Abstract
Natural killer (NK) cells constitute a first line of defense against viral infections; their function is governed by the integration of signals from multiple activating and inhibitory surface receptors. We hypothesized that since NKs become rapidly activated by cytokines, response to anti-HCV therapy would be predicted by the phenotype and function of NKs. We used a cohort of 101 patients (56 African-, 45 Caucasian-American) who received Pegylated-IFN and Ribavirin for 48 weeks. Multiparameter FACS analysis was used to examine relative expression of 14 different inhibitory/activating receptors. IL-28B genotyping (rs12979860) was also performed. Pre-treatment levels of inhibitory receptors CD158a, CD158b and CD158e were higher in patients who demonstrated poor viral decline within the first 28 days of therapy. Higher expression levels of inhibitory receptors NKG2A, CD158b and CD158e were demonstrable in patients who failed to achieve SVR. Patients carrying the IL-28B T allele had higher NKG2A expression on effector NKs. We created a mathematical regression model incorporating race, viral level, and two inhibitory receptors. The area-under-the curve was 0.88 which is highly predictive of SVR. Moreover, the model performed complementarily with IL-28B across the CC, CT, and TT genotypes. Purified NKG2Aneg NKs treated with pegylated-IFN-α for 4 hours demonstrated higher levels of interferon-γ–inducible protein-10 (IP-10) and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) compared to their NKG2Apos counterparts.
Conclusions
These results provide novel insights into the associations of NK phenotype with IL-28B genotype and gene expression patterns, as well as the role of NKs in mediating IFN-induced viral clearance of chronic HCV infection.
Keywords: Human, NK Receptors, Cytokine SNP
Hepatitis C virus (HCV) is a major causative agent of chronic hepatitis, with an estimated global prevalence of 3%, i.e., 200 million people, and is a leading cause of cirrhosis, liver cancer and indication for liver transplantation in the Western world [1]. Current antiviral treatment, based on the combination of pegylated IFN-alfa (α) and the nucleoside analog ribavirin (PEG/RIB), is associated with significant frequency of side effects and sustained virologic response (SVR), i.e., serum HCV RNA negativity six months after cessation of antiviral therapy, in less than 50% of treated subjects [2]. Understanding the precise mechanisms whereby HCV is controlled has important implications for predicting response and developing novel therapeutic strategies.
Natural killer (NK) cells constitute a first line of defense against viral infections; their function is governed by the integration of signals from a wide spectrum of activating and inhibitory cell surface NK receptors (NKRs) [3]. Furthermore, in contrast to T cells, NK cells do not require priming in order to recognize virus-infected cells. We hypothesized that since NK cells are rapidly activated by cytokine stimulation, the response to anti-HCV therapy (the major component of which is IFN-α) would be predicted by the pre-treatment phenotype and function of NK cells. Here, we show lymphocytes have higher expression levels of inhibitory receptors CD158a (KIR2DL1), CD158b (KIR2DL2/DL3) and CD158e (KIR3DL1) in patients with impaired viral kinetics whereas higher pre-treatment expression levels of the activating receptor NKp44 correlates directly with early viral decline following initiation of antiviral therapy. These and other markers were also predictive of SVR.
Recently, variation within or near the IL-28B gene (encoding interferon-lambda 3) has been associated with SVR [4]. We found that HCV-infected patients carrying the unfavorable T allele at position rs12979860, located on the long arm of chromosome 19, demonstrate higher expression of NKG2A, an inhibitory NKR. A multivariate model was developed to predict SVR comprised of four variables: race, pre-treatment viral level, NKG2A and CD158e (KIR3DL1) expression on lymphocytes. To explore the mechanism by which NK receptor expression in chronic HCV infection is linked to antiviral responsiveness, we characterized the in vitro effect of PEG on purified NK cell subsets. Our data provide novel insights into the associations of NK phenotype with IL-28B genotype and gene expression patterns, as well as the roles of NK cells in mediating therapy-induced viral clearance of this common infection.
Materials and Methods
Study population
This study utilized one hundred and one participants from the Study of Viral Resistance to Antiviral Therapy of Chronic Hepatitis C (Virahep-C), a multicenter study sponsored by the National Institutes of Health aimed at understanding the mechanisms of resistance to antiviral therapy for chronic HCV infection among interferon treatment-naive individuals infected with genotype 1 HCV (1a and 1b), as well as the differences in outcome by race among AAs and CAs [5]. Patients for this study were selected on the basis of having received maximal doses of PRG/RIB. Two patients from the AA and two from the CA group had early discontinuation of therapy, three subjects achieved SVR and had received greater than 6 months of therapy; one patient failed to achieve SVR having received 4 months of therapy. Eight patients had dose reductions in the first 28 days 5 AAs (Day 15-22) and 3 CAs (day 6-28). This research was conducted in accordance with the Helsinki principles: all patients gave informed, written consent prior to their participation in Virahep-C and its associated basic science components, and all components of Virahep-C were approved by their local Institutional Review Boards. Extensive demographic data was available on all patients and the characteristics of the study cohort are shown in Table 1.
Table 1. Demographic details for study cohort.
| Race | African American (AA) | Caucasian (CA) | p value |
|---|---|---|---|
| Number of subjects | n=55 | n=46 | |
| Gender (% male) | 60.0% | 73.91% | ns |
| Median Age (range) | 49 (34-70) | 48 (23-71) | 0.03 |
| Median Viral load (range) | 2.95 × 106 (0.035-19.5) | 2.145 × 106 (0.004-47.1) | ns |
| SVR (% positive) | 27.27% | 60.87% | <0.001 |
| Viral kinetics‡ | <0.001 | ||
| Poor$ | 58.0% | 27.27% | |
| Intermediate | 26.0% | 25.0% | |
| Marked | 16.0% | 47.73% | |
| IL-28B (rs12979860) genotype* | 0.005 | ||
| CC | 15.63% | 50.0% | |
| CT | 40.62% | 34.62% | |
| TT | 43.75% | 15.38% | |
| HCV genotype (1a/1b) | 33/22 | 36/10 | ns |
| Metavir Activity Score† (1-3) | ns | ||
| 1 | 18.52% | 26.09% | |
| 2 | 31.48% | 19.56% | |
| 3 | 50.0% | 54.35% | |
| Metavir Fibrosis Score† (0-4) | ns | ||
| 0 | 11.11% | 13.04% | |
| 1/2 | 74.07% | 69.57% | |
| 3/4 | 14.82% | 17.39% |
Twenty three AA and 21 CA subjects did not give permission for genetic analysis
One AA subject had no biopsy data recorded
Viral kinetic data was not available for 5 AA and 2 CA subjects
ns=not significant
Sample collection and storage
All patients were treatment-naïve prior to enrollment and study samples were collected 2 weeks before beginning of therapy. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by cellular preparation tubes (Becton-Dickinson, Franklin Lakes, New Jersey; anticoagulant sodium citrate). PBMCs were viably frozen in 80% FBS (BioWhittaker, Walkersville, MD), 10% DMSO, and 10% RPMI 1640 Media (Life Technologies, Grand Island, NY) in liquid nitrogen for subsequent analyses.
