Abstract
Among the Chalcidoids, hymenopteran parasitic wasps that have diversified lifestyles, a partial mitochondrial genome has been reported only from Nasonia. This genome had many unusual features, especially a dramatic reorganization and a high rate of evolution. Comparisons based on more mitochondrial genomic data from the same superfamily were required to reveal weather these unusual features are peculiar to Nasonia or not. In the present study, we sequenced the nearly complete mitochondrial genomes from the species Philotrypesis. pilosa and Philotrypesis sp., both of which were associated with Ficus hispida. The acquired data included all of the protein-coding genes, rRNAs, and most of the tRNAs, and in P. pilosa the control region. High levels of nucleotide divergence separated the two species. A comparison of all available hymenopteran mitochondrial genomes (including a submitted partial genome from Ceratosolen solmsi) revealed that the Chalcidoids had dramatic mitochondrial gene rearrangments, involved not only the tRNAs, but also several protein-coding genes. The AT-rich control region was translocated and inverted in Philotrypesis. The mitochondrial genomes also exhibited rapid rates of evolution involving elevated nonsynonymous mutations.
Introduction
In most animals, the mitochondrial genome is maternally inherited, generally nonrecombining with other mitochondrial lineages, and comprised of 13 protein-coding genes, 2 rRNAs and 22 tRNAs. The gene products work with nuclear-encoded mitochondrial proteins in the process of oxidative phosphorylation (OXPHOS) [1]. Due to its vital role in metabolism and relatively small size, the evolution of animal mitochondrial genomes remains intensively investigated. Complete mitochondrial genomes are known from many species of insects, yet few are recorded from the Hymenoptera. Although the mtDNA of the honeybee, Apis mellifera, has been available since 1993 [2], today few other complete hymenopteran genomes are known [3]. This situation may be due to two characteristics: the mitochondrial genome is extremely AT-rich rendering amplification and sequencing difficult and it has unusually high rates of substitution and frequent gene arrangements that confound primer design and amplification [4], [5]. For the Chalcidoidea, only a partial mitochondrial genome is known from Nasonia, and it has an unusually high accelerated rate of evolution and several unique gene rearrangements [6].
Mitochondrial genomes serve as good models for the study of molecular evolution and population genetics [7], [8], [9], [10]. Mitochondrial genome organization provides informative characters in sufficient quantity and quality for inferring phylogeny [1], [11], [12]. The high rate of evolution and genome reorganization of Nasonia may be typical of parasitic lifestyles in the Hymenoptera [5], [13], [14]. Parasitic chalcidoids have diversified lifestyles and mitochondrial genomic data from fig wasps that live inside the compact syconium of figs [15], might reveal features associated with their phyletic positions and lifestyles.
This study is concerned with three species of fig wasp: Ceratosolen solmsi and two species of Philotrypesis (P. pilosa and Philotrypesis sp.), all of which live in the same fig tree, Ficus hispida. Among these species, C. solmsi enjoys a mutualistic association with the fig; pollination occurs as it feeds on floret tissue. In contrast, species of Philotrypesis are parasitic on C. solmsi. Herein, sequences from the mitochondrial genome of C. solmsi (submitted) are compared to those of the parasites. The nearly complete mitochondrial genomes of the two species of Philotrypesis are highly diverged. Further, Philotrypesis also has unusual, dramatic gene rearrangments, not only in tRNAs, but also in several protein-coding genes. Below we discuss the relationship of accelerated mtDNA evolution and the unusual rearrangements with the evolution of fig wasps and the parasitic lifestyles in the Chalcidoidea.
Materials and Methods
Ethics statement
No experiments involving vertebrate samples were performed in this study. An ethics statement is not required for the experiments which only involve insects. The collections of wasps were permitted by the local park in Danzhou.
Specimens and DNA extraction
Specimens of P. pilosa and Philotrypesis sp. were collected in 2008 from Danzhou, Hainan province, China. Wasps were identified and stored in 95% ethanol at −20°C. Images of the wasps used to confirm identification were captured by using a Nikon AZ100 microscope system. Only one individual from each species was chosen for DNA extraction by methods applicable for long PCR [16].
Amplification and sequencing of mitochondrial genome fragments
We used degenerate primers modified from previous studies [17] to amplify the relatively conservative fragments co1–nad3, nad5–cob, and nad1–12s. Subsequently, species-specific primers were designed for the amplification of the regions between these fragments. Primer sequences were summarized in table S1. We used HiFi Taq (TransGen, Beijing, China) following the manufacturer's suggestions for PCR and the amplicons were either purified for direct sequencing or cloned for sequencing. The sequences were deposited in GenBank under the accession numbers JF808722 and JF808723.
