Abstract
Background
Occult hepatitis B infection (OBI) is a form of hepatitis in which there is an absence of detectable HBsAg, despite the presence of HBV-DNA in the peripheral blood of patients. It seems that non-effective or attenuated immune system responses against HBV lead to the development of OBI. Previous studies showed that the Fas/Fas ligand (FasL) system is an important death signaling pathway that is used by cytotoxic T lymphocytes to eradicate HBV from the liver.
Objectives
To investigate polymorphisms in the -670 region of the Fas gene in those with OBI.
Patients and Methods
The plasma samples from 3700 blood donors were tested for HBsAg and anti-HBs by ELISA. The HBsAg-/anti-HBc(+) samples were selected and screened for HBV-DNA by PCR. Those with HBV-DNA were diagnosed as OBI and PCR-RFLP technique was performed to examine polymorphisms within their Fas gene.
Results
352 (9.5%) of 3700 blood samples were HBsAg-/anti-HBc(+). HBV-DNA was detected in 57 (16.1%) of 352 HBsAg-/anti-HBc(+) samples. Therefore, 57 HBsAg-/anti-HBc+/HBV-DNA(+) patients were diagnosed as OBI. Patient and control groups had no significant differences in terms of the studied polymorphisms.
Conclusions
The functional polymorphisms in the promoter region of Fas gene are not associated with OBI. Therefore, it may be concluded that polymorphisms at the -670 position of the Fas gene do not have any critical effects on the immune response against HBV in OBI.
Keywords: Hepatitis B infection, Fas, Polymorphism, HBsAg, HBV, DNA
Background
Occult hepatitis B infection (OBI) is a clinical form of hepatitis B in which there are no detectable HBsAg in patient's serum, despite being positive for HBV-DNA [1]. Some of the patients do not have detectable HBV-DNA in serum, despite the presence of HBV-DNA in hepatocytes [2]. Furthermore, high prevalence rates of OBI in chronic HCV-infected patients [3], hemodialysis patients [1][4], immunocompromised patients [5] and HIV-infected patients [6] have been reported. OBI is clinically important because it may induce chronic liver disease [2] and cryptogenic cirrhosis [7]. This type of hepatitis also poses a threat to blood transfusion services and its detection remains a significant challenge for these agencies. The high prevalence of OBI in Iranian blood donors [8][9][10] may be a critical risk for post-transfusion hepatitis (PTH), and despite appropriate screening of all donated blood and blood components for HBsAg, some cases of PTH B are reported [8][11]. The majority of PTH B infections are caused by OBI [12] which we previously reported in our investigations in Isfahan [9] and Kerman [10], the two central provinces of Iran. The mechanisms responsible for progression of OBI are yet to be clarified however, some investigators have suggested that genetic and immunological parameters may play a significant role in the resistance of some individuals and sensitivity of other patients [11][13][14]. Previous studies showed that the Fas/Fas ligand (FasL) system is an important death signaling pathway that is used by cytotoxic T lymphocytes to eradicate HBV from the liver [15]. Elevated expression of FasL was also reported during HBV infection by some investigators [16][17]. Furthermore, several studies showed that the polymorphisms within the Fas Fas (-670 A→G) gene can alter its expression [18][19].
Objectives
This study was conducted to investigate the relation between OBI and the functional polymorphisms within the promoter region of Fas gene.
Patients and Methods
Patients
Peripheral blood samples were collected from 3700 volunteer blood donors of the Rafsanjan Blood Transfusion Services (Kerman, Iran) and placed in EDTA pre-coated 5.5-mL tubes. The samples were centrifuged at 3700 g for 4 min and the sera were collected. All sera were separated within 24 hrs of collection. If needed, serum samples were stored at 20 ºC for a maximum of two months or at 70 ºC, when longer storage was required for further processing. For analysis of polymorphisms a 2-mL aliquot was collected from patients with OBI (57 cases) and 100 healthy controls (HBsAg-/anti-HBc+/HBV-DNA+). The study protocol was approved by the Ethical Committee of Rafsanjan University of Medical Sciences. All of the participants completed and signed an informed consent form which was designed based on the objectives of the study.
Detection of serological HBV markers
HBsAg screening tests were performed by enzyme-linked immuno-sorbent assay (ELISA) (Behring, Germany). Anti-HBc screening tests were performed by a manual microplate enzyme immunoassay using an anti-HBc commercial kit (RADIM, Italy). The present method is based on a competitive enzyme immunoassay (EIA). All of the samples were also screened by ELISA (RADIM, Italy) for possible HCV, HIV and HTLV-1 infections.