HCV viral assessment
Quantitative measurements of viral levels were obtained using the Roche Amplicor assay version 2 at baseline (2 weeks before initiation of treatment), during treatment, at the end of treatment (24 weeks) and at the end of follow-up (48 weeks following the start of treatment). The primary endpoints were early viral kinetics (early viral response, EVR) and sustained virological response (SVR) defined as undetectable HCV RNA 24 weeks following end of treatment. Kinetics were defined as Marked (responders had a decline in HCV titers greater than 3.5 log10 or to undetectable between baseline and day 28 of therapy), Intermediate (responders had declines of 3.5–1.4 log10) and Poor (responders had declines of less than 1.4 log10).
Liver histology
Participants had a liver biopsy within 18 months of study enrollment. Liver biopsies were scored by a single pathologist, who was blinded to patient outcome and clinical status. Biopsies were assessed for the severity of hepatitis C by grading inflammation and staging of fibrosis using Metavir activity and fibrosis score.
Antibodies for analysis of cell surface antigen expression/FACS analysis
Four-color multiparameter flow cytometry was performed using a BD FACSCanto II or BD FACScan instrument (BD Biosciences, San Jose, CA) compensated with single fluorochromes and analyzed using Diva™ or CellQuest™ software (BD). Lymphocyte populations were identified by their characteristic forward scatter/side scatter properties. Fluorochrome-labeled (PerCP/APC) monoclonal antibodies (MAb) specific for CD3/CD56 (BD) were used to identify NKs (CD3-CD56+) and NTs (CD3+CD56+) within the overall lymphocyte population. Anti-NK receptor (NKR) antibodies (FITC/PE) CD161, CD94, CD16, CD158a, CD158b, CD158e and NKG2D were obtained from BD Biosciences. Anti-NKG2C-PE and TRAIL-PE MAbs were supplied by R&D systems (Minneapolis, MN). Anti-NKG2A-PE, NKp30-PE, NKp44-PE, and NKp46-PE were obtained from Immunotech (Beckman Coulter, Fullerton, CA). Thawed PBMCs (1-2 × 106) were stained for cell surface antigen expression at 4°C for 30 minutes. Then, washed twice in 2 ml phosphate-buffered saline (PBS) containing 1% bovine serum albumin and 0.01% sodium azide (Facs Wash) and fixed in 200ul of 1% paraformaldehyde (Sigma-Aldrich, St. Louis, MO). Anti-IFN-α/β R1 antibody was purchased from R&D. Isotype-matched control antibodies were used to determine background levels of staining.
IL-28B Genotyping
Among the 101 individuals included in this study, a subset of 57 consented to participate in host genetics studies and had DNA available for genotyping. Genomic DNA samples were genotyped for SNP rs12979860 at the University of Maryland and Duke University using the TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA) according to the standardized manufacturer's protocol [6] [7]. Replicate genotyping in 8% of the samples resulted in a >98% concordance rate.
Interferon-α response
NK cells were enriched from PBMCs (n=4) using the NK Isolation Kit (negative selection, Miltenyi Biotech, Auburn, CA) according to manufacturer's instructions. Enriched NK fractions were stained for expression of CD56/CD3/NKG2A as described above. FACS sorting (Aria, BD) was used to isolate NKs based on the expression of NKG2A and cultured for 4 hours at one million/ml in the presence or absence of PEG-IFN-α (100ng/ml IFN-α part; Roche). After culture NK cells were washed and RNA extracted from cell pellets using the PicoPure™ RNA Isolation Kit (Arcturus, Applied Biosystems, Carlsbad, CA). cDNA was transcribed using 250ng of RNA in a 20ul reaction using the Quantitect RT Kit (Qiagen, Valencia, CA).
Real-time PCR
Expression of IFN-response genes (ISGs) was assessed using the Step One Plus Real time PCR system using the Fast SYBR Green Master Mix Protocol (Applied Biosystems). QuantiTect primer assays for use with Sybr Green detection were purchased from Qiagen/Superarray (IP-10 #QT01003065 and TRAIL #QT00079212).
Statistical modeling to predict SVR
Continuous variables were summarized as mean +/- standard deviation, and categorical variables as frequency and percentage. Student t-test and chi-square test were applied, as appropriate. Univariate and multivariate logistic regression analyses were conducted to identify significant predictors of SVR. As candidate predictors of SVR, we investigated inhibitory and activating receptor expression levels, IL28B genotype (dichotomized as CC versus CT/TT), as well as clinical factors that have been previously associated with SVR including, gender, race, fibrosis score and pre-treatment HCV viral load. Variables that were significantly associated with SVR on univariate analyses (p<0.05) were included in the multivariate logistic regression models. We also tested for interactions between race and IL-28B genotype, HCV pre-treatment viral load, and the receptor expression levels. We found no evidence of significant racial interactions, with the exception of a borderline significant interaction between race and pre-treatment viral load (log viral load mean [+/- SD] of 6.05 [1.01] for Caucasians and 6.33 [0.66] for African Americans). With the ultimate intention of developing a model that does not require invasive testing (ie, liver biopsy), we excluded fibrosis score from the modeling scenarios. The most robust model, based upon Chi-square, is shown in Table 3 and includes the expression of two NK inhibitory receptors. The sample size allows for inclusion of two other variables in the regression model in addition to race and viral load. To evaluate the diagnostic accuracy of the model, we calculated the concordance (c)-statistic, which is the equivalent of the area under receiver operating characteristic curve. This statistic may range from 0 to 1, with 1 corresponding to perfect discrimination and 0.5 being what may be expected by chance alone. A diagnostic test with a c-statistic ≥ 0.7 is generally considered a clinically useful test.
