Abstract
HIV-1 CRF02_AG accounts for >50% of infected individuals in Cameroon. CRF02_AG prevalence has been increasing both in Africa and Europe, particularly in Italy because of migrations from the sub-Saharan region. This study investigated the molecular epidemiology of CRF02_AG in Cameroon by employing Bayesian phylodynamics and analyzed the relationship between HIV-1 CRF02_AG isolates circulating in Italy and those prevalent in Africa to understand the link between the two epidemics. Among 291 Cameroonian reverse transcriptase sequences analyzed, about 70% clustered within three distinct clades, two of which shared a most recent common ancestor, all related to sequences from Western Africa. The major Cameroonian clades emerged during the mid-1970s and slowly spread during the next 30 years. Little or no geographic structure was detected within these clades. One of the major driving forces of the epidemic was likely the high accessibility between locations in Southern Cameroon contributing to the mobility of the population. The remaining Cameroonian sequences and the new strains isolated from Italian patients were interspersed mainly within West and Central African sequences in the tree, indicating a continuous exchange of CRF02_AG viral strains between Cameroon and other African countries, as well as multiple independent introductions in the Italian population. The evaluation of the spread of CRF02_AG may provide significant insight about the future dynamics of the Italian and European epidemic.
Introduction
The human immunodeficiency virus type 1 (HIV-1) is characterized by an extensive and ever increasing genetic variability. Four major Groups (M, N, O, and P), at least nine different subtypes (A to K) within the major M Group, and 48 circulating recombinant forms (CRFs) have been described so far.1 Recent estimates indicate that 33.4 to 35.8 million people worldwide are infected by HIV-1,2 with sub-Saharan Africa as the most heavily affected region accounting for 67% of all infections. The sub-Saharan African epidemic, however, varies significantly from country to country in both scale and scope. Adult national HIV-1 prevalence is below 2% in several countries of West and Central Africa, as well as in the horn of Africa, but it exceeds 5% in most Central and East African countries including Cameroon, the Central African Republic, Gabon, Malawi, Mozambique, Uganda, and the United Republic of Tanzania.2
Cameroon hosts one of the broadest genetic arrays of HIV viruses suggesting that the country may be one of the epicenters of the African epidemic. In addition to each of the Group M subtypes and several CRFs, HIV-1 Groups N, O, and P as well as HIV-2 strains have been identified in the country.3–17 The first AIDS case in Cameroon was diagnosed in 1985.18 Since then, about 540,000 cases have been officially reported. Cameroon is currently facing a generalized epidemic, with adult (aged 15 to 49 years) prevalence in the range of 3.9–6.2%.2 Recent studies show a consistent increase in prevalence of several CRFs including CRF02_AG, CRF01_AE, CRF06_cpx, CRF09_cpx, CRF11_cpx, CRF13-cpx, CRF18-cpx, CRF22-cpx, CRF25_cpx, and CRF37_cpx.9,11,12,15 In 2002, the phylogenetic characterization of isolates obtained from subjects living in the cities of Yaoundé, the capital, and Douala showed that 60% of samples were CRF02_AG.13 Recent data from Yaoundé also indicate that the CRF02_AG strain represents up to 50% of the total infection.15 CRF02_AG prevalence has been increasing not only in West and West-Central Africa,8,13–15,19–23 but also in different countries of Europe,24–27 such as Italy,28–32 because of migrations from the sub-Saharan region. This viral variant is one of the most prevalent recombinant forms of HIV-1 in the world, responsible for at least 5% of infections.33
In the present study we sought to investigate the origin and demographic history of CRF02_AG in Cameroon by employing phylogenetic and population genetic (phylodynamic) analysis in conjunction with viral gene-flow estimates from genetic data (phylogeography). Moreover, as Italy's position in the Mediterranean Sea makes it a strategic migration route in all Europe, we also analyzed the relationship between African and Italian HIV-1 CRF02_AG lineages circulating in Italy and those prevalent in different African geographic regions in order to understand the link between both epidemics.
Materials and Methods
Datasets
The analysis was performed on reverse transcriptase (RT, amino acid positions analyzed: 36–213) from HIV-1 CRF02_AG pol sequences. All available African sequences were downloaded from the Los Alamos HIV databases [http://www.hiv.lanl.gov/] or generated for clinical routine testing in Italy at the Monitoring Unit of Antiretroviral Therapies, INMI, Lazzaro Spallanzani, Rome, or in the Division of Infectious Disease center, Bergamo. More than 2000 sequences were retrieved from the HIV databases. However, since the main focus of the present work was to analyze CRF02_AG molecular epidemiology, only viral sequences satisfying specific inclusion criteria were included in the final data set:34 (1) sequences had already been published in peer-review journals (except for the new sequences described below), (2) there was no uncertainty about the subtype assignment of each sequence, (3) sequences were not epidemiologically linked by direct donor–recipient transmission, (4) only one sequence per individual could be randomly selected, and (5) city/state of origin and sampling date were known and clearly established in the original publication. The final data set included 824 sequences, including 53 new sequences and 771 reference sequences (Table 1). Among the new sequences, 11 were from patients followed in Cameroon and 42 were from patients living in a small geographic area of Northern Italy (36 of African origin and 6 of Italian origin). The full list of reference sequences analyzed with accession numbers is given in Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/aid).
Table 1.