Genome annotation
The protein-coding and rRNA genes were identified by Blast searches in GenBank and aligned to the orthologous mitochondrial genes of Nasonia and Apis mellifera. Positional confirmation and annotation of the tRNAs was accomplished by using the online software of tRNAscan-SE 1.21 [18]. The detection of repeats used Tandem repeats finder [19].
Genetic Divergence and Phylogeny
The combined sequences were aligned by using ClustalW and the software package DnaSP 5.0 [20] was used to compute nucleotide divergence, the ratio of Ka (the number of synonymous substation per synonymous site) and Ks (the number of nonsynonymous substation per nonsynonymous site). Based on genetic divergence, Co1 and co2 were inferred to be the most conserved protein-coding genes. They were employed for hypothesizing the phylogenetic relationships of the hymenoptera by using MrBayes 3.12 [21]. Twenty-four mitochondrial genome sequences (accession numbers for the downloaded genomes: EU746610-EU746612, NC_011923, NC_010967, NC_004529, NC_014295, NC_001566, NC_014272, NC_014278, NC_012708, NC_014677, NC_014669, NC_014672, NC_015075, NC_011520, NC_013238, NC_008323, NC_012688, NC_014485, NC_012689.) were used for phylogenetic inference and gene rearrangement analyses. Orussus occidentalis was chosen as the outgroup. We used The MrModeltest 2 to select the best-fit model for Bayesian analysis. Bayesian calculations used 1 million generations while sampling a tree every 100 generations. A 50% majority rule consensus tree was calculated from the sampled trees.
Results
Mitochondrial genomes of Philotrypesis
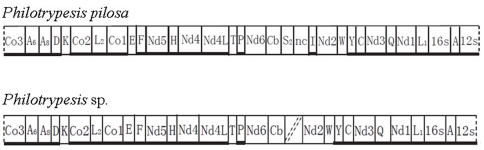
For P. pilosa, 15,122 bp fragment of the mitochondrial genome was sequenced as follows: 13 protein-coding genes (co3 incomplete), 16 tRNAs, 2 rRNAs (12s incomplete), and 1670 bp non-coding region between trnS2 and trnI. The overall AT bias was 84.14%. For Philotrpesis. sp., we obtained two fragments of 8,567 bp and 3,330 bp. An unsequenced region occurred between cob and nad2. The sequenced fragments contained 13 protein-coding genes (co3, cob, and nad2 incomplete), 14 tRNAs (missing trnS2 and trnI compared to P. pilosa), and 2 rRNAs (12s incomplete) (Figure 1 and Table 1). The overall AT bias was 81.7%, slightly lower than in P. pilosa.
Figure 1. The mitochondrial genome of Philotrypesis pilosa and Philotrypesis sp.
An unsequenced gap is located between cob and nad2 in Philotrypesis sp.
Table 1. Gene annotation and features for both genomes.
| Gene | Strand | Phipotrypesis pilosa | Philotrypesis sp. | ||||||
| Length | Start | Stop | Space | Length | Start | Stop | Space | ||
| co3 | - | incomplete | ATA | 0 | incomplete | ATA | 12 | ||
| atp6 | - | 675 | ATG | TAA | -7 | 675 | ATG | TAA | -7 |
| atp8 | - | 165 | ATT | TAA | 1 | 165 | ATT | TAA | 0 |
| D | - | 65 | 9 | 70 | 6 | ||||
| K | + | 69 | 4 | 70 | 4 | ||||
| co2 | - | 673 | ATG | T- | 69 | 675 | ATT | TAA | 68 |
| L2 | - | 66 | -4 | 66 | 92 | ||||
| co1 | - | 1545 | ATG | TAA | 6 | 1539 | ATG | TAA | 3 |
| E | + | 69 | -1 | 68 | 0 | ||||
| F | - | 66 | 0 | 65 | 0 | ||||
| nad5 | - | 1666 | ATT | T- | 0 | 1675 | ATT | T- | 0 |
| H | - | 67 | 1 | 67 | 3 | ||||
