Abstract
Background
Under drought, plants accumulate the signaling hormone abscisic acid (ABA), which induces the rapid closure of stomatal pores to prevent water loss. This event is trigged by a series of signals produced inside guard cells which finally reduce their turgor. Many of these events are tightly regulated at the transcriptional level, including the control exerted by MYB proteins. In a previous study, while identifying the grapevine R2R3 MYB family, two closely related genes, VvMYB30 and VvMYB60 were found with high similarity to AtMYB60, an Arabidopsis guard cell-related drought responsive gene.
Results
Promoter-GUS transcriptional fusion assays showed that expression of VvMYB60 was restricted to stomatal guard cells and was attenuated in response to ABA. Unlike VvMYB30, VvMYB60 was able to complement the loss-of-function atmyb60-1 mutant, indicating that VvMYB60 is the only true ortholog of AtMYB60 in the grape genome. In addition, VvMYB60 was differentially regulated during development of grape organs and in response to ABA and drought-related stress conditions.
Conclusions
These results show that VvMYB60 modulates physiological responses in guard cells, leading to the possibility of engineering stomatal conductance in grapevine, reducing water loss and helping this species to tolerate drought under extreme climatic conditions.
Background
Grapevine (Vitis vinifera L.) is a fruit crop traditionally subjected to moderate or severe water stress, as this is an efficient strategy to improve fruit and wine quality (reviewed in [1,2]). Vitis species adapt well to drought conditions due to good osmotic adjustment, large and deep root systems, efficient control of stomatal aperture and xylem embolism [3,4]. The strength and timing of these responses varies between different cultivars and major differences in water stress tolerance can be found when compared to other species or hybrids from the Vitis genus [5]. Although these genotype-related variations involve different aspects of the physiology of the plant, they are largely linked to differences in stomatal conductance (gs) [6]. Stomata are microscopic pores distributed on the surface of leaves and stems, surrounded by two highly specialized guard cells. The opening and closure of the pore, in response to internal signals and environmental cues, allows the plant to cope with the conflicting needs of ensuring adequate uptake of CO2 for photosynthesis and preventing water loss by transpiration [7]. Under drought, abscisic acid (ABA) is accumulated, inducing rapid stomatal closure to limit water loss.
Increasing evidence indicates a role for transcription factors belonging to the R2R3 MYB subfamily as key modulators of physiological responses in stomata [8,9]. In particular, AtMYB60 has been shown to be differentially expressed in guard cells in response to ABA, and the loss-of function atmyb60-1 mutant displays constitutive reduction of light-induced stomatal opening and enhanced tolerance to dehydration [10]. Guard cell-specific MYB genes are thus focal points in understanding stomatal regulation in plants and represent molecular targets to modulate guard cell activity to improve crop survival and productivity during drought.
The grapevine genome has been estimated to contain a total of 279 MYB genes [11], of which 108 belong to the R2R3 subfamily [12]. A phylogenetic tree, constructed with the complete grape, Arabidopsis and rice R2R3MYB subfamilies, showed that many genes sharing similar functions were clustered in the same phylogenetic groups. Some of these clades were conserved in gene copy number (e.g. those related to trichome development) while in those controlling flavonoid synthesis several expansions events may have occurred [12].
In this work, we report the identification of two close homologues of the guard cell-related AtMYB60 gene in the grape genome, namely VvMYB60 and VvMYB30. Analysis of gene expression in grape tissues revealed that both VvMYB60 and VvMYB30 were expressed in green tissues and developing seeds. As opposite to VvMYB30, VvMYB60 transcript abundance was greatly reduced by ABA and osmotic stress. A GUS reporter gene approach in Arabidopsis showed that activity of the VvMYB60 promoter was restricted to stomatal guard cells and was down-regulated by ABA. Comparative analysis of regulatory regions revealed the presence of common guard cell-specific motifs in the promoters of the grape and Arabidopsis MYB60 genes. Finally, VvMYB60, unlike VvMYB30, fully complemented the stomatal defects of the atmyb60-1 mutant, thus indicating that VvMYB60 is a functional ortholog of the Arabidopsis AtMYB60 stomatal regulator.