HBV-DNA extraction from plasma samples
Viral DNA was purified from 200-µL of plasma samples. Briefly, each plasma sample was incubated at 72 ºC for 10 min and then cooled down to 4 ºC for 5 min in 200 µL proteinase K (200 µg/mL) (Cinnagen, Iran). Following phenol/chloroform extraction (1:1), the viral DNA was precipitated with ethanol and the pellet was re-dissolved in DNase-free, deionized water (Cinnagen, Iran) and stored at 20 ºC for further use.
HBV-DNA polymerase chain reaction (PCR) and gel eletrophoresis
PCR was carried out in a 25-µL mixture containing 10 mM tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 5 units recombinant TaqDNA polymerase, 200 µM of each dNTP, 0.6 µM of each primer, and 5 µL of the DNA extracted from 200 µL of plasma. The sequences of all primers used in this study are shown in Table 1. For HBV analysis, the primers are designed to amplify a 500-bp amplicon of the surface antigen or S gene of the HBV genome. Fast temperature cycling was performed using a thermal cycler (C1000, Bio-Rad, USA). PCR amplification was performed using the touchdown method which included one cycle of 93 ºC for 60 sec, 60 ºC for 20 sec and 72 ºC for 40 sec, then five cycles of 93 ºC for 20 sec, 60 ºC to 56 ºC for 20 sec and 72 ºC for 40 sec followed by 30 cycles of 93 ºC for 20 sec, 55 ºC for 20 sec and 72 ºC for 40 sec. HBV genomic DNA provided by the Cinnagen company (Iran) and a negative patient sample were used as positive and negative controls, respectively. For the analysis of the PCR amplification, 10 µL of the amplified DNA were run on 2% agarose gel after addition of 4 µL of loading buffer using a chemi-doc-XRS system (Bio-Rad, USA). The presence of a 500-bp fragment indicated a positive result. In parallel with samples, a 100-bp DNA ladder was also run on the gels to estimate the molecular weight of DNA fragments in the gel.
Table 1. The sequence of the primers used in the study as well as the appropriate annealing temperatures and expected PCR product sizes.
| Target Genes | Primers | Annealing temperature | Product size (bp) |
| S gene (HBV) | F: 5' TCGTGGTGGACTTCTCTC 3' | 60 °C | 500 |
| R: 5' ACAGTGGGGGAAAGCCC 3' | |||
| Fas (‑670) | Fas-670 F:5' CTACCTAAGAGCTATCTACCGTTC 3' | 58 °C | 233 |
| Fas ‑670 R: 5' GGCTGTCCATGTTGTG GCTGC 3' |
Genomic DNA extraction
Peripheral blood was collected in EDTA tubes and genomic DNA was extracted using a commercial kit (Bioneer, Korea) following the recommended procedures. Extracted DNA was aliquoted (for each patient sample) and stored at 20 ºC for further use.
Detection of polymorphisms
The gene polymorphisms were analyzed by the PCR-restricted fragment length polymorphism (PCR-RFLP) method. PCR of the Fas gene was performed in a volume of 50 µL containing 250 ng of DNA template, 200 µM of each dNTP (Cinnagen, Iran), 0.5 U Taq DNA polymerase (Cinnagen, Iran), 1x PCR buffer (Cinnagen, Iran), 3 mM MgCl2, and 5 pM of each specific primer (Table 1). The PCR conditions were; an initial denaturation at 95 ºC for 5 min, followed by 35 cycles of melting at 95 ºC for 50 sec, suitable annealing temperature for 50 sec (Table 1), and extension at 72 ºC for 50 sec, with a final extension step of 5 min at 72 ºC using a thermal cycler (C1000, Bio-Rad, USA). The expected size of the amplified PCR product used to detect the -670 amplicon of Fas was 233 bp. The ScrFI restriction enzyme (Fermantase, Finland) was used to distinguish the Fas -670 A→G polymorphisms, which resulted in 189 plus 44-bp fragments in the case of the -670 G allele. More than 10% of the samples were randomly selected and retested by appropriate PCR-RFLP techniques for confirmation; the results were 100% concordant. The digested products were run on a 2.5% agarose gel (Cinnagen, Iran) and analyzed using a chemi-doc-XRS system (Bio-Rad, USA) after staining with ethidium bromide.