CRF02_AG Reverse Transcriptase Sequences Analyzed in the Study
| Country | New sequences (N)a | Sequences from Los Alamos HIV sequence database (N)b | Overall |
|---|---|---|---|
| Algeria | — | 15 | 15 |
| Burkina Faso | 1 | 4 | 5 |
| Cameroon | 11 | 280 | 291 |
| Central African Republic | — | 3 | 3 |
| Cote D'Ivoire | 7 | 82 | 89 |
| Democratic Republic of Congo | — | 6 | 6 |
| Dijbouti | — | 5 | 5 |
| Gabon | — | 21 | 21 |
| Ghana | 12 | 31 | 43 |
| Italy | 6 | — | 6 |
| Kenya | — | 17 | 17 |
| Liberia | — | 1 | 1 |
| Libyan Arab Jamahiriya | — | 34 | 34 |
| Madagascar | — | 5 | 5 |
| Mali | — | 205 | 205 |
| Marocco | 1 | — | 1 |
| Nigeria | 7 | 5 | 12 |
| Repubblic of Angola | — | 6 | 6 |
| Senegal | 7 | 49 | 56 |
| Seychelles | — | 2 | 2 |
| Sierra Leone | 1 | — | 1 |
| Overall | 53 | 771 | 824 |
Among the new sequences, all the 11 sequences from Cameroon are from patients living in this country, while the other sequences are from 42 patients living in the North of Italy (36 with African origin and 6 with Italian origin).
The full list of reference sequences analyzed with accession numbers is given in Supplementary Table S1.
RNA extraction, amplification, sequencing, and genetic subtyping
The new 53 sequences analyzed were generated by HIV genotype analysis on 1 ml plasma samples by means of a commercially available kit (ViroSeq HIV-1 genotyping system; Abbott Laboratories). Briefly, RNA was extracted using a commercially available kit (QIAmp Viral RNA mini-kit, Qiagen Inc., USA), retrotranscribed by murine leukemia virus RT, and amplified with Amplitaq-Gold polymerase enzyme by using two different sequence-specific primers for 40 cycles. RT-PCR was regularly launched with a positive and a negative PCR control. Pol-amplified products (containing the entire protease and the first 335 amino acids of the RT open reading frame, 1302 nucleotides) were full-length sequenced in sense and antisense orientations by an automated sequencer (ABI 3130) by using seven different overlapping sequence-specific primers.35 The sequences were analyzed using SeqScape-v.2.5 software. The quality endpoint for each individual was ensured by a coverage of the protease and RT sequence by at least two sequence segments. Sequences having a mixture of wild-type and mutant residues at single positions were considered to have the mutant(s) at that position. HIV-1 subtypes were determined by phylogenetic analysis of pol region sequences, as previously described.36
Phylogenetic analysis
Multiple sequence alignments were obtained with the Clustal algorithm37 and manually edited for optimization. Maximum likelihood (ML) phylogenetic trees were inferred with the PhyML program [http://www.atgc-montpellier.fr/phyml/],38 using the GTR+G+I nucleotide substitution model, which was selected with the hierarchical likelihood ratio test described by Swofford and Sullivan.39 Neighbor-joining (NJ) trees were also obtained using the same nucleotide substitution model with the program PAUP* version 4.0 written by David L. Swofford. The reliability of specific clades in the inferred trees was evaluated by using the SH-like approximate likelihood ratio test (aLRT), which compares the likelihoods of the best and the second best alternative arrangements around the branch of interest. According to type I error rate (test significant|branch is not corrected) analysis, the aLRT of an interior branch is almost exact for a cut-off value ≥0.9 and is considered well supported for a cut-off value >0.75.40
Bayesian genealogies were also inferred with the BEAST v.1.5.3 software package [http://evolve.zoo.ox.ac.uk/beast/]41 using the HKY substitution model, a relaxed molecular clock (see next section), and a constant population size coalescent prior. A Markov Chain Monte Carlo (MCMC) was run for 100,000,000 generations with sampling every 10,000th generation. The results were visualized with Tracer v1.4.1 [http://beast.bio.ed.ac.uk/Tracer]. The effective sample size (ESS) value for each parameter was >500 indicating sufficient mixing of the Markov chain. The maximum clade credibility (MCC) tree was then selected from the posterior tree distribution using TreeAnnotator v.1.4.8 available within the BEAST software package. Final trees were visualized and annotated with FigTree v.1.2.2 [http://tree.bio.ed.ac.uk/software/figtree/].
Molecular clock analysis
To obtain a Bayesian estimate of the origin of the major CRF02_AG subepidemics in Cameroon, sequences belonging to each highly supported clade were constrained to be monophyletic. The evolutionary rate (nucleotide substitutions per site per year) and the time of the most recent common ancestor (TMRCA, years) were inferred using sequences sampled at different time points by the MCMC approach implemented in BEAST. The analyses were performed with the same nucleotide substitution model and coalescent prior described in the previous section assuming a strict or a relaxed molecular clock.42 Separate analyses were performed using either the root height of the tree or uniform root height, setting up the lower and upper values to 1908 and 1933, respectively, as assumed to be the 95% confidence interval for the HIV-1 group M origin.43 An MCMC was run for 100,000,000 generations with sampling every 10,000 generation. The results were visualized with Tracer. The ESS value for each parameter was >500, indicating sufficient mixing of the Markov chain.
