogenet. 1990;46:129–31. doi: 10.1016/0165-4608(90)90019-7. [DOI] [PubMed] [Google Scholar]
- 39.Cunningham JM, Shan A, Wick MJ, McDonnell SK, Schaid DJ, Tester DJ, Qian J, Takahashi S, Jenkins RB, Bostwick DG, Thibodeau SN. Allelic imbalance and microsatellite instability in prostatic adenocarcinoma. Cancer Res. 1996;56:4475–82. [PubMed] [Google Scholar]
- 40.Liang H, Samanta S, Nagarajan L. SSBP2, a candidate tumor suppressor gene, induces growth arrest and differentiation of myeloid leukemia cells. Oncogene. 2005;24:2625–34. doi: 10.1038/sj.onc.1208167. [DOI] [PubMed] [Google Scholar]
- 41.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51. [PubMed] [Google Scholar]
- 42.Kudo J, Nishiwaki T, Haruki N, Ishiguro H, Shibata Y, Terashita Y, Sugiura H, Shinoda N, Kimura M, Kuwabara Y, Fujii Y. Aberrant nuclear localization of beta-catenin without genetic alterations in b eta-catenin or Axin genes in esophageal cancer. World J Surg Oncol. 2007;5:21. doi: 10.1186/1477-7819-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizushima T, Nakagawa H, Kamberov YG, Wilder EL, Klein PS, Rustgi AK. Wnt-1 but not epidermal growth factor induces beta-catenin/T-cell factor-dependent transcription in esophageal cancer cells. Cancer Res. 2002;62:277–82. [PubMed] [Google Scholar]
- 44.Nakajima M, Fukuchi M, Miyazaki T, Masuda N, Kato H, Kuwano H. Reduced expression of Axin correlates with tumour progression of oesophageal squamous cell carcinoma. Br J Cancer. 2003;88:1734–9. doi: 10.1038/sj.bjc.6600941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo W, Lin SC. Axin: a master scaffold for multiple signaling pathways. Neurosignals. 2004;13:99–113. doi: 10.1159/000076563. [DOI] [PubMed] [Google Scholar]
- 46.Salahshor S, Woodgett JR. The links between axin and carcinogenesis. J Clin Pathol. 2005;58:225–36. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 48.Wharton KA, Jr, Zimmermann G, Rousset R, Scott MP. Vertebrate proteins related to Drosophila Naked Cuticle bind Dishevelled and antagonize Wnt signaling. Dev Biol. 2001;234:93–106. doi: 10.1006/dbio.2001.0238. [DOI] [PubMed] [Google Scholar]
- 49.Yan D, Wallingford JB, Sun TQ, Nelson AM, Sakanaka C, Reinhard C, Harland RM, Fantl WJ, Williams LT. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc Natl Acad Sci USA. 2001;98:3802–7. doi: 10.1073/pnas.071041898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uren A, Fallen S, Yuan H, Usubütün A, Küçükali T, Schlegel R, Toretsky JA. Activation of the canonical Wnt pathway during genital keratinocyte transformation: a model for cervical cancer progression. Cancer Res. 2005;65:6199–206. doi: 10.1158/0008-5472.CAN-05-0455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.