Statistical analysis
Hardy-Weinberg equilibrium was assessed using genotype data. Allele and genotype frequencies were calculated in patients and healthy controls by direct gene counting. Statistical analysis of the differences between groups was determined by x(2) test using EPI 2000 and SPSS® ver 13. A p < 0.05 was considered statistically significant.
Results
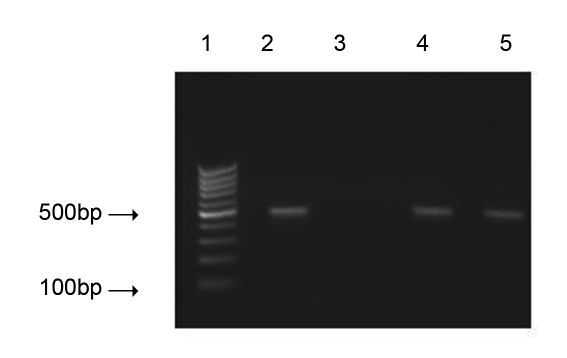
This study was performed on 3700 blood samples collected from patients attending the Rafsanjan blood transfusion services. All of the samples were found to be negative for HBsAg, anti-HCV, anti-HTLV-1 and anti-HIV antibodies. Out of 3700 samples, 352 (9.5%) were positive for anti-HBc out of whom 57 were found positive for HBV-DNA (Figure 1); These 57 HBsAg-/anti-HBc(+)/HBV-DNA(+) patients were diagnosed as OBI. Results of this study indicated that 16.1% of HBsAg-negative but anti-HBc-positive samples had detectable HBV-DNA which is 1.54% (57 of 3700) of the total collected samples. The mean±SD age of patients and controls were 28±6 and 28±8 years, respectively; there was no statistically significant difference in age between the two groups (Table 2). Three (3%) of the control group members were female and 97 (97%) were male while two of the patients (4%) were female and 55 (96%) were male. In addition, analysis of socio-economic conditions showed that there was also no significant difference between the patient and control groups (Table 2). Evaluation of the polymorphisms at 670 position of Fas showed that the frequency of ‘A' and ‘G' alleles were 69 (60.5%) and 45 (39.5%) in patients, respectively; the values were 124 (62%) and 76 (38%) in controls, respectively. Statistical analysis of these alleles indicated that the differences were not statistically significant (p = 0.810) (Table 3). Our results also showed that the prevalence of A/A genotype within the 670 region of Fas was 18 (32%) in patients and 40 (40%) in controls; the frequency of A/G genotype was 33 (58%) and 44 (44%) in patients and controls, respectively; and the frequency of G/G genotype in patients was 6 (11%) and in controls was 16 (16%) (Table 3). Statistical analysis of our data could not show any significant difference between the two groups regarding the frequencies of these genotypes (p = 0.232).
Figure 1.
HBV-DNA PCR. An ethidum bromide stained agarose gel showing typical results of HBV-DNA PCR screening of collected samples which were HBsAg negative and anti-HBc positive. Lane 1: DNA ladder marker; lane 2: A positive control showing the expected 500-bp product; lane 3: A negative control; lanes 4 and 5: Positive samples.
Table 2. Demographic and socioeconomic conditions of OBI patients and controls.
| Variant | Healthy control (n = 100) | Patient (n = 57) | |
| Age (Mean ± SD)(Year) | 28 ± 8 | 28 ± 6 | |
| Sex | Female | 3 (3%) | 2 (3.5%) |
| Male | 97 (97.8%) | 55 (96.5%) | |
| Socio-economic status | Weak | 22 (22%) | 12 (21%) |
| Medium | 47 (47%) | 28 (49%) | |
| High | 31 (31%) | 17 (30%) | |
Table 3. The frequency of the 670 A→G polymorphisms within the Fas gene promoter of OBI patients and controls.