| nad4 | - | 1341 | ATG | TAA | -7 | 1341 | ATG | TAG | -7 |
| nad4l | - | 285 | ATT | TAA | 1 | 285 | ATT | TAA | 1 |
| T | + | 68 | 0 | 63 | 0 | ||||
| P | - | 66 | 4 | 66 | 2 | ||||
| nad6 | + | 564 | ATG | TAA | 2 | 561 | ATG | TAA | 3 |
| cob | + | 1140 | ATG | TAA | -2 | incomplete | ATG | ||
| S2 | + | 66 | -1 | ||||||
| nc | 1670 | 0 | |||||||
| I | - | 67 | 62 | ||||||
| nad2 | + | 969 | ATA | TAA | 8 | incomplete | T- | 0 | |
| W | + | 66 | 0 | 64 | 5 | ||||
| Y | - | 66 | 67 | 65 | 65 | ||||
| C | - | 65 | 1 | 64 | 0 | ||||
| nad3 | - | 334 | ATT | T- | 0 | 334 | ATT | T- | 0 |
| Q | - | 70 | 0 | 71 | 0 | ||||
| nad1 | - | 919 | ATA | T- | 0 | 919 | ATA | T- | 0 |
| L1 | - | 63 | 0 | 64 | 0 | ||||
| 16s | - | 1309 | 0 | 1283 | 0 | ||||
| A | - | 65 | 0 | 69 | 0 | ||||
| 12s | - | incomplete | incomplete | ||||||
Table 1 compared the features of the mitochondrial genomes of both species. The two genomes had the same gene orientations, with 10 protein-coding genes and most of the tRNAs and both rRNAs located on the light strand. Only minor differences occurred. The lengths of most protein-coding gene were identical, except for co1, nad5 and nad6. The size of 16s rRNA genes also differed. Only three tRNAs (trnL2, trnH, and trnP) of the 14 tRNAs had the same size. All predicted initiation codons translated to either methionine or isoleucine, as with most other bilateral animals [2]. Co2 was the only gene to differ between the two species' start codons: P. pilosa had methionine (ATG) and Philotrypesis sp. isoleucine (ATT). Several genes had incomplete stop codons, a single T, which is common in animals. The products can be completed by posttranscriptional polyadenylation [6], [22]. As characteristic of mitochondrial genomes, inter-gene spaces were usually very short, yet several exceptions of up to 65–92 bp in length occurred in both genomes. The genome of P. pilosa had a 1670 bp non-coding region between trnS2 and trnI. The corresponding region remained unknown in Philotrypesis sp.; we were unsure whether the latter species also had similar sequences or not. All of the predicted tRNAs had similar secondary structures (Figure S1).
Accelerated rate of evolution
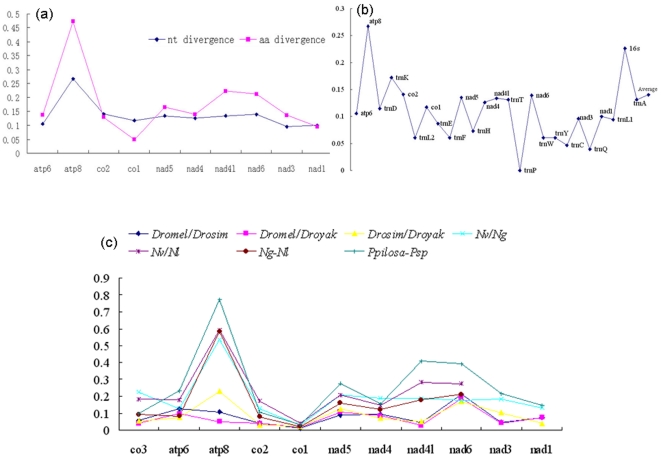
The mitochondrial sequences diverge dramatically in the two species (Figure 2a and 2b). The average sequence divergence of the two genomes is 0.140. Protein-coding genes were more divergent than tRNAs; the highest divergence occurred in atp8 (0.267). In contrast, trnP was identical in the two genomes. The large ribosomal RNA gene, 16s, had the second largest divergence (0.226). This pattern indicated that the mitochondrial genomes of Philotrypesis either evolved rapidly or that the species diverged long ago. The former explanation seemed more likely because: 1) some of the genes were similar or even identical between the two species; 2) previous phylogenetic results based on the combination of mitochondrial (cob) and nuclear (ITS2) markers indicated that the two species may be sister taxa [23]; 3) morphological comparisons show that the two species were similar (Figure S2).
Figure 2. Genetic diversity of the two mitochondrial genomes of Philotrypesis.