Table 3. Multivariate logistic regression.
| SVR | Coefficient | Std. Error | P value |
|---|---|---|---|
| Race | -1.671114 | 0.6117374 | 0.006 |
| Log10 Viral Load | -1.149117 | 0.4266382 | 0.007 |
| NKG2A+ (% Lymphocytes) | -0.2338825 | 0.0895245 | 0.009 |
| CD158e+ (% Lymphocytes) | -0.3352191 | 0.1962866 | 0.088 |
| Constant | 10.62911 | 2.989614 | <0.001 |
For all analyses, a two-tailed p-value <0.05 was considered statistically significant. ROC curves were generated using JMP (Version 7. SAS Institute Inc., Cary, NC, 1989-2007) and logistic regression analyses were conducted using STAT 11.1 (StataCorp. 2009. Stata Statistical Software: Release 11. College Station, TX).
Results
Pretreatment PBMC were analyzed from a total of 101 HCV genotype 1-infected patients who subsequently underwent antiviral treatment with PEG/RIB. As shown in Table 1, the study group was comprised of 55 African-Americans (AAs) and 46 Caucasian-Americans (CAs); there were no differences in the two racial groups with regards to gender, viral level or histologic severity at baseline. In keeping with prior reports [5], a higher proportion of AAs demonstrated poor early viral kinetics and diminished likelihood of SVR.
Association of inhibitory and activating NK receptor expression with early viral kinetics
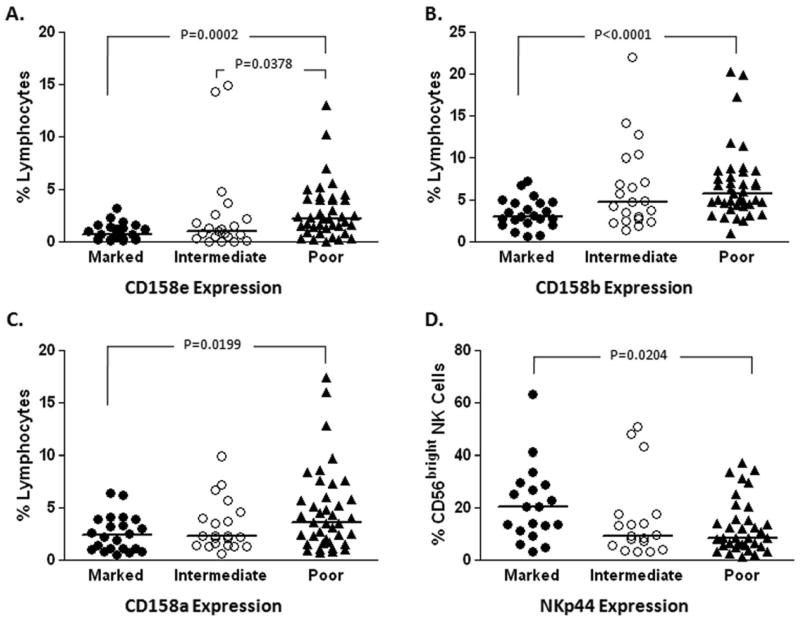
Impaired IFN responsivessness has been detected as early as 24 to 48 hrs after the first dose of PEG/RIB, implicating the innate rather than adaptive immune response in mediating defective early viral kinetic responses [8]. In this study, “marked” responders had a decline in HCV titers of >3.5 log10 or to undetectable levels between baseline and day 28 of therapy, “intermediate” responders had declines of 1.4 to 3.5 log, and “poor” responders had declines of <1.4 log, as described previously [9]. IFN-α, a type I interferon, has been shown to activate NK cells which in turn might mediate early viral kinetics. In keeping with this hypothesis, multiparameter FACS analysis identified statistically higher expression of inhibitory receptors CD158a (KIR2DL1), CD158b (KIR2DL2/DL3) and CD158e (KIR3DL1) on pre-treatment lymphocytes from patients who subsequently demonstrated poor viral decline after initiation of antiviral therapy and lower levels of the activating natural cytotoxicity receptor (NCR) NKp44 on the CD56bright (immature/regulatory) NK subset (Figure 1).
Figure 1.
NK receptor expression correlates with early viral kinetics.
Multi-parameter flow cytometric analysis was used to assess the expression of a range of NK receptors on lymphocyte subsets in patient PBMC samples collected two weeks before initiation of standard therapy for chronic HCV infection. Higher expression of inhibitory killer immunoglobulinlike receptors CD158e (A), CD158b (B) and CD158a (C) was observed on total lymphocytes from these patients who demonstrated poor early viral decline (declines less than 1.4 log10 by day 28 of therapy) following initiation of antiviral therapy. Activating natural cytotoxicity receptor NKp44 expression was decreased on the immature/regulatory (CD56bright) NK cell subset in subjects who demonstrate poor early viral kinetics (D).