Bayesian estimate of demographic histories
For each well-supported Cameroon clade in the CRF02_AG genealogy, demographic curves of effective viral population size change over time were estimated according to both parametric (constant and exponential) and nonparametric (Bayesian Skyline Plot, BSP) models. For the BSP calculation, a Bayesian skyline coalescent tree prior was used under a constant skyline model with 10 groups. Parametric and nonparametric curves, and the parameters of each model (including upper and lower 95% high posterior density, HPD intervals), were estimated by a MCMC run for 100,000,000 generations with sampling every 10,000th generation. The results were visualized with Tracer v.1.3. Convergence of the Markov chain was assessed by calculating the ESS for each parameter. All ESS values were >500, indicating sufficient sampling.
Bayesian model selection
Different molecular clock and demographic models were compared by calculating the Bayes Factor (BF), which is the ratio of the marginal likelihoods (marginal with respect to the prior) of the two models being compared.44 We calculated approximate marginal likelihoods for each coalescent model via importance sampling (1000 bootstraps) using the harmonic mean of the sampled likelihoods (with the posterior as the importance distribution). The difference (in loge space) of marginal likelihood between two models is the loge of the Bayes Factor, loge(BF). Evidence against the null model (i.e., the one with lower marginal likelihood) is indicated by 2>[2·loge(BF)] >6 (strong) and >10 (very strong). BF calculations were performed with Tracer v1.4.1.
Phylogeographic analysis
For the phylogeography analysis, sequences from each one of the Cameroon clades were analyzed separately. The hypothesis of metapopulation structure, i.e., the existence within each clade of different subpopulations linked to different Cameroon geographic regions, was tested with a modified version of the Slatkin and Maddison test45,46 using the MCC trees. A one-character matrix was obtained from the original dataset by assigning to each taxon in the tree a one-letter code indicating its geographic region of origin. The putative origin of each ancestral sequence in the tree was then inferred by finding the most parsimonious reconstruction (MPR) of the ancestral character using either the ACCTRAN or DELTRAN option. The final tree length, i.e., the number of observed migrations in the genealogy, was computed and compared to the tree length distribution of 10,000 trees obtained by random joining-splitting. Observed genealogies significantly shorter than random trees (p<0.01) indicate the presence of subdivided populations with restricted gene flow. Calculations were carried out with MacClade v.4.06.47 The viral gene-flow (migrations) among different regions was traced using the State changes and stasis tool (MacClade software), which counts the number of changes in a tree for each pairwise character state. Viral gene-flow counts were traced for each of the four datasets and then averaged.
Geographic information system (GIS) data acquisition
Accessibility maps were drawn with ArcGIS software with data obtained from the Africover Initiative (FAO-UN). An accessibility map shows the travel time to the nearest city of population >100,000 people, using road/track-based travel. This accessibility is computed using a cost-distance algorithm, which computes the "cost" of travelling between two locations on a regular raster grid48 based principally on road network data extracted from the Vector Map Level 0 (VMap0) released by the National Imagery and Mapping Agency (NIMA). The cost landscape was derived from road and railway network data, navigable rivers and major water bodies, shipping lanes, national borders, land cover, urban areas, elevation, and slope. The full methodology is described here at http://gem.jrc.ec.europa.eu/gam/sources.htm. Demographic data on the number of African immigrants living in Italy between 2002 and 2008 were obtained from the Italian National Institute of Statistics (http://demo.istat.it/).
Results
Phylogeny of HIV-1 CRF02_AG in Cameroon and Italy
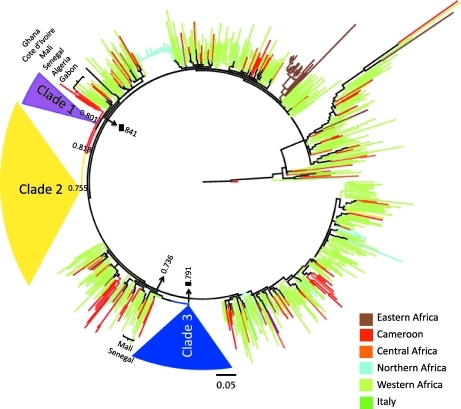
Among the 291 sequences from Cameroon analyzed, about 30% appeared to be intermixed with other African sequences in the ML tree (Fig. 1), indicating a continuous exchange of CRF02_AG viral strains between Cameroon and other African countries. The remaining Cameroon sequences clustered within three well-supported (aLRT, p>0.75) monophyletic clades, henceforth referred to as clades 1, 2, and 3. The presence of three well-supported major Cameroonian clades was confirmed in the NJ (data not shown). Clades 1 and 2 shared a common ancestor and appeared to be related to sequences from West Africa. Clade 3 belonged to a distinct lineage related to strains from Gabon, Ivory Coast, Mali, and Senegal (the tree with fully labeled tips is given in Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/aid). HIV-1 strains in clade 1 were isolated mostly from Eastern Cameroonian cities between 1996 and 2007, but no city appeared to be significantly more represented within a specific clade (Supplementary Fig. S2; Supplementary Data are available online at www.liebertonline.com/aid).
FIG. 1.
Maximum likelihood (ML) phylogenetic analysis of HIV-1 CRF02_AG. The ML tree includes 824 strains of African and Italian origin. The color of a tip branch represents the geographic region from where the strain originated, according to the legend given in the figure. Branch lengths were scaled in nucleotide substitutions per site as indicated by the bar at the bottom. Approximate likelihood-ratio test (aLRT) SH-like p-values for supported clades (p>0.75) are also indicated. For display purposes, the three supported Cameroon monophyletic clades, 1, 2, and 3, have been collapsed and colored in purple, yellow, and blue, respectively. A tree with fully labeled tips is given in Supplementary Fig. S1.