Results
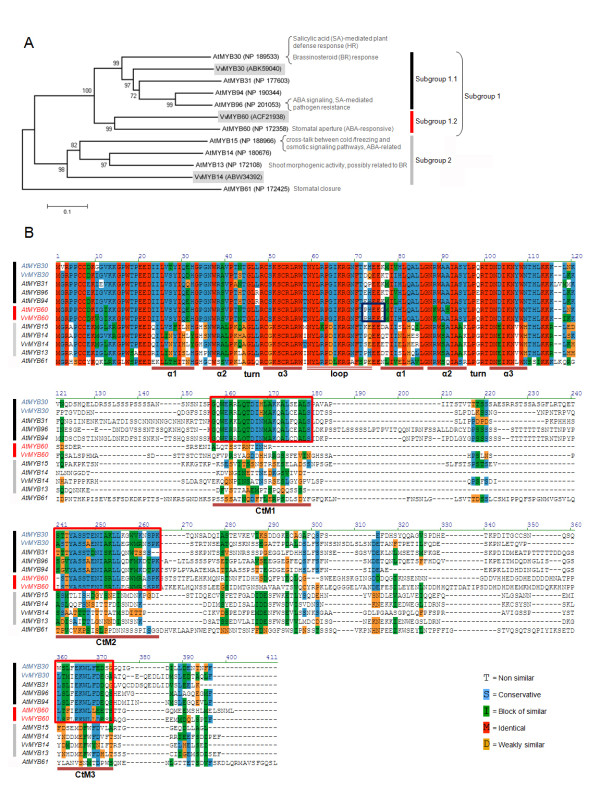
Phylogenetic relationships of MYB60 homologues
As a first approach to identify grape homologues of the AtMYB60 transcription factor, we searched the 108 R2R3 MYB proteins identified in the Vitis vinifera PN40024 genome [12], for the presence of a distinctive C-terminal motif (CtM2, YaSST/AeNIA/SR/KLl), found in members of Subgroup 1 of the Arabidopsis MYB family [13]. This subgroup includes: AtMYB60, regulating light-induced stomatal aperture [10,14]; AtMYB30, related to the regulation of brassinosteroid-induced gene expression [15] and to the biosynthesis of very-long-chain fatty acids involved in hypersensitive cell death [16]; AtMYB96, an ABA/auxin cross-talker, mediating ABA signaling during drought stress and involved in promoting pathogen resistance [17,18] and AtMYB94, whose function is still unknown.
Our search yielded two grape close homologues in the grape genome version 12x: the annotated gene models GSVIVT01008005001 (protein accession ABK59040) and GSVIVT01029904001 (protein accession ACF21938). A parsimony consensus tree was constructed to investigate the phylogenetic relationships within these grape proteins and members of Arabidopsis R2R3 MYB Subgroup 1. Subgroup 2 was also included as some of its members are involved in drought responses and ABA signaling [19,20]. From this subgroup, a grape MYB14 homologue had also been previously isolated [12]. AtMYB61, regulating stomatal activity [21], but not belonging to any of these subgroups, was included as an out-group. As shown in Figure 1A, the two grape proteins ACF21938 and ABK59040 clustered with members of the Arabidopsis Subgroup 1. Interestingly, AtMYB60, the most distant member of Subgroup 1, was more closely related to the grape protein accession ACF21938 than to the other members of the subgroup (AtMYB30, 31, 94 and 96). On the other hand, the grape accession protein ABK59040 was closely related to AtMYB30 and AtMYB31 and to a lesser extent to AtMYB94 and AtMYB96 (Figure 1A). Hereafter, we will refer to ACF21938 and ABK59040 as VvMYB60 and VvMYB30, respectively. Based on these results, we further divided Subgroup1 into Subgroup 1.1, (AtMYB30, AtMYB31, AtMYB94 and AtMYB96) and Subgroup 1.2 (AtMYB60 and VvMYB60) (Figure 1A).
Figure 1.
Analysis of grape and Arabidopsis MYB homologues within Subgroup 1. (A), Phylogenetic relationships between Arabidopsis and grape subgroups 1 and 2 of R2R3 MYB factors, as described by Kranz et al., [13]. A consensus rooted tree was inferred using the Maximum Parsimony method, constructed with MEGA4® software. (B) Alignment of deduced amino acid sequences of subgroup 1 and 2 R2R3 MYB homologues from Arabidopsis and grape. The R2 and R3 repeats lie between the three alpha helices of each repeat. Boxes represent the C-terminal motifs CtM1, CtM2 and CtM3 (red boxes) conserved in members of subgroup 1 and the PHEEG signature (blue box), distinctive of AtMYB60 and VvMYB60 (subgroup 1.2). Amino acid residues are shaded in different colors, as indicated in the legend. Dots represent gaps introduced to improve the alignment.