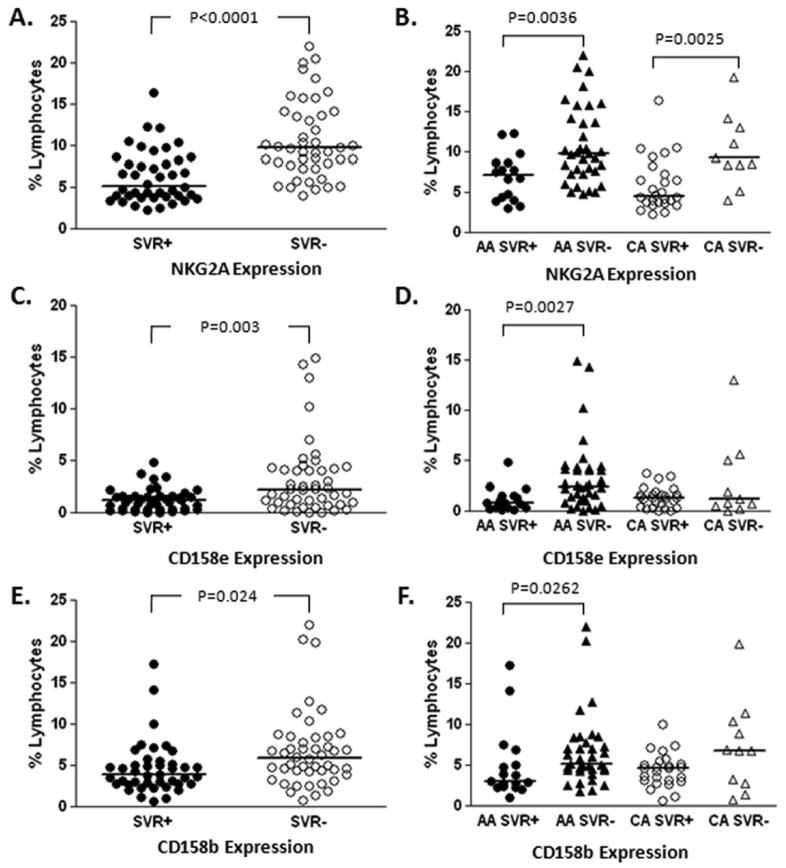
Differential expression of activating and inhibitory receptors associated with long-term virologic response to treatment
As shown in Table 2, higher expression levels of inhibitory receptors NKG2A, CD158e (KIR3DL1) and CD158b (KIR2DL2/DL3) on total lymphocyte and NK populations were demonstrable in patients who failed to eradicate HCV with combination therapy. Figure 2 demonstrates representative distribution of these receptors according to SVR and racial categories; importantly, these receptors did not vary according to race but were associated with subsequent treatment response. Flow-cytometric analysis also demonstrated an association with SVR and increased expression of NK activating receptors. Pre-treatment NKRp44 and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression levels were higher on NK cell populations in those who subsequently experienced SVR with combination therapy. CD161, a co-stimulatory molecule, was found at statistically higher levels on pre-treatment NK cells from patients who achieved SVR as compared to those with resistance to antiviral therapy. Supplemental Figure 1 shows that these associations of activating receptors with SVR were independent of race.
Table 2. Univariate analysis of NK cell receptors associated with SVR.
| Tx Outcome | |||||
|---|---|---|---|---|---|
| Receptor | SVR+ | SVR- | Odds Ratio | 95% C.I. | p value* |
| NKG2A | |||||
| % Lymphocytes | 5.14 (2.23-16.43)‡ | 9.73 (3.98-62.97) | 0.7363408 | 0.6317558 - 0.8582393 | <0.001 |
| % CD3+ T cells | 2.12 (0.60-13.88) | 3.85 (0.54-26.03) | 0.8165125 | 0.6903253 - 0.9657659 | 0.018 |
| % CD56+ cells | 39.63 (12.13-88.68) | 48.07 (18.24-83.40) | 0.9701283 | 0.9450686 - 0.9958524 | 0.023 |
| CD158e (KIR3DL1) | |||||
| % Lymphocytes | 1.20 (0-4.82) | 2.25 (0-14.87) | 0.6119121 | 0.4411554 - 0.8487632 | 0.003 |
| % CD56+ cells | 5.6 (0-25.09) | 13.47 (0-50.28) | 0.9197449 | 0.8705965 - 0.9716679 | 0.003 |
| % NK cells | 8.42 (0-24.59) | 14.17 (0-57) | 0.9406372 | 0.8960014 - .9874966 | 0.014 |
| % CD56dim NK cells | 9.0 (0-32.0) | 14.96 (0-50) | 0.9435168 | 0.9016136 - 0.9873674 | 0.012 |
| CD158b (KIR3DL2/DL3) | |||||
| % Lymphocytes | 3.84 (0.61-17.27) | 5.93 (0.81-61.01) | 0.8534621 | 0.7435093 - 0.9796751 | 0.024 |
| TRAIL† | |||||
| % NK cells | 4.19 (0.68-29.0) | 2.64 (0.96-18.30) | 1.146269 | 1.012023 - 1.298323 | 0.032 |
| NKp44 (NCR2) | |||||
| % NK cells | 4.15 (1.11-42.44) | 3.39 (0.87-32.32) | 1.086335 | 1.004672 - 1.174635 | 0.038 |
| % CD56bright NK cells | 15.7 (1.31-63.07) | 8.19 (2.04-50.76) | 1.079094 | 1.025104 - 1.135927 | 0.004 |
| CD161 (NKR-PA1) | |||||
| % NK cells | 51.24 (10.71-86.35) | 45.52 (0-79.64) | 1.026324 | 1.003427 - 1.049743 | 0.024 |
P > |z|
Median (Range)
Tumor Necrosis Factor–Related Apoptosis-Inducing Ligand
Figure 2.
Lymphocyte inhibitory NK receptor expression correlates with lack of response to treatment.
Multi-parameter flow cytometric analysis was used to assess the expression of a range of NK receptors on lymphocyte subsets in patient PBMC samples collected two weeks before initiation of therapy for chronic HCV infection. Patients who failed to achieve sustained virological response (SVR) demonstrated relatively increased levels of expression of NK inhibitory receptors NKG2A, CD158e and CD158b. Expression is shown with respect to SVR +/-, (A, C and E) and further broken down as a function of race (African American [AA] or Caucasian American [CA], B, D and F).
Expression of inhibitory receptor NKG2A correlates with IL-28B genotype
Although there is compelling evidence that the host immune response largely determines whether HCV is eradicated spontaneously or becomes persistent as seen in the majority of patients [10], the role of immunity in predicting outcome of antiviral therapy was open to question until recently. However, genome-wide association studies identified single nucleotide polymorphisms (SNPs) on chromosome 19 within or near the IL-28B gene encoding interferon-lambda 3 (IFN-λ3) as highly predictive of antiviral success [4] [11]. Patients homozygous for the C allele at the rs12979860 SNP have a more than two-fold greater chance of cure, compared to those with the TT genotype. Moreover, the advantageous allele is significantly more frequent in Caucasian and Asian populations relative to AAs and is estimated to explain approximately half of the difference in SVR rates between AAs and CAs (see Table 1).