(a) Comparison of the protein-coding genes on both nucleotide and translated amino acid sequences; (b) Genetic divergence patterns throughout the genome. Pairwise sequence divergences are calculated with Bioedit and displayed as images suing Microsoft Excel. Genes on the x-axis are ordered according to their position in the genome of Philotrypesis. Numbers on the y-axis indicate the gene sequence divergence between the two species; 0.1 = 10% divergence or 90% similarity. (c) Ratio of Ka and Ks for 11 mitochondrial genes. Values of Ka and Ks are estimated with DnaSP v5 and corrected by the JC method.
We compared the nucleotide and amino acid sequences of 10 protein-coding genes to further confirm that the mitochondrial genome of Philotrypesis was evolving rapidly (Figure 2a). Except for co1, co2 and nad1, divergences in the amino acid sequences were greater than those of the nucleotide sequences. The greatest divergence occurred in atp8. The nucleotide divergence was 0.267, while the amino acid divergence was as high as 0.473. For genes showing greater amino acid sequence divergences, a greater number of nonsynonymous substitutions may have been accumulated, which would have accelerated their evolution by either positive or relaxed selection. In contrast, co1 had a lower level of amino acid divergence (0.049) than nucleotide divergence (0.117), which indicated purifying selection. Further intra-specific comparisons may help determine whether the mitochondrial genomes have been evolving under neutral or positive selection.
A comparison of Ka/Ks ratios for 11 protein-coding genes was given for species in the genera Drosophila, Nasonia and Philotrypesis in Figure 2c and Table S2. Nasonia was reported to have dramatic higher Ka/Ks ratios than Drosophila, yet the two species of Philotrypesis had even higher ratios for most genes. This pattern indicated elevated evolutionary rates in the two species of Philotrypesis.
Gene rearrangements
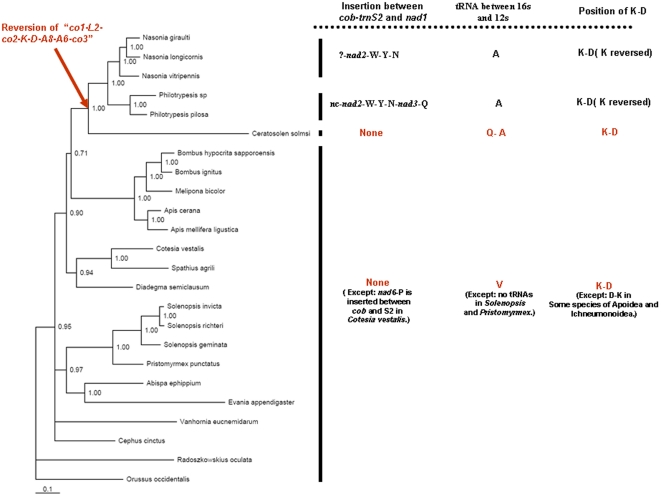
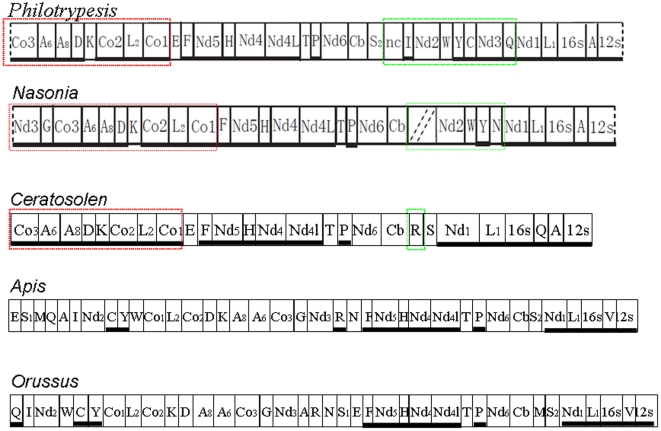
For a comparison of the gene orders, we made a NCBI search and downloaded all available hymenopteran mitochondrial genomes, and then constructed a phylogeny based on co1 plus co2 sequences. Orussus occidentalis was indicative of the ancestral state and used as the outgroup for all other hymenoptera mitochondrial genomes (Figure 3). The gene orders of five genomes are listed in Figure 4 (Philotrypesis, Nasonia, Ceratosolen, Apis, and Orusses), of which Ceratosolen was from the Agaonidae, and Nasonia and Philotrypesis represented the Pteramalidae. The later three genera were from the Chalcidoidea in the Proctotrupomorpha. Hymenopterans were reported to have many tRNA rearrangement events in their mitochondrial genomes [3]. However, the organizations of most of the species across the Apoidea, Ichneumnoidea, Vespoidea, Cephoidea, Evanioidea, and Proctotrupoidea were similar to that of the outgroup, Orussoidea, thus indicating little change. In comparison, chalcidoids had a series of mitochondrial gene rearrangements, the most striking being a large inversion in the region of at least five protein-coding genes. Nasonia had positional changes in at least seven protein-coding genes, and the large inversion affected six protein-coding genes [6]. The inversion affected only five genes in Ceratosolen and Philotrypesis.