As expected, all the proteins included in the tree disclosed a highly conserved R2R3 DNA binding domain (Figure 1B). The identity between the R2R3 domain of AtMYB60 and VvMYB60 and VvMYB30 was 99%, and 90%, respectively. In addition, AtMYB60 and VvMYB60 disclosed a distinctive PHEEG signature, encompassing the two highly conserved glutamic acid residues, located in the loop connecting the R2 and R3 repeats (Figure 1B). The complete protein sequence of AtMYB60 showed 51% amino acid identity to VvMYB60, and 48% identity to VvMYB30. All of these proteins share two C-terminal motifs (CtM2 and CtM3) which are only found in subgroup 1. In addition, AtMYB30, 31, 96 and 94 possess a third MYB domain (CtM1), which is absent in AtMYB60 and VvMYB60 (Figure 1B). The function of these C-terminal domains is still unknown although they might reflect the functional differences between subgroups 1.1 and 1.2. We determined the precise gene structure of both VvMYB30 and VvMYB60, by comparing the complete coding sequence with the full length cDNA sequence, amplified from Pinot noir PN40024 genomic DNA and leaf cDNA, respectively (Additional file 1). It was interesting to note that in the 12x version of the grape genome, GSVIVP01008005001, representing the VvMYB60 gene model, was misannotated in terms of exon number. Indeed, our results indicate the presence of three exons, as opposed to the five exons predicted by the gene model, thus revealing a conserved exon/intron organization for VvMYB30, VvMYB60 and AtMYB60 (Additional file 1). Based on gene structure, MYB genes have been classified in four different groups [12]. VvMYB30, VvMYB60 and AtMYB60 all belong to Group I, which contains genes with a characteristic R2 domain split between exons 1 and 2, and a R3 domain split between exons 2 and 3 (Additional file 1). The biggest differences in lengths were found in the first intron and the third exon, which were longer in the grape genes compared to AtMYB60.
Expression of VvMYB30 and VvMYB60 in grape tissues and in response to hormonal and stress factors
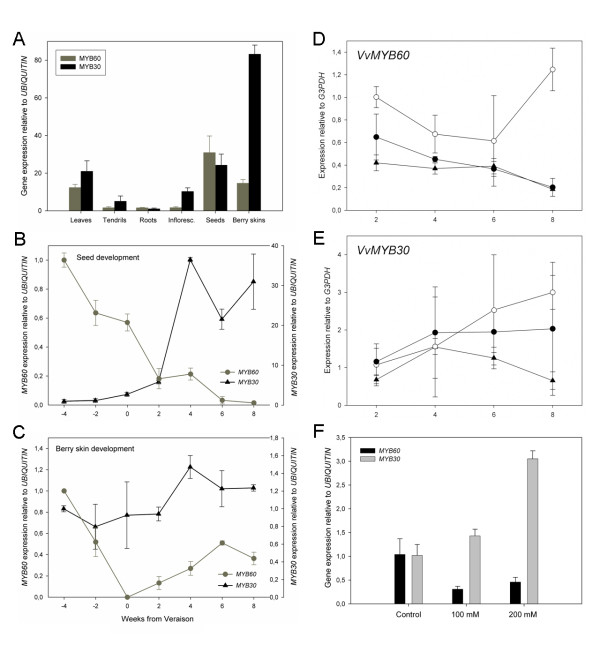
qPCR analysis of gene expression in different grape organs indicated that VvMYB30 and VvMYB60 transcripts were most abundant in leaves, seeds and ripened berry skins (Figure 2A). Interestingly, VvMYB30 and VvMYB60 revealed completely opposite expression patterns during seed development; while VvMYB60 expression was gradually down-regulated towards the onset of ripening (veraison), VvMYB30 expression was rapidly activated (Figure 2B). During berry skin development, VvMYB60 expression also showed a dramatic decrease to full repression at veraison, followed by a slight increase towards ripening (Figure 2C). In this tissue, VvMYB30 was mostly constantly expressed throughout the green and ripening stages (Figure 2C).
Figure 2.
Gene expression profiles of VvMYB60 and VvMYB30 in different plant tissues and in response to ABA. (A) Expression in grapevine organs. (B) and (C) Expression throughout berry seed and skin development (X-axis corresponds to weeks from veraison). Each gene was independently normalized. (D) and (E) Expression in response to applied ABA in leaf disks. X-axis corresponds to hours after ABA application. White circle: Mock solution, black circle: 50 μM ABA, black triangle: 100 μM ABA. (F) Expression changes of VvMYB60 and VvMYB30 in grapevine plantlets subjected to salt stress conditions. Each gene was independently normalized against its control treatment (standard MS, with 3 mM NaCl). Means and SD are the result of three independent replicates. Reference genes (UBIQUITIN and GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE) were differently selected according to the experimental condition in which they presented less variation among samples and assuming they behaved similarly as described in [43].