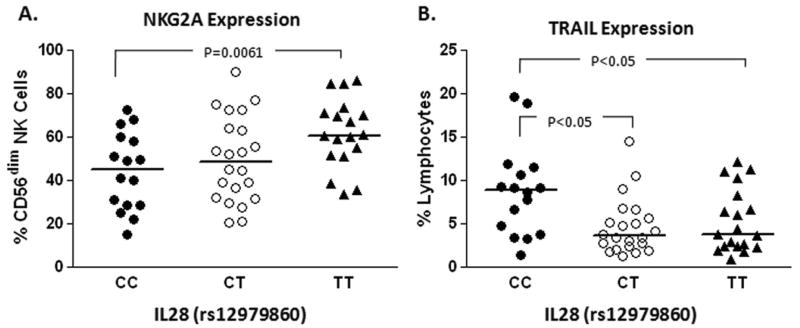
It has been hypothesized that IL-28B genotype somehow regulates the innate immune response to HCV and modifies the threshold for virologic control [12], but data supporting this concept have been lacking. We found that patients expressing two copies of the T allele show statistically higher levels of NKG2A on the CD56dim (effector) NK cell population (Figure 3A). CD56dim NK cells are more innately cytotoxic whereas CD56bright NK cells are IFN-γ-producing cells [13]. Moreover, the IL-28B T allele was associated with a reduced frequency of lymphocytes expressing TRAIL (Figure 3B). We did not see similar associations with the other receptors which were associated with SVR; however, the sample size may be limiting. In the model, there was no statistical interaction between NKG2A and race. Thus, the expression of inhibitory NK receptor(s) may represent phenotypic correlates of genetic variation in the IL-28B gene independent of race.
Figure 3.
NK receptor expression correlates with IL-28 genotype.
NK receptor expression where available was categorized according to IL-28 genotype for each individual patient who had consented for genetic analyses (n=52). The favorable CC genotype was associated with relatively decreased NKG2A inhibitory receptor expression compared to subjects having the unfavorable TT genotype (A) and conversely with relatively increased expression of the activating TRAIL receptor (B).
Multivariate model to predict SVR
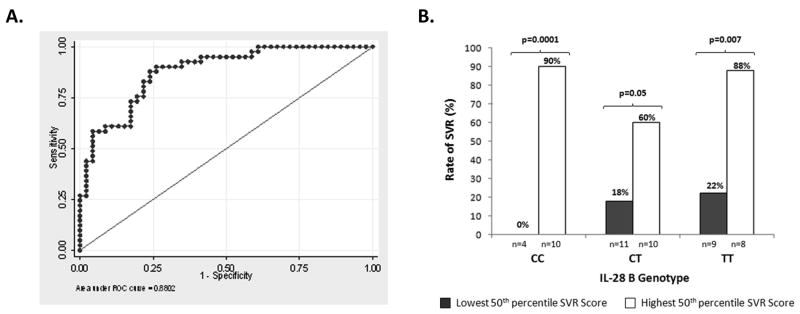
Next, we entered the variables associated with SVR at the univariate level into multivariate regression models, including factors previously reported as predictive such as race and pre-treatment viral level. The most robust model is shown in Table 3 and includes the expression of two NK inhibitory receptors. The likelihood of SVR was calculated by combining the four variables as follows: -1.67 (race) − 1.149 (log10VL) - 0.234 (% NKG2A expression on lymphocytes) − 0.335 (%CD158e on expression on lymphocytes) + 10.629 (constant) with race coded as 1 for AA and 0 for CA. In order to evaluate the diagnostic accuracy of the model, concordance (c-statistic), which is equivalent to the area under the (ROC) curve, was calculated. By convention, a c-statistic of 0.7 or greater is considered to be clinically useful; the c-statistic for our multivariate model was 0.88 (Figure 4A).
Figure 4.
Multivariate model to predict SVR.
The likelihood of SVR was calculated by combining four variables: race, log10 viral load and % of lymphocytes expressing NKG2A and CD158e as described in table 3. Receiver operating characteristics (ROC) of SVR model is shown (A). The ROC curve returns a c-statistic of 0.88. The SVR score model enhances the predictive value of IL-28 genotype. There were N=57 individuals with consent to use IL-28B data. Of these, n=52 had relevant data allowing calculation of SVR model score (B).
Within our population, for which the prevalence of SVR was 48%, the negative predictive value (NPV) of this model was estimated at 81% and positive predictive value at 77%. Other reports indicate that IL-28B has excellent positive predictive value, but relatively low NPV, and thus, should not be used to withhold antiviral therapy [12]. Because the current SVR model demonstrates strong NPV, we determined whether it performed complementarily with IL-28B genotype. As shown in Figure 4B, the SVR model score enhanced the predictive value of IL-28B genotype; for example, among CC patients with SVR model scores above the 50th percentile, 90% had a chance of virologic cure whereas none of the patients with scores in the bottom 50th percentile experienced SVR. Among patients with the TT genotype, patients with higher SVR model scores demonstrated four-fold higher likelihood of sustained cure. Thus, the new model provides additional discriminative information to the IL-28B genotype.