Figure 3. Bayesian estimation of phylogenetic relationships with mapped genome rearrangements for all hymenopterans having whole or partial mitochondrial genomic data.
The major mitochondrial genome rearrangement events are compared and mapped out for Ceratosolen, Nasonia and Philotrypesis.
Figure 4. Mitochondrial genome organization in five genera.
Red blocks indicate the large inversion specific to the Chalcidoidea, and the green blocks show the different and dramatically changed regions in the Chalcidoidea.
Another dramatic change was located downstream of cob–trnS2. Compared to the Pteramalidae, Ceratosolen has a relatively ancient gene composition and order for cob–trnR–trnS2–nad1, with no insertion between trnS2 and nad1. A large insertion occurred in members of the Pteramalidae. In Nasonia, at least nad2, trnW, trnY, and trnN were inserted before nad1. It was possible that additional genes are inserted in the region but that they did not amplify [6]. In Philotrypesis, a large insertion occurred between trnS2 and nad1. The insertion was comprised of two protein-coding genes, five tRNAs and a non-coding region of 1670 bp.
Two additional apomorphic rearrangements occurred with tRNAs in the Pteramalidae relative to Ceratosolen. First, trnK was positioned in a ‘hot spot’ for rearrangements in the Hymenoptera [11]. Subsequent to the large inversion of co1 and co3 (or nad3 in Nasonia), the orientation of trnK reversed in Pteramalidae but did not change in Ceratosolen. Second, among tRNAs occurring in the middle of the two rRNAs, the position of trnV appeared to be plesiomorphic because it occurred here in other arthropods [1]. As shown in Figure 3, the changes progressed from trnV in ancestral hymenoptera, to trnQ-trnA in Ceratosolen, and then to trnA in the Pteramalidae. When mapped onto the phylogeny of the hymenopterans (Figure 3), the apomorphic rearrangements clearly depicted the relationships of species in the Chalcidoidea.
Non-coding sequences in Philotrypesis pilosa
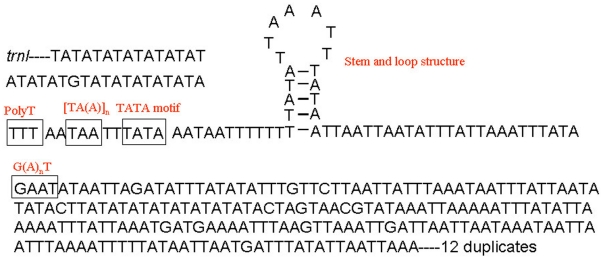
A 1670 bp fragment of non-coding sequences was resolved in the genome of P. pilosa. This fragment had an AT composition of 81.88%, a little less than the average AT bias in the entire genome. The fragment was located between trnS2 and trnI and it was comprised of 12 duplicates plus a partial one for a total of 112 bp. Each of the 12 full duplicates had from one to five site mutations. The partial duplicate contained only 32 bp of the 5′ duplicate (Text S1). An AT-rich region followed the duplicates and it had an AT composition of 95.1%. This AT-rich region had five characteristic elements of the mitochondrial AT-rich control region believed to be involved in the regulation of transcription and control of DNA replication as follows: (1) a polyT stretch at the 5′end of the AT-rich region; (2) a [TA(A)]n-like stretch following the polyT stretch; (3) a stem-loop structure; (4) a TATA motif; and (5) a G(A)nT motif [24] (Figure 5). This control region was reversed and located on the light strand, a common characteristic of the Hymenoptera [25]. This inversion may have been associated with the inversion of large fragments of protein-coding genes, along with an inversion of the initiation of transcription sites.
Figure 5. Structural elements of the AT-rich region in Philotrypesis pilosa shown as reverse and its complement.
All the five elements characterized for the control region are indicated.