In Arabidopsis, it has been shown that the expression of the AtMYB60 gene is rapidly down-regulated following treatment with ABA [10]. We thus analysed the expression of the grape genes in leaves treated with 50 or 100 μM ABA (Figure 2D and 2E). As reported in Figure 2D, VvMYB60 showed a significant decrease in expression levels in both 50 and 100 μM ABA treated samples, when compared to the mock treated leaves. Conversely, VvMYB30 did not show any change in expression after exposure to the hormone under these conditions (Figure 2E). To further investigate the expression of VvMYB60 and VvMYB30 in response to osmotic stresses, we designed an in vitro long-term salt stress experiment. Nodal explants were placed vertically on sterile MS media supplemented with 3 (standard), 100 or 200 mM NaCl. Explants were maintained for a month in a growth chamber until roots and/or leaves were visible and fully expanded. At the end of the experiment, plantlets from the 100 mM NaCl treatment had a small radicule and high leaf anthocyanin accumulation, as a clear sign of stress in the plant, while plants at 200 mM showed more severe symptoms, including systemic wilting and brown pigmentation (Additional file 2). Under these conditions, VvMYB60 and VvMYB30 showed opposite responses to the increasing salt concentrations; while VvMYB60 expression was reduced five-fold at both concentrations when compared to the control treatment, VvMYB30 expression increased three-fold on addition of 200 mM NaCl (Figure 2F).
Activity of the VvMYB30 and VvMYB60 promoters in Arabidopsis transgenic lines
We employed a reporter gene approach in the heterologous model system Arabidopsis thaliana to investigate the activity of both VvMYB30 and VvMYB60 promoters. A region of approximately 2 kb located upstream of the ATG codon of VvMYB30 and VvMYB60 was fused to the β-glucuronidase (GUS) reporter gene and the resulting pVvMYB30:GUS and pVvMYB60:GUS constructs were introduced in Arabidopsis by Agrobacterium-mediated transformation [22].
We assessed the cell and tissue specificity of reporter gene expression in ten independent T3 transgenic lines for each promoter:GUS combination. Fifteen-day-old pVvMYB30:GUS seedlings displayed expression of the reporter at the shoot apex, at the base of trichomes located on leaf primordia, and in the emerging lateral roots (Figure 3A, B and 3C). At the same developmental stage, pVvMYB60:GUS seedlings showed GUS expression exclusively in guard cells distributed on cotyledons, hypocotyls and developing leaves (Figure 3D and 3E). No expression of the reporter gene was detected in rosette leaves from mature pVvMYB30:GUS plants, even after prolonged incubation of plant tissues in the GUS solution (data not shown). On the other hand, we observed guard cell-specific signals in mature leaves of pVvMYB60:GUS plants, consistent with the GUS profile observed in young seedlings (Figure 3F).
Figure 3.
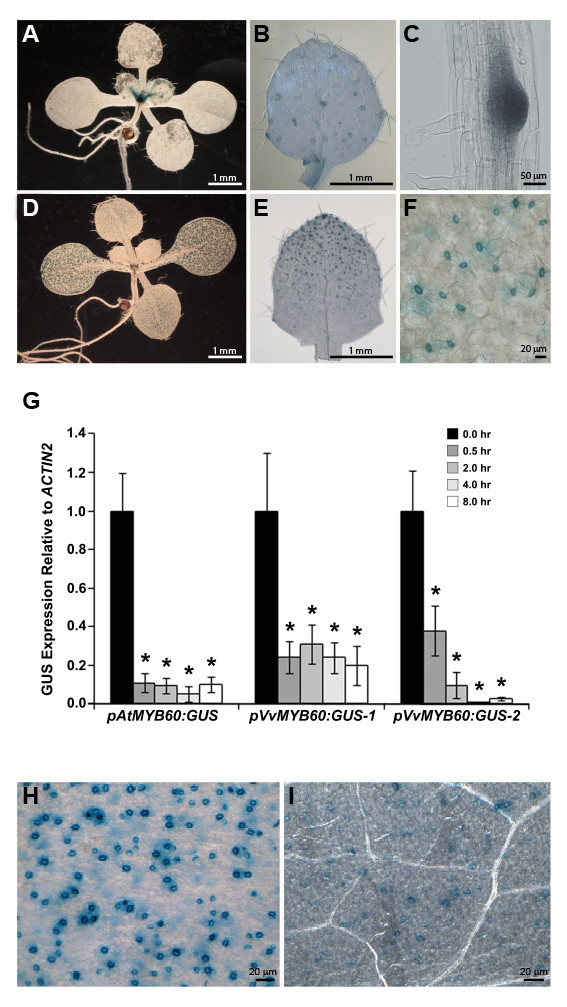
Activity of the grape VvMYB60 promoter is localized to guard cells in Arabidopsis, and is down-regulated by ABA. (A) 15-day-old pVvMYB30:GUS seedling. (B) Magnification of leaf primordia in (A), showing staining at the base of trichomes. (C) Detail of an emerging later root. (D) 15-day-old pVvMYB60:GUS seedling. (E) Magnification of leaf primordia in (D), showing staining of differentiating stomata. (F) Detail of a pVvMYB60:GUS mature leaf, showing staining of fully differentiated stomata. (G) qPCR analysis of GUS expression in response to 100 μM ABA, in two independent pVvMYB60:GUS lines (mean ± SD, n = 3). A transgenic line carrying the 1.3 kb Arabidopsis MYB60 promoter fused to GUS (pAtMYB60:GUS) was used as a control. Total RNA samples were extracted at the time points indicated (hours). Relative GUS transcript levels were determined using gene-specific primers and normalized to the expression of the AtACTIN2 gene (At3g18780). Asterisks indicate values significantly different from the untreated control (P < 0.001, t-test). (H) and (I) Histochemical analysis of GUS expression in pVvMYB60:GUS leaves in response to ABA. (H) GUS staining of stomata in a control leaf. (I), GUS staining of stomata following 6 hours of exposure to 100 μM ABA.