| PatientsNo.(%) | ControlNo.(%) | p-value | ||
| Genotype | A/A n | 38 (67%) | 40 (40%) | p = 0.232 |
| A/G n | 33 (58%) | 44 (44%) | ||
| G/G n | 6 (11%) | 16 (16%) | ||
| Alleles | A n | 69 (60.5%) | 124 (62%) | p = 0.810 |
| G n | 45 (39.5%) | 76 (38%) | ||
Discussion
It is now well established that following viral hepatitis, some of the infected hepatocytes express the Fas and FasL system [15][20]. It is also stated that the rate of expression of FasL is correlated with the kind of clinical presentation and also correlates with the different stages of the HBV infection and associated liver disease [21]. Furthermore, Fas/FasL system may play a key role in apoptosis of the infected hepatocytes [22]. For example, over-chronic HBV and HCV infection [15] and the fulminant form of HBV infection [24] were reported. It seems that over-expression of Fas/FasL led to hepatocyte injury in hepatitis [23][24]. However, despite evidence suggesting a potential correlation between Fas/FasL system and disease status, our results showed that the frequency of evaluated alleles and genotypes was not different between OBI patients and healthy controls. Therefore, it can be concluded that these polymorphisms are not associated with OBI. Previous studies showed that the level of expression of Fas and FasL was associated with the clinical pattern of the disease [15][25]. For example, Bortolami et al. reported more expression of Fas and FasL in HBV-infected hepatocytes from patients with cirrhosis than in patients with chronic hepatitis [15]. Studies also reported that polymorphisms in the promoter regions of Fas and FasL influence the pattern of their expression [26]. Since OBI patients are unable to clear HBV completely, it could be suggested that the expression of Fas and FasL in OBI patients may be compromised by mechanisms other than polymorphisms within the promoter region. Despite evidence linking a functional role for Fas to progression of viral hepatic disease, our data showed that the polymorphisms, which are known to influence Fas expression levels, were not statistically different between OBI patients and healthy controls. Further studies should be done on the expression of Fas at the protein level to confirm this. To our knowledge, this is the first report which investigates the involvement of Fas polymorphisms with OBI. However, in related research, Jung et al. also showed that there were no significant associations between FasL (844 C/T) polymorphism and HBV clearance in chronic hepatitis patients [22]. Interestingly, similarly to our own findings, Sung and colleagues could not find a significant difference between the healthy donors and patients with hepatocellular carcinoma [21]. Similarly, several studies reported a correlation between Fas and FasL polymorphisms during HCV infection [27][28][29]. One reason for the discrepancy between our results and these studies could be explained by the different types of hepatitis infection (HCV) and there may also be genetic differences in the populations studied. In addition, there may also be subtle differences in the type of disease from our studied population. It is not clear through what mechanisms OBI patients are unable to completely overcome the viral contamination, however, based on the current studies it seems that the polymorphisms within the promoter region of the Fas were not correlated with OBI. Finally, due to the complexity of OBI, other aspects of the disease are needed to be examined. For instance our previous study showed that the serum level of IL-17, an inflammatory cytokine, was increased, whereas the serum level of IL-10 has decreased in OBI patients [11]. Therefore, our future studies will explore polymorphisms and the expression levels of these and other important cytokines and their receptors within the OBI patients.
Acknowledgments
The authors would like to acknowledge all the OBI patients and healthy controls who contributed to this research. This work was supported by a grant from Rafsanjan University of Medical Sciences.
Footnotes
Implication for Health policy/practice/research/medical education: The role of gene polymorphisms in patients with Hepatitis B virus infection is discussed in this study. Reading this article is suggested to all virolo¬gists, genetic specialists in the field of hepatology.
Please cite this paper as: Arababadi MK, Mohammadzadeh A, Pourfathollah AA, Kennedy D. Polymorphisms within Fas gene are not associated with occult HBV infection. Hepat Mon. 2011;11(1):23-26.