Discussion
In this study, we report the successful sequencing of the almost complete mitochondrial genomes from two species of Philotrypesis that shelter in the same figs. The major results are: (1) a high level of genetic divergence occurs between the two species; (2) like other mitochondrial genomes from the Chalcidoidea, Philotrypesis has dramatic gene rearrangments, not only in tRNAs, but also in several protein-coding genes; and (3) the AT-rich control region is translocated and inverted in Philotrypesis.
Rapid genetic evolution in the Chalcidoidea and adaptation for endoparasitoids
Two species of Philotrypesis live sympatrically inside figs on the same tree, and they are phylogenetically and ecologically tightly associated with one another. They have an average mtDNA nucleotide divergence of 0.140. Most of the protein-coding genes have higher divergences in amino acid sequences than their corresponding nucleotide sequences. Both of these characteristics indicate that their mitochondrial genomes evolve rapidly. Oliveira et al. show that the mitochondrial genomes of closely related species of Nasonia are also very divergent and evolving very quickly [6]. Similarly, species of Ceratosolen have very divergent co1 nucleotide sequences, and this is the most conserved gene in the group's mtDNA genome (unpublished data).
Insects in the Chalcidoidea have diverse parasitic lifestyles and perhaps their lifestyles are associated with the rapid rate of evolution of their mitochondrial genomes. This is suggested for parasitic hymenopterans [5], [13], [14]. Accelerated evolution of the mitochondrial genome may be associated with either the increased rate of speciation in parasitic Hymenoptera, adaptive radiations, or specific aspects of the endoparasitoid biology of the wasps [13].
Dramatic gene rearrangements
Mitochondrial gene rearrangement, especially for tRNAs, is common in invertebrates [11]. However, changes in the relative positions of protein-coding genes are rare. Nasonia is the first species discovered to have a large inversion spanning at least six protein-coding genes [6].
The mitochondrial genomes of C. solmsi (submitted) and two species of Philotrypesis are now known. Similar to Nasonia, these species also have dramatic mitochondrial gene rearrangements and the extent of rearrangement is greatest in Nasonia and Philotrypesis. Members of the Pteramalidae have a translocation of the protein-coding gene nad2, the relative inversion of trnK. They also have a change of the tRNA between two rRNAs. These rearrangements are consistent with the observation that mitochondrial gene rearrangments can be used in phylogenetic reconstructions, just like genome ‘morphology’ [12], because C. solmsi is in the Agaonidae and Nasonia and Philotrypesis are in the Pteramalidae (Figures 3 and 4).
A comparison of gene order among Orussus, Apis, Ceratosolen, and the Pteramalidae reveals that Ceratosolen has more plesiomorphic character states than species in the Pteramalidae (Figure 4). The most striking gene reorganization occurs downstream of cob, where Ceratosolen has an insertion of only trnR while the Pteramalidae has a large inserted fragment comprising several tRNAs plus one or two protein-coding genes. Further, whereas the orientation of trnK is not changed in Ceratosolen, it is reversed in the Pteramalidae and subsequent to the large inversion event of co1 to co3 (or to nad3 in Nasonia) in the Chalcidoidea. A third shift involves the tRNAs between the two rRNAs. The trnV in the outgroup taxa Orussus and Apis shifts to trnQ-trnA in Ceratosolen, and then to trnA in the Pteramalidae. Phylogenetic studies indicate that fig wasps do not share a common ancestor: Different lineages of chalcids are involved in many independent colonization events and the family Agaonidae may be older than all other families of fig wasps [26], [27]. Our data on mitochondrial gene rearrangement supports an older age for the Agaonidae relative to Philotrypesis. The latter appears to have colonized figs after the origin of the fig-wasp association (Figure 3).
The pattern of accelerated gene rearrangement may be correlated with parasitic lifestyles, though this is still debated [3], [28]. The rate of gene rearrangements is correlated with mitochondrial genetic diversity [29], [30] and our data show that chalcids have a rapid rate of genetic evolution. We emphasize that among the three genera we examined from the Chalcidoidea, Ceratosolen has the least amount of rearrangements, and Nasonia has fewer rearrangements than Philotrypesis. For example, nad3 is not translocated in Nasonia but it changes in Philotrypesis. With respect to lifestyles, Ceratosolen is a galler that feeds on the floret tissues of the fig and acts as one partner in the mutualism system of fig and wasp. In contrast, Nasonia and Philotrypesis are both endoparasites that feed on other insects. Indeed, Philotrypesis is parasitic to Ceratosolen in the floret inside the syconium, a compact and dark world, and it lives in a distinctly different oxygen environment from Nasonia.