IFN-alpha responsiveness varies according to NK inhibitory receptor expression
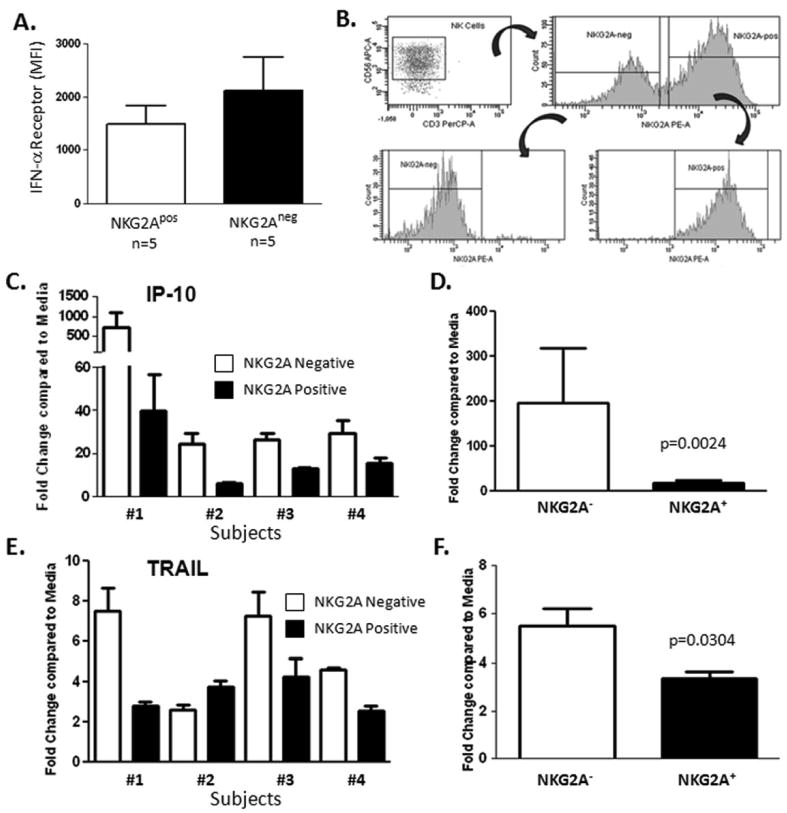
Because levels of NKG2A were associated with lack of antiviral response, we measured the expression of IFN-α receptor on NK cells and found an inverse relationship with NKG2A as a potential mechanism for impaired responsiveness to PEG IFN-α (Figure 5A). In experiments aimed at elucidating the cause of unresponsiveness to IFN-α, we sort-purified NK cells based on their expression NKG2A (the sorting strategy is shown in Figure 5B) and tested gene responses following in vitro stimulation with PEG-IFN-α. As shown in Figure 5C and 5D, exogenous IFN-α induced mRNA levels of the antiviral gene interferon-γ–inducible protein-10 (IP-10) to a much greater extent in the NKG2A-negative fraction. Recently, the expression of TRAIL was shown to inversely correlate with HCV RNA levels during the early phase of antiviral therapy and that IFN-α -stimulated NK cells kill an HCV replicating cell in a TRAIL-dependent manner [13]. In the current study, we found that TRAIL induction was statistically lower in NKG2A-positive cells (Figure 5E and 5F). Our data are consistent with another study which demonstrated that chronic HCV patients had significant impairment of IFN-α -stimulated NK cell degranulation as compared to normal controls [14]. On aggregate, these data show that surface expression of the inhibitory receptor NKG2A may demarcate intracellular differences in IFN signaling and correlate with cell surface expression of TRAIL.
Figure 5.
The NKG2A-negative NK cell subset shows enhanced response to PEG-IFN-α in vitro.
IFN-α receptor expression is increased in NKG2Aneg NK cells relative to their NKG2Apos counterparts (A). Staining for flow cytometric analysis was performed on 5 chronic HCV patients. NK cells were sort-purified based on their expression NKG2A following the gating strategy shown (B). Gene responses were assessed using real-time RT-PCR following 4-hour in vitro stimulation with PEG-IFN-α (100ng/ml). Exogenous IFN-α induced mRNA levels of the antiviral gene interferon-γ–inducible protein-10 (IP-10) to a much greater extent in the NKG2A-negative fraction (C and D). TRAIL induction was statistically lower in NKG2A-positive cells (E and F).
Discussion
NK cells and their dysfunction have been implicated in all stages of HCV infection [15]. Recently, aberrant expansion of functionally impaired CD56-negative NK cells has been demonstrated to be a harbinger of anti-HCV treatment failure [16]. NK cells are known to become rapidly activated by cytokine stimulation. In order to test the premise that the nature and function of NK cells are associated with the antiviral effects of IFN-α, we used the well-characterized, longitudinal Virahep-C cohort [5]. We found that functionally impaired CD56-negative NK cells circulated at higher levels in chronically infected HCV patients (compared to normals), correlated with poor early viral kinetics following initiation of antiviral therapy, and were highest in those resistant to treatment (Supplemental Figure 2).
The dominant “default” feature of NK class I receptor-mediated function is inhibition [17]. Although it is accepted that the balance of activating and inhibitory signals control NK cell activity, the relative importance of these signals in mediating antiviral efficacy to IFN in vivo had not been defined. We compared the expression of these markers as a predictor of early virologic response, i.e., within 28 days following initiation of combination therapy, and found pre-treatment differences in multiple inhibitory and activating receptors. Moreover, by univariate analysis, we found pre-treatment levels of stimulatory receptors NKRp44, TRAIL, CD161 were positively associated with SVR, whereas the expression levels of inhibitory receptors NKG2A, CD158e (KIR3DL1) and CD158b (KIR2DL2/DL3) were negatively correlated with SVR. By multivariate modeling, the expression of two inhibitory receptors (NKG2A and CD158e) was predictive of treatment failure. The robustness of the model ROC indicates that it provides useful discriminative clinical information.
A recent study found an association between the IL-28B SNP and an inhibitory KIR gene, KIR2DL3 [18]. In the current report, we found that expression levels of another inhibitory receptor NKG2A were higher in patients homozygous for the poor response T allele of the IL-28B (IFN-λ 3) SNP. Little is known about the interaction between type I and type III IFNs; they utilize unique signaling receptors but share common downstream signaling [19]. We examined potential mechanisms whereby NKG2A expression on NK cells might contribute to the differential response of IFN-α -based therapy. We provide experimental evidence that the surface expression of NKG2A demarcate significant intracellular gene expression differences. Using the Virahep-C cohort, Taylor and colleagues had previously demonstrated blunted transcription of ISGs in whole PBMC of poor-response patients [20]. The importance of examining the cell of origin with regards to ISG expression was underscored recently by the finding that ISG up-regulation was more pronounced in hepatocytes in nonresponders to IFN therapy, but in Kupffer cells in responders [21]. Reduced expression of IFN-α receptor on NKG2A-positive NK cells compared to their NKG2A negative counterparts suggests one mechanism contributing to the differential response of IFN-α -based therapy.