References
- 1.Hollinger FB, Habibollahi P, Daneshmand A, Alavian SM. Occult Hepatitis B Infection in Chronic Hemodialysis Patients: Current Concepts and Strategy. Hepat Mon. 2010;10(3):199–204. [Google Scholar]
- 2.Hollinger FB, Sood G. Occult hepatitis B virus infection: a covert operation. J Viral Hepat. 2010;17(1):1–15. doi: 10.1111/j.1365-2893.2009.01245.x. [DOI] [PubMed] [Google Scholar]
- 3.Levast M, Larrat S, Thelu MA, Nicod S, Plages A, Cheveau A, Zarski JP, Seigneurin JM, Morand P, Leroy V. Prevalence and impact of occult hepatitis B infection in chronic hepatitis C patients treated with pegylated interferon and ribavirin. J Med Virol. 2010;82(5):747–54. doi: 10.1002/jmv.21695. [DOI] [PubMed] [Google Scholar]
- 4.Arababadi MK, Hassanshahi G, Yousefi H. HBV-DNA in hemodialysis patients infected by HCV. Saudi J Kidney Dis Transpl. 2009;20(3):398–401. [PubMed] [Google Scholar]
- 5.Jardim RN, Goncales NS, Pereira JS, Fais VC, Goncales Junior FL. Occult hepatitis B virus infection in immunocompromised patients. Braz J Infect Dis. 2008;12(4):300–5. doi: 10.1590/s1413-86702008000400008. [DOI] [PubMed] [Google Scholar]
- 6.Bagaglio S, Porrino L, Lazzarin A, Morsica G. Molecular characterization of occult and overt hepatitis B (HBV) infection in an HIV-infected person with reactivation of HBV after antiretroviral treatment interruption. Infection. 2010;38(5):417–21. doi: 10.1007/s15010-010-0032-1. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal N, Naik S, Aggarwal R, Singh H, Somani SK, Kini D, Pandey R, Choudhuri G, Saraswat VA, Naik SR. Occult hepatitis B virus infection as a cause of cirrhosis of liver in a region with intermediate endemicity. Indian J Gastroenterol. 2003;22(4):127–31. [PubMed] [Google Scholar]
- 8.Arababadi MK, Pourfathollah AA, Jafarzadeh A, Hassanshahi G, Rezvani ME. Association of exon 9 but not intron 8 VDR polymorphisms with occult HBV infection in south-eastern Iranian patients. J Gastroenterol Hepatol. 2010;25(1):90–3. doi: 10.1111/j.1440-1746.2009.05950.x. [DOI] [PubMed] [Google Scholar]
- 9.Jafarzadeh A, Arababadi M, Pourazar M. Occult hepatitis B virus infection among blood donors with antibodies to hepatitis B core antigen. Acta Medica Iranica. 2008;46(1):27–32. [Google Scholar]
- 10.Arababadi M, Pourazar A, Salehi M. Evaluation of occult HBV infection in HBsAg Negative and anti-HBc positive blood donors. J Shahid Sadoughi Yazd Univ Med Sci. 2007;15(1):74–8. [Google Scholar]
- 11.Arababadi MK, Pourfathollah AA, Jafarzadeh A, Hassanshahi G. Serum Levels of IL-10 and IL-17A in Occult HBV-Infected South-East Iranian Patients. Hepat Mon. 2010;10(1):31–5. [PMC free article] [PubMed] [Google Scholar]
- 12.Arababadi MK, Pourfath E, Hasanshahi G, Pouladvand V, Yaghini N, Shamsizadeh A. Evaluation the relationship between alleles of+ 1188 in region of il-12 with serum level of Cytokine in patients with occult HBV infection. J Guilan Univ Med Sci. 2010;18(72):39–46. [Google Scholar]
- 13.Arababadi MK, Pourfathollah A, Jafarzadeh A, Hassanshahi G. Detection of the ccr5-δ 32 mutation in patients infected with occult hepatitis b. Tabib-e-shargh. 2009;11(2):41–8. [Google Scholar]
- 14.Arababadi MK, Pourfathollah AA, Jafarzadeh A. Evaluation of serum level of IL-12 in patients with occult HBV infection. J Mazandaran Univ Med Sci. 2009;19(72):81–3. [Google Scholar]
- 15.Bortolami M, Kotsafti A, Cardin R, Farinati F. Fas / FasL system, IL-1beta expression and apoptosis in chronic HBV and HCV liver disease. J Viral Hepat. 2008;15(7):515–22. doi: 10.1111/j.1365-2893.2008.00974.x. [DOI] [PubMed] [Google Scholar]
- 16.Jun EJ, Han JY, Sun HS. [Clinical significances of serum soluble fas and soluble fas ligand in chronic hepatitis B] Korean J Hepatol. 2006;12(4):507–14. [PubMed] [Google Scholar]