In conclusion, our study presents two new mitochondrial genomes from chalcidoids including the two species of Philotrypesis. It evaluates these genomes with respect to those of other insects in the Chalcidoidea. This comparison leads to the discovery of rapid rates of evolution involving elevated nonsynonymous mutations and unusual, dramatic gene rearrangements. These changes may be correlated with parasitic lifestyles including the evolution of fig wasps in the peculiar syconia environment.
Supporting Information
Primers used in the amplification of the mitochondrial genomes.
(DOC)
Sequence divergence estimated by Ka/Ks for 11 mitochondrial genes.
(DOC)
Predicted secondary structure of the tRNAs in both mitochondrial genomes.
(DOC)
The morphological comparisons of the two Philotrypesis species. a-c: Philotrypesis pilosa; d-f: Philotrypesis sp. (a,d: body of the female; b,e: body of the male; c,f: dorsum of male's head). The two species are very similar except some minor differences As follows: female Philotrypesis pilosa, ratio of seventh and eighth Gastral tergum length about 3, and ovipositor length twice body length; female Philotrypesis sp., ratio of the seventh and eighth Gastral tergum length about 6, and ovipositor length 3 times body length; male Philotrypesis pilosa, malar space obviously shorter than length of eyes; male Philotrypesis sp., malar space larger or approximately equal to length of eyes.
(TIF)
The 1670 bp non-coding region between tRNA-S2 and tRNA-I in Philotrypesis pilosa .
(DOC)
Acknowledgments
We thank Dr. Li-Ming Niu for helping to collect specimens, Dr. Wen Xin for providing reagents for the experiments, and the anonymous reviewers for their valuable comments and suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by National Natural Science Foundation of China (NSFC grant no. 31090253, 30900137), partially by Major Innovation Program of Chinese Academy of Sciences (KSCX2-EW-Z-2, KSCX2-YW-Z-0908), by Program of Ministry of Science and Technology of the Republic of China (2006FY110500), by National Science Fund for Fostering Talents in Basic Research (Special subjects in animal taxonomy, NSFC-J0930004), and by a grant (No. O529YX5105) from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences. Manuscript preparation was supported by a Visiting Professorship for Senior International Scientists from the Chinese Academy of Sciences and by the National Sciences and Engineering Research Council (Discovery Grant A3148) to RM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boore JL. Animal mitochondrial genomes. Nucleic Acids Research. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting MF. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Molecular Biology and Evolution. 2009;26:1607–1617. doi: 10.1093/molbev/msp072. [DOI] [PubMed] [Google Scholar]
- 4.Dowton M, Campbell NJH. Intramitochondrial recombination - is it why some mitochondrial genes sleep around? Trends in Ecology and Evolution. 2001;16:269–271. doi: 10.1016/s0169-5347(01)02182-6. [DOI] [PubMed] [Google Scholar]
- 5.Dowton M, Austin AD. Increased genetic diversity in mitochondrial genes is correlated with the evolution of parasitism in the Hymenoptera. Journal of Molecular Evolution. 1995;41:958–965. doi: 10.1007/BF00173176. [DOI] [PubMed] [Google Scholar]
- 6.Oliveira DCSG, Raychoudhury R, Lavrov DV, Werren JH. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Molecular Biology and Evolution. 2008;25:2167–2180. doi: 10.1093/molbev/msn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 8.Reyes A, Gissi C, Pesole G, Saccone C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Molecular Biology and Evolution. 1998;15:957–966. doi: 10.1093/oxfordjournals.molbev.a026011. [DOI] [PubMed] [Google Scholar]
- 9.Cameron S, Lambkin C, Barker S, Whiting M. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Systematic Entomology. 2007;32:40–59. [Google Scholar]
- 10.Cameron S, Whiting M. Mitochondrial genomic comparisons of the subterranean termites from the Genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae). Genome. 2007;50:188–202. doi: 10.1139/g06-148. [DOI] [PubMed] [Google Scholar]
- 11.Dowton M, Austin A. Evolutionary dynamics of a mitochondrial rearrangement "hot spot" in the Hymenoptera. Molecular Biology and Evolution. 1999;16:298–309. doi: 10.1093/oxfordjournals.molbev.a026111. [DOI] [PubMed] [Google Scholar]