On the other hand, clades 2 and 3 included strains isolated mostly from Yaoundé, as well as other cities in central Cameroon, between 1996 and 2007. Overall, the result suggested the presence of at least three separate subepidemics, two of which (clades 1 and 2) possibly originated from a common introduction from Western Africa and the other (clade 3) from Northwestern Africa. Five of the 11 new sequences from patients infected and residing in Cameroon appeared to be intermixed with other African strains (Supplementary Fig. S1). The remaining ones were distributed within the three monophyletic clades: sequence CM39x07 clustered within clade 1, CM98FT07 and CM88FK07 clustered within clade 2, while sequences CM95F06, CM85B06, and CM91FT07 clustered within clade 3. The new sequences from African patients residing in Italy were all intermixed in the tree and did not cluster within any well-supported clade. Most of the new sequences from Italian patients appeared to be only distantly related to each other. Two highly supported clades, comprising two Italian strains each, clustered with one strain from Mali (aLRT, p=0.67) and strains from Cameroon and Mali (aLRT, p=0.81), respectively. The remaining two sequences were significantly related to strains from Ivory Coast (aLRT, p=1.0) and Mali (aLRT, p=0.78). Overall, the results strongly suggest at least four independent events leading to infection of Italian subjects with African CRF02_AG.
Phylodynamics of CRF02_AG Cameroon clades
The evolutionary rate and the TMRCA of each of the three Cameroon monophyletic clades were estimated by molecular clock analysis. Separate Bayesian genealogies were obtained for strains belonging to each clade and the molecular clock was calibrated by employing the known sampling time of each strain. As expected, the relaxed molecular clock fitted the data significantly better than the strict molecular clock for each clade (Supplementary Table S2; Supplementary Data are available online at www.liebertonline.com/aid).
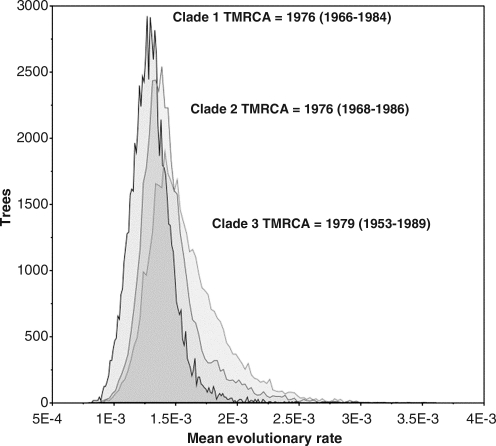
The median estimate of the evolutionary rate resulted in 1.3×10–3 (95% HPD=0.7×10–3−2.3×10–3), 1.4×10–3 (95% HPD=0.8×10–3−2.6×10–3), and 1.6×10–3 (95% HPD=0.7×10–3−3.7×10–3) for clades 1, 2, and 3, respectively. The marginal density of the rates obtained from the Bayesian analysis, which represents the variance of the molecular clock, was also largely overlapping and all three clades appeared to have emerged at about the same time during the mid to late 1970s (Fig. 2).
FIG. 2.
Marginal density of evolutionary rate estimates for each Cameroon CRF02_AG monophyletic clade. Distribution of mean evolutionary rate estimates (x-axis) from 10,000 Markov chain Monte Carlo (MCMC) sampled trees (minus 10% burn-in). The time of the most recent common ancestor (TMRCA) with 95% high density posterior (95% HPD) intervals is also indicated.
To investigate further the population dynamic patterns of each CRF02_AG Cameroon clade, we compared different demographic models of effective population size (Ne) change over time. Surprisingly, the constant size model could not be rejected when compared to the exponential growth model or the nonparametric Bayesian Skyline Plot (Supplementary Table S3; Supplementary Data are available online at www.liebertonline.com/aid).
Bayesian estimates of median Ne for the constant model were about 1.5 times larger for clades 2 and 3 than for clade 1, but since the 95% HPD appeared to be completely overlapping, the hypothesis that Ne was not significantly different for different clades could not be rejected (Table 2). Overall, the data indicate that clades 1, 2, and 3 emerged simultaneously and have been spreading at a relatively low but similar rate within three distinct Cameroon epidemiological networks.
Table 2.
TMRCA and Effective Population Size Medians with 95% HPD Interval Estimates (Relaxed Molecular Clock Model) of Cameroon Monophyletic Clades
| |
Nea |
|||
|---|---|---|---|---|
| Dataset | Selected clock model | Selected demographic model | Median | 95% HPD |
| Clade 1 | Relaxed | Constant | 38.4 | 2–252 |
| Clade 2 | Relaxed | Constant | 60.7 | 20.8–156.8 |
| Clade 3 | Relaxed | Constant | 55.3 | 14.3–188 |
The effective population size (Ne) represents the number of genomes effectively contributing to the next generation and is related to the number of effective infections.
TMRCA, time of the most recent common ancestor; HPD, high posterior density.