- 17.Hayashi N, Mita E. involvement of Fas system-mediated apoptosis in pathogenesis of viral hepatitis. J Viral Hepat. 1999;6(5):357–65. doi: 10.1046/j.1365-2893.1999.00175.x. [DOI] [PubMed] [Google Scholar]
- 18.Nasi M, Pinti M, Bugarini R, Troiano L, Lugli E, Bellodi C, Mussini C, Borghi V, Trenti T, Balli F, Esposito R, Cossarizza A. Genetic polymorphisms of Fas (CD95) and Fas ligand (CD178) influence the rise in CD4+ T cell count after antiretroviral therapy in drug-naive HIV-positive patients. Immunogenetics. 2005;57(9):628–35. doi: 10.1007/s00251-005-0031-z. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Wang LE, Sturgis EM, El-Naggar AK, Hong WK, Amos CI, Spitz MR, Wei Q. Polymorphisms of FAS and FAS ligand genes involved in the death pathway and risk and progression of squamous cell carcinoma of the head and neck. Clin Cancer Res. 2006;12(18):5596–602. doi: 10.1158/1078-0432.CCR-05-1739. [DOI] [PubMed] [Google Scholar]
- 20.Shin EC, Shin JS, Park JH, Kim H, Kim SJ. Expression of fas ligand in human hepatoma cell lines: role of hepatitis-B virus X (HBX) in induction of Fas ligand. Int J Cancer. 1999;82(4):587–91. doi: 10.1002/(sici)1097-0215(19990812)82:4<587::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Kim SS, Hong SJ, Ahn YG, Kim BS, Yuh YJ, Han KY, Lee HJ, Chung JH, Yim SV, Cho J, Park YH. Apo-1/Fas (CD95) Gene Polymorphism in Korean Hepatocellular Carcinoma Patients. Korean J Physiol Pharmacol. 2003;7(1):29–31. [Google Scholar]
- 22.Jung YJ, Kim YJ, Kim LH, Lee SO, Park BL, Shin HD, Lee HS. Putative association of Fas and FasL gene polymorphisms with clinical outcomes of hepatitis B virus infection. Intervirology. 2007;50(5):369–76. doi: 10.1159/000109751. [DOI] [PubMed] [Google Scholar]
- 23.Lee SH, Shin MS, Lee HS, Bae JH, Lee HK, Kim HS, Kim SY, Jang JJ, Joo M, Kang YK, Park WS, Park JY, Oh RR, Han SY, Lee JH, Kim SH, Lee JY, Yoo NJ. Expression of Fas and Fas-related molecules in human hepatocellular carcinoma. Hum Pathol. 2001;32(3):250–6. doi: 10.1053/hupa.2001.22769. [DOI] [PubMed] [Google Scholar]
- 24.Zou Y, Chen T, Han M, Wang H, Yan W, Song G, Wu Z, Wang X, Zhu C, Luo X, Ning Q. Increased killing of liver NK cells by Fas/Fas ligand and NKG2D/NKG2D ligand contributes to hepatocyte necrosis in virus-induced liver failure. J Immunol. 2010;184(1):466–75. doi: 10.4049/jimmunol.0900687. [DOI] [PubMed] [Google Scholar]
- 25.Dmitrieva EV, Moskaleva E, Kogan EA, Bueverov AO, Belushkina NN, Ivashkin VT, Severin ES, Paltsev MA. [The role of Fas/FasL system in induction of hepatocyte apoptosis in chronic viral hepatitides] Arkh Patol. 2003;65(6):13–7. [PubMed] [Google Scholar]
- 26.Takahashi T, Tanaka M, Inazawa J, Abe T, Suda T, Nagata S. Human Fas ligand: gene structure, chromosomal location and species specificity. Int Immunol. 1994;6(10):1567–74. doi: 10.1093/intimm/6.10.1567. [DOI] [PubMed] [Google Scholar]
- 27.Aguilar-Reina J, Ruiz-Ferrer M, Pizarro MA, Antinolo G. The -670A > G polymorphism in the promoter region of the FAS gene is associated with necrosis in periportal areas in patients with chronic hepatitis C. J Viral Hepat. 2005;12(6):568–73. doi: 10.1111/j.1365-2893.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Ferrer M, Antinolo G, Aguilar-Reina J. Analysis of the -844C > T polymorphism in the promoter region of FASLgene in a cohort of Spanish HCV patients. J Viral Hepat. 2007;14(4):293–4. doi: 10.1111/j.1365-2893.2006.00821.x. [DOI] [PubMed] [Google Scholar]
- 29.Mcllroy D, Theodorou I, Ratziu V, Vidaud D, Pellet P, Debré P, Poynard T. FAS promoter polymorphisms correlate with activity grade in hepatitis C patients. Eur J Gastroenterol Hepatol. 2005;17(10):1081–8. doi: 10.1097/00042737-200510000-00012. [DOI] [PubMed] [Google Scholar]