- 12.Dowton M, Castro LR, Austin AD. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome ‘morphology’. Invertebrate Systematics. 2002;16:345–356. [Google Scholar]
- 13.Castro LR, Austin AD, Dowton M. Contrasting rates of mitochondrial molecular evolution in parasitic Diptera and Hymenoptera. Molecular Biology and Evolution. 2002;19:1100–1113. doi: 10.1093/oxfordjournals.molbev.a004168. [DOI] [PubMed] [Google Scholar]
- 14.Jermiin LS, Crozier RH. The cytochrome b region in the mitochondrial DNA of the ant Tetraponera rufoniger: Sequence divergence in Hymenoptera may be associated with nucleotide content. Journal of Molecular Evolution. 1994;38:282–294. doi: 10.1007/BF00176090. [DOI] [PubMed] [Google Scholar]
- 15.Weiblen GD. How to be a fig wasp. Annual Review of Entomology. 2002;47:299–330. doi: 10.1146/annurev.ento.47.091201.145213. [DOI] [PubMed] [Google Scholar]
- 16.Hu M, Jex AR, Campbell BE, Gasser RB. Long PCR amplification of the entire mitochondrial genome from individual helminths for direct sequencing. Nature Protocols. 2007;2:2339–2344. doi: 10.1038/nprot.2007.358. [DOI] [PubMed] [Google Scholar]
- 17.Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics. 2006;37:545–579. [Google Scholar]
- 18.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 21.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 22.Ojala D, Merkel C, Gelfand R, Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980;22:393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- 23.Jiang ZF, Huang D-W, Chen LL, Zhen WQ, Fu YG, et al. Rampant host switching and multiple female body colour transitions in Philotrypesis (Hymenoptera: Chalcidoidea: Agaonidae). Journal of Evolutionary Biology. 2006;19:1157–1166. doi: 10.1111/j.1420-9101.2006.01087.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang D-X, Hewitt GM. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochemical Systematics and Ecology. 1997;25:99–120. [Google Scholar]
- 25.Wei S-j, Shi M, Sharkey M, van Achterberg C, Chen X-x. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to holometabolous insects. BMC Genomics. 2010;11:371. doi: 10.1186/1471-2164-11-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jousselin E, van Noort S, Berry V, Rasplus J-Y, Ronsted N, et al. One fig to bind them all: host conservatism in a fig wasp community unravelled by cospeciation analyses among pollinating and non-pollinating fig wasps. Evolution. 2008;62:1777–1797. doi: 10.1111/j.1558-5646.2008.00406.x. [DOI] [PubMed] [Google Scholar]
- 27.Rasplus J-Y, Kerdelhue C, Le Clainche I, Mondor G. Molecular phylogeny of fig wasps. Agaonidae are not monophyletic. Comptes Rendus De l'Academie Des Sciences Serie III, Sciences De La Vie Sciences. 1998;321:517–526. doi: 10.1016/s0764-4469(98)80784-1. [DOI] [PubMed] [Google Scholar]
- 28.Shao R, Campbell NJH, Barker SC. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera). Molecular Biology and Evolution. 2001;18:858–865. doi: 10.1093/oxfordjournals.molbev.a003867. [DOI] [PubMed] [Google Scholar]
- 29.Wei X, Daniel J, Bin T, Paul GH. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. Journal of Molecular Evolution. 2006;63:375–392. doi: 10.1007/s00239-005-0246-5. [DOI] [PubMed] [Google Scholar]
- 30.Shao R, Dowton M, Murrell A, Barker SC. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Molecular Biology and Evolution. 2003;20:1612–1619. doi: 10.1093/molbev/msg176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in the amplification of the mitochondrial genomes.
(DOC)
Sequence divergence estimated by Ka/Ks for 11 mitochondrial genes.
(DOC)
Predicted secondary structure of the tRNAs in both mitochondrial genomes.
(DOC)
The morphological comparisons of the two Philotrypesis species. a-c: Philotrypesis pilosa; d-f: Philotrypesis sp. (a,d: body of the female; b,e: body of the male; c,f: dorsum of male's head). The two species are very similar except some minor differences As follows: female Philotrypesis pilosa, ratio of seventh and eighth Gastral tergum length about 3, and ovipositor length twice body length; female Philotrypesis sp., ratio of the seventh and eighth Gastral tergum length about 6, and ovipositor length 3 times body length; male Philotrypesis pilosa, malar space obviously shorter than length of eyes; male Philotrypesis sp., malar space larger or approximately equal to length of eyes.
(TIF)
The 1670 bp non-coding region between tRNA-S2 and tRNA-I in Philotrypesis pilosa .
(DOC)