Phylogeography of CRF02_AG Cameroon clades
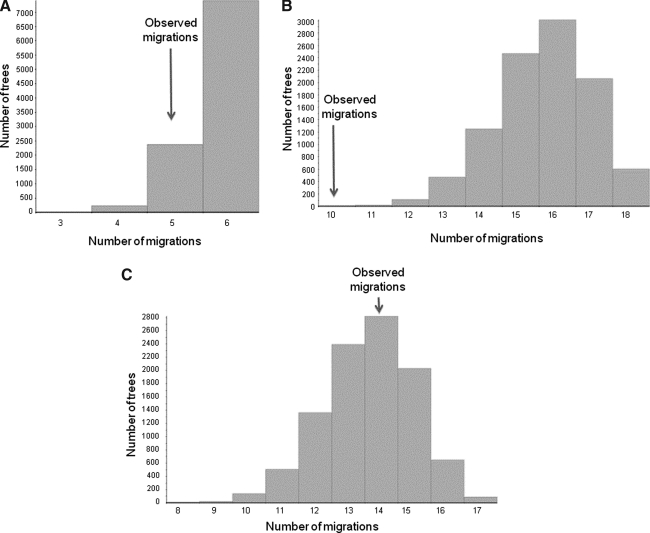
The next step of the analysis was to ascertain whether specific phylogeographic trends existed in different clades. Clades 1 and 3 did not show any significant metapopulation structure (p>0.05; Fig. 3). A weak metapopulation structure was observed for clade 2 (p=0.0001; Fig. 3) where two distinct subpopulations, one including strains sampled from Western cities and the other sequences from Central Cameroon, were evident in the Bayesian genealogy (Supplementary Fig. S3; Supplementary Data are available online at www.liebertonline.com/aid).
FIG. 3.
Phylogeographic structure of Cameroon CRF02_AG clades. Distribution of observed migrations (x-axis) in 10,000 random trees generated using the sequences of each of the three well-supported monophyletic CRF02_AG Cameroon clades highlighted in Fig. 1. The arrow indicates the number of migrations observed in the Bayesian maximum clade credibility (MCC) tree including only the sequences from a specific clade (MCC trees are given in Supplementary Fig. S3). (A) Cameroon clade 1 sequences. (B) Cameroon clade 2 sequences. (C) Cameroon clade 3 sequences.
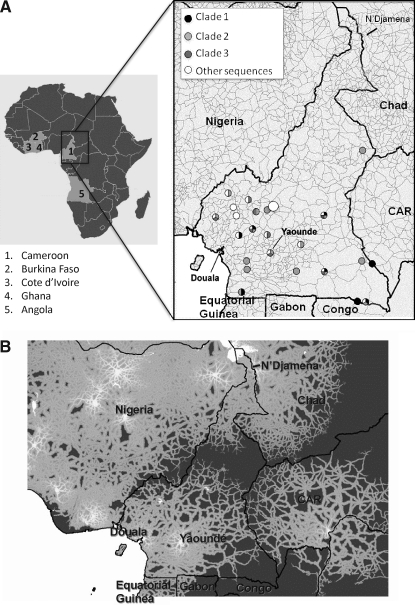
To better characterize the geographic distribution and spread of HIV-1 CRF02_AG strains within the country, the location of all Cameroonian strains was superimposed on the country map and compared with accessibility data (Fig. 4). The sampling sites were most densely distributed in the Southwestern part of Cameroon (Fig. 4A). The accessibility map (Fig. 4B) suggested a potential correlation between the strong accessibility network in the south and the dissemination of discrete subepidemics. The map displayed the estimated time to travel from any location to the nearest major urban center, defined as an urban area with >100,000 inhabitants. Cities interconnectivity was very strong in the Litoral and Central regions, around Douala, the most populated city in Cameroon, and Yaoundé, but became progressively lower toward the Northern region. Indeed, Northern Cameroon appeared to be largely disconnected from the south and more connected with N'Djamena, the capital of Chad.
FIG. 4.
CRF02_AG geographic distribution and Cameroon accessibility map. (A) On the left a map of Africa is displayed with the countries involved in the CRF02_AG flow to Cameroon, and an expanded Cameroon map is shown to the right. Circles indicate the sampling locations of Cameroonian sequences. Strains clustering within clade 1, 2, 3 or intermixed in the tree are represented according to the legend on the right top of the map. (B) Cameroon accessibility map. The gray gradient of colors, according to the color bar in the figure, indicates the estimated travel time to the nearest city of population >100,000 people, with white at one extreme indicating low travel times (<30 min) and dark gray at the other extreme indicating long travel times (>10 h).
Discussion
The present study characterized new CRF02_AG strains sampled from Cameroonian subjects, as well as strains from both Italian and African individuals residing in Italy. First, the molecular epidemiology of this subtype in West-Central African countries was investigated, with particular focus on Cameroon, an epicenter of the African epidemic. The phylogenetic analysis showed a continuous exchange of viral strains between Cameroon and other African countries, as well as the presence of three different monophyletic clades within Cameroon, all of which originated around the mid-1970s. All clades were related to strains from different West African countries, none of which, however, is geographically adjacent to Cameroon. A potential explanation is that the French occupation of Burkina Faso, Ivory Coast, and Cameroon until the 1960s led to a founder effect in Cameroon, arising from connections among countries within the French sphere of influence.