Next, we investigated expression of the reporter in flowers and siliques from adult plants. Prior to pollination, pVvMYB30:GUS flowers revealed a diffuse staining of carpels and stigmatic tissues (Additional file 3A). We did not observe GUS expression in pre- and post-fertilization flowers from most pVvMYB60:GUS lines. In two transgenic lines, a weak staining was occasionally detected in stamens, at the interface of filaments and anthers (Additional file 3B). Finally, we did not detect expression of the reporter in developing seeds from either pVvMYB30:GUS or pVvMYB60:GUS transgenic lines (Additional file 3C).
Expression of both the endogenous Arabidopsis and grape MYB60 genes is rapidly down-regulated following treatment with ABA [10] (Figure 2D). We thus investigated changes in GUS expression in the pVvMYB60:GUS lines in response to exogenous applications of this hormone, using both qPCR and histochemical analyses. A previously described transgenic line carrying a transcriptional fusion between the Arabidopsis AtMYB60 promoter and the reporter GUS (pAtMYB60:GUS) was used as a control for the experiment [10]. As expected, qPCR analysis of GUS expression revealed a significant and rapid decrease in the accumulation of GUS transcripts in the control pAtMYB60:GUS plants following exposure to ABA (P < 0.001) (Figure 3G). We observed a comparable reduction in GUS expression in two independent pVvMYB60:GUS lines, that were randomly selected for the qPCR experiment (P < 0.001) (Figure 3G). Staining of rosette leaves excised from all the ten pVvMYB60:GUS lines, before and after treatment with ABA, confirmed the negative effect of the hormone on the activity of the VvMYB60 promoter (Figure 3H and 3I). Conversely, treatment of pVvMYB30:GUS plants with ABA did not significantly affect the expression of the reporter (data not shown).
Occurrence of guard cell-specific motifs in the VvMYB60 promoter
The conserved activity of the Arabidopsis and grape MYB60 promoters, as emphasized by the analysis of the corresponding promoter:GUS transgenic lines, suggests that these two regulatory regions might share common cis-elements responsible for the guard cell-specific expression of the reporter. Previous evidence indicates a role for DNA consensus sequences for DOF-type transcription factors ([A/T]AAAG) as guard cell-specific cis-active enhancers [23]. Specifically, clusters of at least three [A/T]AAAG motifs located on the same strand within a region of at most 100 bp were identified as putative guard cell-specific cis-regulatory elements [24].
The Arabidopsis AtMYB60 promoter contains multiple [A/T]AAAG clusters, of which the most proximal to the translation start codon (-143 bp), is necessary and sufficient to drive expression in guard cells (Cominelli, unpublished results) (Additional file 4). We thus searched the grape VvMYB30 and VvMYB60 promoters for the occurrence of [A/T]AAAG oligonucleotides, in a region of 300 bp upstream of the translation start site. We identified a cluster of three [A/T]AAAG motifs in the VvMYB60 promoter, located at -169 bp from the ATG codon of the endogenous gene, a distance comparable to the position of the guard cell regulatory element found in the promoter of AtMYB60. Consistent with the lack of activity in Arabidopsis guard cells no [A/T]AAAG clusters were identified in the promoter of the grape VvMYB30 gene (Additional file 4).
The cellular specificity of gene expression has been investigated for a very limited number of grape genes. Among these, VvSIRK, encoding a K+ channel, has been reported to be specifically expressed in guard cells [25]. Interestingly, we discovered an [A/T]AAAG cluster upstream of the translation start codon (-200 bp) of VvSIRK, in the opposite orientation relative to the direction of transcription (Additional file 4).