Higher serum and hepatic levels of interferon-γ–inducible protein-10 (IP-10), a CXC chemokine (CXCL10), have been associated with nonresponse to HCV therapy [22] [23]. However, the fold induction in IP-10 has been associated with early viral kinetics in patients who are IFN-responders [24], as well as SVR [25]. Our data indicate that NK cells are a source of IP-10 following stimulation with IFN-α. Indeed, NK cells depleted of NKG2A-positive NK cells produced significantly greater IP-10. Prior studies have demonstrated that antiviral effector cells, including CTLs and NK, express CXCR3 and are responsive to IP-10 in chemotaxis [26]; further work is warranted to examine the role of NKG2A expression on chemotaxis within the hepatic compartment.
We found statistically higher pretreatment levels TRAIL on total NK cells in patients who responded to treatment as compared to nonresponders. Recent reports demonstrated that 4 hrs after IFN-α treatment, TRAIL (TNFSF10) mRNA is markedly upregulated in liver biopsies from HCV patients, and on circulating NK cells correlated inversely with serum HCV-RNA levels [13]. Moreover, in the latter study, HCV recovered patients had superior up-regulation of TRAIL after in vitro IFN-α stimulation than non-responder patients. In keeping with these results, we found that PEG IFN-α upregulated expression of TRAIL on NK cells, but that NKG2A expression attenuated this effect.
Taken together, our data indicate that NKG2A-positive cells are increased in frequency in patients with chronic HCV infection who are more likely to fail treatment and demonstrate impaired response to exogenous PEG IFN-α. As a single variable, when NKG2A expression on lymphocytes was greater than 8.0%, the sensitivity and specificity for futility (lack of SVR) was 71.7% and 75.6%, respectively; the ROC curve (c-statistic of 0.80) is shown in supplemental Figure 3. Because PEG-IFN-α and ribavirin is likely to remain the backbone of antiviral therapy for the foreseeable future, our results may affect how clinicians use direct antiviral agents such as protease or polymerase inhibitors. For example, for patients with NK receptor expression associated with poor early viral kinetics, response to IFN-α therapy is slower and thus are, in theory, more likely to develop resistance to direct antiviral agents [12]. Furthermore, IL-28B genotype has now been incorporated into treatment algorithms to predict patients' responses to anti-HCV therapy; however, because of low negative predictive value for SVR, IL-28B genotype cannot be used to determine whether to withhold treatment in an individual patient [12]. Our results indicate that a simple model incorporating inhibitory NK receptor expression levels can accurately discern likelihood of SVR across different IL-28B genotypes. These results have implications for improving prediction of treatment response and development of new approaches that target NK inhibitory pathways.
Supplementary Material
Supplemental Figure 1: Activating NK receptor expression correlates with response to treatment.
Multi-parameter flow cytometric analysis was used to assess the expression of a range of NK receptors on lymphocyte subsets in patient PBMC samples collected two weeks before initiation of therapy for chronic HCV infection. Patients who failed to achieve sustained virological response (SVR) demonstrated relatively decreased levels of expression of NK activating receptors TRAIL and NKp44 as well as lower levels of the co-stimulatory molecule CD161. Data is shown is shown with respect to sustained virological response (SVR +/-, A, C and E) and further broken down as a function of race (African American [AA] or Caucasian American [CA], B, D and F).
Supplemental Figure 2: CD16posCD56neg NK Cells.
Circulating CD56-negative functionally impaired NK cells are increased in chronic HCV-infected patients compared to uninfected controls with the highest levels seen in those who fail to respond to standard PEG/RIB therapy (A). Higher levels of these dysfunctional NK cells also correlate with poor early viral kinetics (viral decline < 1.4 log10 in the first 28 days of treatment) (B).
Supplemental Figure 3. Receiver operating characteristics (ROC) of NKG2A.
Receiver operating characteristics (ROC) of NKG2A expression on lymphocytes (cutoff 8.0%) as predictor of nonresponse; the PPV is 76.8% and NPV is 70.3%, respectively.
Acknowledgments
We thank the members of Virahep-C group for contributing to the study and the patients for their willingness to participate.
Grant Information: This work is supported by RO1 DK071560 (to HRR). This study was funded as a cooperative agreement by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the Intramural Research Program of the National Cancer Institute (NCI) with further support under a Cooperative Research and Development Agreement (CRADA) with Roche Laboratories, Inc. A complete listing of participants in the Virahep-C study is given in reference 5. Grant numbers: U01 DK60329, U01 DK60340, U01 DK60324, U01 DK60344, U01 DK60327, U01 DK60335, U01 DK60352, U01 DK60342, U01 DK60345, U01 DK60309, U01 DK60346, U01 DK60349, U01 DK60341. Dr. Clark received funding support from the National Health and Medical Research Council of Australia, the Duke Clinical Research Institute, the Richard Boebel Family Fund, the National Centre in HIV Epidemiology and Clinical Research (now The Kirby Institute for Infection and Immunity in Society), University of New South Wales, Australia.
Abbreviations
- AA
African American
- CA
Caucasian American
- EVR
Early Virologic Response
- HCV
Hepatitis C Virus
- ISG
Interferon-Stimulated Gene
- KIR
Killer Immunoglobulin-like Receptor
- MAb
Monoclonal Antibody
- NCR
Natural Cytotoxicity Receptor
- NK
Natural Killer cell
- NKRs
Natural Killer cell Receptors
- NT
Natural T
- PEG/RIB
Combination of Pegylated IFN-α and the nucleoside analog Ribavirin
- SVR
Sustained Virologic Response
- TRAIL
Tumor necrosis factor-related apoptosis-inducing ligand
Footnotes
Author contributions: LGM, planned, performed experiments; KMB, analyzed data and created mathematical model; LC, performed experiments; MWT, analyzed data, advised on genes to focus on; NA, CH, PJC, contributed IL-28B data and provided feedback on manuscript; LGM and HRR, planned experiments and wrote the paper.
The authors declare no conflict of interest.