The lack of metapopulation structure within the Cameroonian epidemic, in which none of the three major clades was significantly associated with a specific geographic area, is consistent with GIS data. Accessibility maps indicated that Southern Cameroon is characterized by developed road networks and harbor areas that may have significantly fostered HIV-1 spread after initially limited introduction from other African countries. This is in agreement with the hypothesis, recently suggested by Gray et al.,49 that accessibility plays a major role in the emergence and spread of viral regional epidemics. This hypothesis is also supported by data showing that the most vulnerable groups in Cameroon include truck drivers, mobile populations, and military personnel.50
The phylogenetic analysis also showed that CFR02_AG strains from Italian individuals, as well as from non-Cameroonian African immigrants residing in a small locale of northern Italy, were intermixed throughout the tree. In particular, most of the Italian sequences were only distantly related in the phylogeny, which was indicative of at least four independent events leading to infection of Italian subjects. Additional studies including sequences from multiple regions in Italy are needed to assess the frequency and extent of CRF02_AG spillover into the country. However, given that the HIV strains analyzed were sampled from a relatively small geographic area, it is remarkable that several independent introductions were already observed.
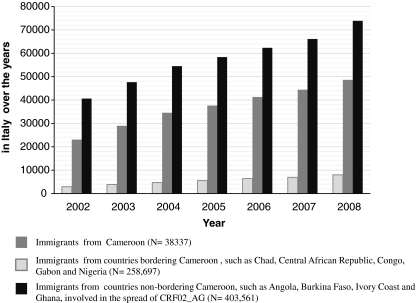
Data from the Italian National Institute of Statistics showed that in recent years African immigrants have constantly been increasing in the country (Fig. 5). In particular, from 2002 to 2008 the number of Cameroonians residing in Italy has almost tripled, from 2926 (8% of immigrants living in Italy) in 2002 to 7994 (21% of immigrants living in Italy) in 2008. A similar trend could be observed for immigrants from other African countries with a significant AG epidemic both bordering and not bordering Cameroon. Taken together these findings suggest that conditions may be present for the development of a generalized epidemic of this recombinant form in Italy that might significantly impact HIV-1 molecular epidemiology thus far predominantly characterized by subtype B infections.
FIG. 5.
Citizens from Cameroon and other African countries (bordering and not bordering Cameroon) involved in the spread of CRF02_AG and residing in Italy: demographic balance over the years 2002–2008. For each year, the demographic balance is updated to December 31. N: Overall number of immigrants residing in Italy over the years 2002–2008.
In the past years the migration trends from Africa to Western Europe have been changing the face of the AIDS epidemic in terms of subtype distribution/prevalence. Italy's position in the Mediterranean Sea makes it a strategic migration route. Therefore, understanding the CRF02_AG epidemic from Africa to Italy may also play a fundamental role in assessing the potential spread of this viral strain within Europe and North America, especially given the enormous exchange of persons and goods between the two continents.
Understanding HIV molecular epidemiology and the potential future spread of different non-B subtypes also has clinical relevance. It is already known that differences among HIV-1 genetic forms may impact clinical management and surveillance of drug resistance, particularly as treatment is expanded to HIV-1 non-B strains.51–55 Moreover, HIV-1 subtypes are relevant for vaccine design. Although cross-clade immune reactivity has been detected among individuals and vaccine recipients, it is reasonable to expect that a vaccine with an antigenic composition including CRFs may induce a more effective response.56
Supplementary Material
Acknowledgments
This work was financially supported by grants from the Italian National Institute of Health, the Ministry of University and Scientific Research, Current and Finalized Research of the Italian Ministry of Health, by the European Commission Framework 7 Programme (CHAIN, the Collaborative HIV and Anti-HIV Drug Resistance Network, Integrated Project no. 223131), PHS R01 AI065265, PHS T32 CA09126, the Center for Research for Pediatric Immune Deficiency, and the Laura McClamma Fellowship and Stephany W. Holloway University Chair for AIDS Research. We thank the Organizers of the XV Workshop in Virus Evolution and Molecular Epidemiology for the training and support that made this study possible. Nazle Mendonca Collaço Véras and Maria Mercedes Santoro contributed equally to this work.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.HIV Sequence Database. http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html
- 2.AIDS epidemic update: Joint United Nations Programme on HIV/AIDS (UNAIDS) and World Health Organization (WHO) 2009. http://www.unaids.org/en/default.asp http://www.unaids.org/en/default.asp
- 3.Nkengasong JN. Janssens W. Heyndrickx L, et al. Genotypic subtypes of HIV-1 in Cameroon. AIDS. 1994;8:1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Mauclère P. Loussert-Ajaka I. Damond F, et al. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Simon F. Mauclere P. Roques P, et al. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 6.Takehisa J. Zekeng L. Ido E, et al. Human immunodeficiency virus type 1 intergroup (M/O) recombination in Cameroon. J Virol. 1999;73:6810–6820. doi: 10.1128/jvi.73.8.6810-6820.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Triques K. Bourgeois A. Vidal N, et al. Near-full-length genome sequencing of divergent African HIV type 1 subtype F viruses leads to the identification of a new HIV type 1 subtype designated K. AIDS Res Hum Retroviruses. 2000;16:139–151. doi: 10.1089/088922200309485. [DOI] [PubMed] [Google Scholar]