Functional complementation of the Arabidopsis atmyb60-1 mutant by VvMYB60
A null allele of the Arabidopsis AtMYB60 gene (atmyb60-1) displays constitutive reduction of the opening of the stomatal pores and reduced water loss during drought [10]. Interestingly, despite its increased tolerance to dehydration relative to the wild type, the atmyb60-1 mutant does not show obvious alterations in the sensitivity of guard cells to ABA [10].
We used the atmyb60-1 allele to investigate the role of VvMYB60 in the regulation of stomatal activity and to explore the conservation of the MYB60 gene function between grape and Arabidopsis. To this end, we introduced the full length VvMYB60 cDNA in transgenic mutant plants (atmyb60-C60 lines) to assess the ability of the grape gene to rescue the stomatal defects of the atmyb60-1 allele. As a control for the complementation, we generated a second series of transgenic plants, in which we transformed the full length VvMYB30 cDNA in the atmyb60-1 background (atmyb60-C30 lines). It is important to note that the two VvMYB30 and VvMYB60 promoters displayed very different patterns of activity in Arabidopsis (Figure 3A, B, C, D, E and 3F). Hence, for a more robust and reliable comparison of the two grape genes in the atmyb60-1 background we used the 1.3 kb AtMYB60 promoter [10] to drive the expression of VvMYB30 and VvMYB60 in guard cells. Three independent transgenic mutant lines with a single insertion locus and comparable levels of expression of the transgene were selected for further analysis of each grape gene (Additional file 5).
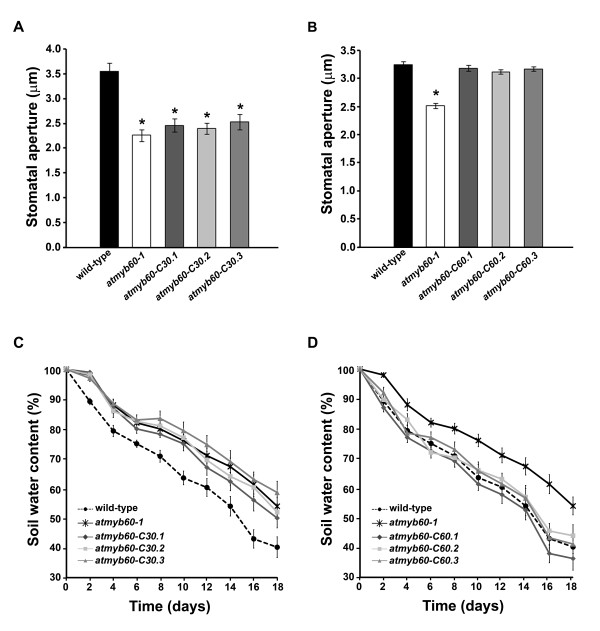
We performed an in-vitro assay to evaluate the aperture of the stomatal pore in epidermal strips excised from mutant and transgenic lines. In agreement with a previous report [10], light-induced stomatal opening was reduced in the atmyb60-1 mutant compared to the wild-type (Figure 4A and 4B). Mutant lines expressing the VvMYB30 gene did not display significant differences in the aperture of the stomatal pores compared to atmyb60-1 (Figure 4A). Conversely, all the mutant lines transformed with the VvMYB60 gene displayed a wild-type response in terms of light-induced stomatal opening, indicating full complementation of the atmyb60-1 mutation (Figure 4B).
Figure 4.
The grape VvMYB60 gene complements stomatal defects of the Arabidopsis atmyb60-1 mutant. (A) and (B) Stomatal aperture assay in wild-type, atmyb60-1 and three independent transgenic mutant lines carrying the VvMYB30 (A) or the VvMYB60 (B) gene, under the control of the guard cell-specific AtMYB60 promoter. Measurements were performed on epidermal strips excised from dark-adapted plants and exposed to light for 4 hr. Each bar indicates mean ± SD of three separate experiments (n = 100 stomata per bar). The asterisk indicates values significantly different from wild-type (P < 0.001, t-test). (C) and (D) Changes in soil water content during drought stress treatment of wild-type, atmyb60-1 and three independent mutant lines complemented with the VvMYB30 gene (C), or the VvMYB60 gene (D). Plants grown under normal watering conditions for 20 days were drought stressed by complete termination of irrigation. For clarity the responses of the atmyb60-C30 and atmyb60-C60 transgenic lines have been plotted in two different graphs. Each point indicates mean ± SD (n = 20).