References
- 1.Davis GL, Albright JE, Cook SF, Rosenberg DM. Projecting future complications of chronic hepatitis C in the United States. Liver Transpl. 2003;9:331–338. doi: 10.1053/jlts.2003.50073. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH, Seeff LB. Peginterferon and ribavirin for chronic hepatitis C. N Engl J Med. 2006;355:2444–2451. doi: 10.1056/NEJMct061675. [DOI] [PubMed] [Google Scholar]
- 3.Kohler K, Xiong S, Brzostek J, Mehrabi M, Eissmann P, Harrison A, Cordoba SP, et al. Matched sizes of activating and inhibitory receptor/ligand pairs are required for optimal signal integration by human natural killer cells. PLoS One. 2010;5:e15374. doi: 10.1371/journal.pone.0015374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 5.Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, Afdhal N, Brown RS, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129 e118. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 7.McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, Tillmann HL, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai AW, Chung RT. Racial differences in response to interferon-based antiviral therapy for hepatitis C virus infection: a hardwiring issue? J Infect Dis. 2009;199:1101–1103. doi: 10.1086/597385. [DOI] [PubMed] [Google Scholar]
- 9.Donlin MJ, Cannon NA, Yao E, Li J, Wahed A, Taylor MW, Belle SH, et al. Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy. J Virol. 2007;81:8211–8224. doi: 10.1128/JVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med. 2005;201:1709–1714. doi: 10.1084/jem.20050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 12.Balagopal A, Thomas DL, Thio CL. IL28B and the Control of Hepatitis C Virus Infection. Gastroenterology. 2010;139:1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegmann KA, Bjorkstrom NK, Veber H, Ciesek S, Riese P, Wiegand J, Hadem J, et al. Interferon-alpha-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology. 2010;138:1885–1897. doi: 10.1053/j.gastro.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 14.Miyagi T, Takehara T, Nishio K, Shimizu S, Kohga K, Li W, Tatsumi T, et al. Altered interferon-alpha-signaling in natural killer cells from patients with chronic hepatitis C virus infection. J Hepatol. 2010;53:424–430. doi: 10.1016/j.jhep.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Cheent K, Khakoo SI. Natural killer cells and hepatitis C: action and reaction. Gut. 2011;60:268–278. doi: 10.1136/gut.2010.212555. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez VD, Falconer K, Bjorkstrom NK, Blom KG, Weiland O, Ljunggren HG, Alaeus A, et al. Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: correlation with outcome of pegylated IFN-alpha and ribavirin treatment. J Immunol. 2009;183:6612–6618. doi: 10.4049/jimmunol.0901437. [DOI] [PubMed] [Google Scholar]
- 17.Golden-Mason L, Rosen HR. Natural killer cells: primary target for hepatitis C virus immune evasion strategies? Liver Transpl. 2006;12:363–372. doi: 10.1002/lt.20708. [DOI] [PubMed] [Google Scholar]
- 18.Dring MM, Morrison MH, McSharry BP, Guinan KJ, Hagan R, Irish HCV Research Consortium. O'Farrelly C, Gardiner CM. Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc Natl Acad Sci U S A. 2011;108:5736–5741. doi: 10.1073/pnas.1016358108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor MW, Tsukahara T, Brodsky L, Schaley J, Sanda C, Stephens MJ, McClintick JN, et al. Changes in gene expression during pegylated interferon and ribavirin therapy of chronic hepatitis C virus distinguish responders from nonresponders to antiviral therapy. J Virol. 2006;81:3391–3401. doi: 10.1128/JVI.02640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, Anand N, et al. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010;138:1123–1133. doi: 10.1053/j.gastro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 22.Darling JM, Aerssens J, Fanning G, McHutchison JG, Goldstein DB, Thompson AJ, Shianna KV, et al. Quantitation of pretreatment serum interferon-gamma-inducible protein-10 improves the predictive value of an IL28B gene polymorphism for hepatitis C treatment response. Hepatology. 2011;53:14–22. doi: 10.1002/hep.24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Askarieh G, Alsio A, Pugnale P, Negro F, Ferrari C, Neumann AU, Pawlotsky JM, et al. Systemic and intrahepatic interferon-gamma-inducible protein 10 kDa predicts the first-phase decline in hepatitis C virus RNA and overall viral response to therapy in chronic hepatitis C. Hepatology. 2010;51:1523–1530. doi: 10.1002/hep.23509. [DOI] [PubMed] [Google Scholar]
- 24.Feld JJ, Lutchman GA, Heller T, Hara K, Pfeiffer JK, Leff RD, Meek C, et al. Ribavirin improves early responses to peginterferon through improved interferon signaling. Gastroenterology. 2010;139:154–162. doi: 10.1053/j.gastro.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeremski M, Markatou M, Brown QB, Dorante G, Cunningham-Rundles S, Talal AH. Interferon gamma-inducible protein 10: a predictive marker of successful treatment response in hepatitis C virus/HIV-coinfected patients. J Acquir Immune Defic Syndr. 45:262–268. doi: 10.1097/QAI.0b013e3180559219. [DOI] [PubMed] [Google Scholar]
- 26.Taub DD, Sayers TJ, Carter CR, Ortaldo JR. Alpha and beta chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol. 1995;155:3877–3888. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Activating NK receptor expression correlates with response to treatment.
Multi-parameter flow cytometric analysis was used to assess the expression of a range of NK receptors on lymphocyte subsets in patient PBMC samples collected two weeks before initiation of therapy for chronic HCV infection. Patients who failed to achieve sustained virological response (SVR) demonstrated relatively decreased levels of expression of NK activating receptors TRAIL and NKp44 as well as lower levels of the co-stimulatory molecule CD161. Data is shown is shown with respect to sustained virological response (SVR +/-, A, C and E) and further broken down as a function of race (African American [AA] or Caucasian American [CA], B, D and F).
Supplemental Figure 2: CD16posCD56neg NK Cells.
Circulating CD56-negative functionally impaired NK cells are increased in chronic HCV-infected patients compared to uninfected controls with the highest levels seen in those who fail to respond to standard PEG/RIB therapy (A). Higher levels of these dysfunctional NK cells also correlate with poor early viral kinetics (viral decline < 1.4 log10 in the first 28 days of treatment) (B).
Supplemental Figure 3. Receiver operating characteristics (ROC) of NKG2A.
Receiver operating characteristics (ROC) of NKG2A expression on lymphocytes (cutoff 8.0%) as predictor of nonresponse; the PPV is 76.8% and NPV is 70.3%, respectively.