- 8.Carr JK. Torimiro JN. Nathan DW. Mpoudi NE, et al. Novel HIV-1 forms are common in Cameroon. Virology. 2001;286:168–181. doi: 10.1006/viro.2001.0976. [DOI] [PubMed] [Google Scholar]
- 9.Carr JK. Wolfe ND. Torimiro JN, et al. HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: Evolutionary relics? Retrovirology. 2010;28:7–39. doi: 10.1186/1742-4690-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konings FA. Haman GR. Xue Y, et al. Genetic analysis of HIV-1 strains in rural eastern Cameroon indicates the evolution of second-generation recombinants to circulating recombinant forms. J Acquir Immune Defic Syndr. 2006;42:331–341. doi: 10.1097/01.qai.0000219784.81163.2e. [DOI] [PubMed] [Google Scholar]
- 11.Powell RL. Zhao J. Konings FA, et al. Circulating recombinant form (CRF) 37_cpx: An old strain in Cameroon composed of diverse, genetically distant lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23:923–933. doi: 10.1089/aid.2007.0040. [DOI] [PubMed] [Google Scholar]
- 12.Powell RL. Zhao J. Konings FA, et al. Identification of a novel circulating recombinant form (CRF) 36_cpx in Cameroon that combines two CRFs (01_AE and 02_AG) with ancestral lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23:1008–1019. doi: 10.1089/aid.2006.0289. [DOI] [PubMed] [Google Scholar]
- 13.Brennan CA. Bodelle P. Coffey R, et al. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J Acquir Immune Defic Syndr. 2008;49:432–439. doi: 10.1097/QAI.0b013e31818a6561. [DOI] [PubMed] [Google Scholar]
- 14.Ndembi N. Abraha A. Pilch H, et al. Molecular characterization of HIV-1 and HIV-2 in Yaoundé, Cameroon: Evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol. 2008;46:177–184. doi: 10.1128/JCM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torimiro JN. D'Arrigo R. Takou D, et al. Human immunodeficiency virus type 1 intersubtype recombinants predominate in the AIDS epidemic in Cameroon. New Microbiol. 2009;32:325–332. [PubMed] [Google Scholar]
- 16.Yamaguchi J. Ndembi N. Ngansop C, et al. HIV type 1 group M subtype G in Cameroon: Five genome sequences. AIDS Res Hum Retroviruses. 2009;25:469–473. doi: 10.1089/aid.2008.0296. [DOI] [PubMed] [Google Scholar]
- 17.Vallari A. Bodelle P. Ngansop C, et al. Four new HIV-1 group N isolates from Cameroon: Prevalence continues to be low. AIDS Res Hum Retroviruses. 2010;26:109–115. doi: 10.1089/aid.2009.0178. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Calleja JM. Zekeng L. Louis JP, et al. HIV infection in Cameroon: 30 months' surveillance in Yaounde. AIDS. 1992;6:881–882. [PubMed] [Google Scholar]
- 19.Peeters M. Recombinant HIV sequences: Their role in the global epidemic. In: Kuiken CL, et al., editors. HIV Sequence Compendium. Theoretical Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM: 2000. 2000. pp. I-39–54. [Google Scholar]
- 20.Fischetti L. Opare-Sem O. Candotti D. Lee H. Allain JP. Higher viral load may explain the dominance of CRF02_AG in the molecular epidemiology of HIV in Ghana. AIDS. 2004;18:1208–1210. doi: 10.1097/00002030-200405210-00017. [DOI] [PubMed] [Google Scholar]
- 21.Fischetti L. Opare-Sem O. Candotti D. Sarkodie F. Lee H. Allain JP. Molecular epidemiology of HIV in Ghana: Dominance of CRF02_AG. J Med Virol. 2004;73:158–166. doi: 10.1002/jmv.20070. [DOI] [PubMed] [Google Scholar]
- 22.Sarr AD. Eisen G. Gueye-Ndiaye A, et al. Viral dynamics of primary HIV-1 infection in Senegal, West Africa. J Infect Dis. 2005;191:1460–1467. doi: 10.1086/429409. [DOI] [PubMed] [Google Scholar]
- 23.Njai HF. Gali Y. Vanham G, et al. The predominance of human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in West Central Africa may be related to its replicative fitness. Retrovirology. 2006;3:1–11. doi: 10.1186/1742-4690-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortiz M. Muñoz L. Bernal A, et al. Molecular characterization of non-B HIV type 1 subtypes from Africa in Spain. AIDS Res Hum Retroviruses. 2000;16:1967–1971. doi: 10.1089/088922200750054693. [DOI] [PubMed] [Google Scholar]
- 25.Parreira R. Pádua E. Piedade J. Venenno T. Paixão MT. Esteves A. Genetic analysis of human immunodeficiency virus type 1 nef in Portugal: Subtyping, identification of mosaic genes, and amino acid sequence variability. J Med Virol. 2005;77:8–16. doi: 10.1002/jmv.20408. [DOI] [PubMed] [Google Scholar]
- 26.Falkensammer B. Doerler M. Kessler HH, et al. Subtype and genotypic resistance analysis of HIV-1 infected patients in Austria. Wien Klin Wochenschr. 2007;119:181–185. doi: 10.1007/s00508-006-0745-4. [DOI] [PubMed] [Google Scholar]
- 27.Thomson MM. Delgado E. Herrero I, et al. Diversity of mosaic structures and common ancestry of human immunodeficiency virus type 1 BF intersubtype recombinant viruses from Argentina revealed by analysis of near full-length genome sequences. J Gen Virol. 2002;83:107–119. doi: 10.1099/0022-1317-83-1-107. [DOI] [PubMed] [Google Scholar]