To substantiate the results obtained in vitro, we investigated the effect of both VvMYB30 and VvMYB60 in vivo, by estimating whole-plant transpiration under stress conditions. Wild-type, atmyb60-1, atmyb60-C30 and atmyb60-C60 plants were grown in soil and pots were covered with tin foil to prevent evaporation, so that water loss occurring through stomatal transpiration could be quantified. Pots were regularly watered for 20 days, and subsequently drought stress was imposed by terminating irrigation. As expected, transpirational water loss, as determined by soil water content measurements, was significantly reduced in atmyb60-1 compared to the wild-type (P < 0.01 at 2, 4 and 10 days, P < 0.001 at 6, 8, 12-18 days) (Figure 4C and 4D). Consistently with results from the in vitro assay, mutant lines expressing the VvMYB30 gene did not show any difference in term of water loss compared to the atmyb60-1 mutant (Figure 4C). Conversely, under the same conditions, the lines expressing the VvMYB60 gene displayed a rate of water loss indistinguishable from the one observed in the wild-type, thus demonstrating complete rescue of the stomatal defects of the atmyb60-1 mutant (Figure 4D).
Discussion
Identification of a Grape ortholog of the AtMYB60 Transcription Factor
The MYB superfamily constitutes the most abundant group of transcription factors found in plants, with at least 198 members in Arabidopsis and 183 in rice [26]. In grape, 108 putative R2R3- MYB family genes were found in the first genome version (8x coverage) [12], whereas more than 125 R2R3 MYB genes can be found using the 12x version (Matus, unpublished results). Plant R2R3-type MYB transcription factors are implicated in several processes related to cell fate, plant development, hormonal responses, pathogen-disease resistance, drought and cold tolerance, light sensing and flavonoid biosynthesis, among many other functions [27]. MYB genes have been intensively investigated in grape, yet most studies have focused on members of the R2R3 clade involved in the regulation of the anthocyanin and pro-anthocyanidin biosynthetic pathway, as the accumulation of these flavonoid compounds in fruit tissues is a key determinant of berry and wine quality [28]. Conversely, MYB genes clustered outside the flavonoid biosynthesis functional group received little attention.
This work shows the identification of the grape VvMYB60 gene, as a functional ortholog of the Arabidopsis AtMYB60 gene, involved in the regulation of light-induced stomatal aperture [10]. Four lines of evidence support this conclusion: i) the aminoacidic sequence of the VvMYB60 and AtMYB60 proteins is highly conserved, ii) the VvMYB60 and AtMYB60 genes show very similar expression profiles, both in terms of tissue- and cell-specificity and response to ABA, iii) the VvMYB60 and AtMYB60 promoters drive expression of reporter genes exclusively in guard cells and share common cis-regulatory elements, iv) the expression of VvMYB60 in the atmyb60-1 mutant background completely rescues the loss of the AtMYB60 function.
The Arabidopsis and grape MYB60 proteins resulted more similar to each other than to any other MYB in grape or Arabidopsis, even inside subgroup 1, reason why we denoted subgroups 1.1 and 1.2 for further classification. Two main features discriminate between the Arabidopsis and grape MYB60 proteins and other closely related proteins from subgroup 1: a distinctive PHEEG signature in the MYB domain, located in the loop connecting the R2 and R3 repeats, and the lack of the first (CtM1) of three C-terminal motifs present in all the other MYB proteins assigned to subgroup 1 (Figure 1B). Notably, both characteristics are conserved in putative MYB60 orthologs that we identified in other plant genomes, including oilseed rape, tomato, cucumber and poplar (data not shown). Even though a role for the PHEEG and CtM1 motifs has not yet been described, it is intriguing to speculate that the presence of the former and the absence of the latter, might contribute to the specificity of the MYB60 function in guard cells.
Expression features of VvMYB60 in grape organs
It has been previously shown that the Arabidopsis AtMYB60 gene is expressed in seedlings, rosette leaves, stems and flowers and its level of expression is rapidly down-regulated by the stress hormone ABA [10]. In addition, publicly available repositories of microarray-based gene profiling experiments indicate that AtMYB60 is transiently expressed during seed development, peaking in stage 7 seeds (walking stick embryos) and rapidly declining in mature seeds (The Bio-Array Resource for Plant Functional Genomics, http://bar.utoronto.ca/).