- 28.Monno L. Brindicci G. Lo Caputo S, et al. HIV-1 subtypes and circulating recombinant forms (CRFs) from HIV-infected patients residing in two regions of central and southern Italy. J Med Virol. 2005;75:483–490. doi: 10.1002/jmv.20300. [DOI] [PubMed] [Google Scholar]
- 29.Tramuto F. Bonura F. Perna AM, et al. Group for HIV-1 Antiretroviral Studies in Sicily. Genetic diversity of HIV-1 non-B strains in Sicily: Evidence of intersubtype recombinants by sequence analysis of gag, pol, and env genes. AIDS Res Hum Retroviruses. 2007;23:1131–1138. doi: 10.1089/aid.2007.0031. [DOI] [PubMed] [Google Scholar]
- 30.Santoro MM. Alteri C. Ronga L, et al. Evaluation of HIV-1 non-B subtypes in Italian treated patients from central Italy over the years 2001–2008; 7th European HIV Drug Resistance workshop; Stockholm, Sweden. Mar 25–27;2009 ; Abstract 130. [Google Scholar]
- 31.Bracciale L. Colafigli M. Zazzi M, et al. Prevalence of transmitted HIV-1 drug resistance in HIV-1-infected patients in Italy: Evolution over 12 years and predictors. J Antimicrob Chemother. 2009;64:607–615. doi: 10.1093/jac/dkp246. [DOI] [PubMed] [Google Scholar]
- 32.Torti C. Lapadula G. Izzo I, et al. Heterogeneity and penetration of HIV-1 non-subtype B viruses in an Italian province: Public health implications. Epidemiol Infect. 2010;29:1–10. doi: 10.1017/S0950268810000166. [DOI] [PubMed] [Google Scholar]
- 33.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 34.Felsenstein J. Accuracy of coalescent likelihood estimates: Do we need more sites, more sequences, or more loci? Mol Biol Evol. 2006;23:691–700. doi: 10.1093/molbev/msj079. [DOI] [PubMed] [Google Scholar]
- 35.Ceccherini-Silberstein F. Erba F. Gago F, et al. Identification of the minimal conserved structure of HIV-1 protease in the presence or absence of drug pressure. AIDS. 2004;18:11–19. doi: 10.1097/01.aids.0000131394.76221.02. [DOI] [PubMed] [Google Scholar]
- 36.Giuliani M. Montieri S. Palamara G, et al. Non-B HIV type 1 subtypes among men who have sex with men in Rome, Italy. AIDS Res Hum Retroviruses. 2009;25:157–164. doi: 10.1089/aid.2008.0175. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD. Higgins DG. Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guindon S. Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Swofford DL. Sullivan J. Phylogeny inference based on parsimony, other methods using PAUP*. In: Lemey P, editor; Salemi M, editor; Vandamme AM, editor. The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. 2nd. Cambridge University Press; New York: 2009. pp. 267–312. [Google Scholar]
- 40.Anisimova M. Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 41.Drummond AJ. Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:1–8. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemey P. Pybus OG. Rambaut A, et al. The molecular population genetics of HIV-1 group O. Genetics. 2004;167:1059–1068. doi: 10.1534/genetics.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Worobey M. Gemmel M. Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–665. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suchard MA. Weiss RE. Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- 45.Slatkin M. Detecting small amounts of gene flow from phylogenies of alleles. Genetics. 1989;121:609–612. doi: 10.1093/genetics/121.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salemi M. Lamers SL. Yu S. de Oliveira T. Fitch WM. McGrath MS. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J Virol. 2005;79:11343–11352. doi: 10.1128/JVI.79.17.11343-11352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maddison DR. Maddison WP. Book MacClade. 4.08. Sinauer Associates, Inc.; Sunderland, MA: 2008. MacClade. [Google Scholar]
- 48.Nelson A. Estimated travel time to the nearest city of 500,000 or more people in year 2000. Global Environment Monitoring Unit–Joint Research Centre of the European Commission, Ispra, Italy. http://gem.jrc.ec.europa.eu. [Feb 17;2009 ]. 2008. http://gem.jrc.ec.europa.eu
- 49.Gray RR. Tatem AJ. Lamers S, et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS. 2009;23:F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ndongmo CB. Pieniazek D. Holberg-Petersen M, et al. HIV genetic diversity in Cameroon: Possible public health importance. AIDS Res Hum Retroviruses. 2006;22:812–816. doi: 10.1089/aid.2006.22.812. [DOI] [PubMed] [Google Scholar]
- 51.Thomson MM. Najera R. Travel and the introduction of human immunodeficiency virus type 1 non-B subtype genetic forms into western countries. Clin Infect Dis. 2001;32:1732–1737. doi: 10.1086/320764. [DOI] [PubMed] [Google Scholar]
- 52.Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis. 2006;19:594–606. doi: 10.1097/QCO.0b013e3280109122. [DOI] [PubMed] [Google Scholar]
- 53.Buonaguro L. Tornesello ML. Buonaguro FM. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: Pathogenetic and therapeutic implications. J Virol. 2007;81:10209–10219. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor BS. Taylor BS. Sobieszczyk ME. McCutchan FE. Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Easterbrook PJ. Smith M. Mullen J, et al. Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J Int AIDS Soc. 2010;13:2–9. doi: 10.1186/1758-2652-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peeters M. Toure-Kane C. Nkengasong JN. Genetic diversity of HIV in Africa: Impact on diagnosis, treatment, vaccine development and trials. AIDS. 2003;17:2547–2560. doi: 10.1097/01.aids.0000096895.73209.89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.