Our survey of VvMYB30 and VvMYB60 expression in grape tissues revealed that both genes are preferentially expressed in leaves, berry skin and seeds (Figure 2A). Similarly to AtMYB60, and opposite to VvMYB30, expression of VvMYB60 in seeds was down-regulated during seed development (Figure 2B). In berry skin VvMYB60 expression was higher before veraison, when the grape berry is photosynthetically active and stomata are functional, and was reduced after veraison, when stomata evolve into non-functional lenticels [29] (Figure 2C). Interestingly, at this stage, the onset of ripening and the accumulation of sugars are correlated to increasing levels of ABA in the berry [30], suggesting a possible negative effect of the hormone on the expression of VvMYB60 in grape tissues. Indeed, treatment of leaves with exogenous ABA resulted in the rapid down-regulation of VvMYB60 expression (Figure 2D). In contrast, the hormone did not have any effect on the accumulation of the VvMYB30 transcripts (Figure 2E). Additionally, osmotic stresses which trigger ABA-mediated responses, as high concentrations of NaCl, caused the rapid down-regulation of VvMYB60 expression in grape tissues (Figure 2F). Interestingly, it has been recently shown that applications of low concentrations of ABA can trigger a transient up-regulation of MYB60 expression in Arabidopsis seedlings [31]. This suggests that the pattern of AtMYB60 expression in response to osmotic stress might be rather complex and dose-dependent. Even though the detailed analysis of the mechanisms that regulate the expression of the VvMYB60 gene extends beyond the scope of this work, it will be intriguing to further investigate the expression profile of VvMYB60 in different grape tissues in response to a wider range of ABA concentrations.
The VvMYB60 promoter specifically drives reporter gene expression in Arabidopsis guard cells
Reporter gene analysis and RT-PCR experiments performed on purified Arabidopsis stomata, clearly demonstrated that in green tissues, AtMYB60 is exclusively expressed in guard cells [8,24].
We produced Arabidopsis lines harboring the GUS marker gene under the control of the VvMYB30 and VvMYB60 promoters to establish the cellular localization of gene expression. Histochemical analysis of GUS expression in several independent lines indicated that the activity of the VvMYB60 promoter is restricted to guard cells (Figure 3D, E and 3F). This result is consistent with the expression of the endogenous VvMYB60 gene in leaves and berry skin, which both contain stomata, and with the lack of expression in roots (Figure 2A). Reporter gene approaches in Arabidopsis provide efficient and reliable tools to investigate the expression of grape genes and to identify gene regulatory elements [25]. Yet, we did not observe reporter activity in developing seeds of the pVvMYB30:GUS and pVvMYB60:GUS lines (Additional file 3C). This finding is in contrast with data from qPCR experiments, which showed that both genes are highly expressed in grape seeds (Figure 2A and 2B). This discrepancy could simply be artefactual, because of the heterologous genetic background. However, it is important to note that we did not detect GUS activity in developing seeds of pAtMYB60:GUS plants, used as a positive control in this study, despite the high expression of the endogenous Arabidopsis gene in these organs [32]. Different hypotheses can be formulated to explain the lack of activity of the AtMYB60 and VvMYB60 promoters in seeds. First, cis-elements responsible for the seed expression of the endogenous genes could be located outside the regulatory genomic regions considered in this work. However, Cominelli and colleagues reported that the complete 5' and 3' AtMYB60 intergenic regions, cloned upstream and downstream of the GUS gene, do not drive expression of the reporter in seeds [10]. Alternatively, expression of AtMYB60 and VvMYB60 in seeds could be mediated by intragenic regulatory elements. Cis-acting motifs, located in introns, have been demonstrated to be required to establish the correct expression domain of transcription factors, such as the MADS-box AGAMOUS gene [33]. Most interestingly, seed-specific enhancers have been mapped in the intronic regions of seed-expressed genes in different plant species [34]. Finally, the finding that the AtMYB60 mRNA is associated with polyribosomes purified from guard cells but not from other plant tissues [35], opens the possibility for a translational level of regulation for MYB60 expression in seeds. Clearly, more work is needed to unravel the nature of the cis-regulatory elements that modulate MYB60 expression in seeds, together with revealing the function of this gene in these organs. Nevertheless, it is reasonable to conclude that the stomata-specific activity of the VvMYB60 promoter in Arabidopsis mirrors the expression of the endogenous gene in grape guard cells.
While the identity of the cis-acting elements required for the expression of AtMYB60 and VvMYB60 in seeds remains elusive, their expression in stomata is most likely regulated, in cis, by DOF recognition DNA motifs. We identified a cluster of [A/T]AAAG DOF target sites in close proximity to the VvMYB60 translational start codon (Additional file 4). Such a cluster has been described as a guard cell-specific cis-regulatory element in different plant species, including Arabidopsis and potato [23,24]. The occurrence of [A/T]AAAG motifs in the guard cell-specific VvMYB60 and VvSIRK grape promoters lends further support to the conservation of the cis- and, possibly, trans-mechanisms that direct expression in guard cells in distantly related plant species. Interestingly, strong conservation across a wide range of flowering plant species has also been reported for other cell-specific cis-motifs, such as the root hair-specific cis-elements (RHEs) [36].
VvMYB60 is a functional ortholog of AtMYB60
The ability of VvMYB60 to fully complement, both in vitro and in vivo, the stomatal defects exhibited by the atmy60-1 mutant unequivocally demonstrates that VvMYB60 is a true ortholog of the Arabidopsis AtMYB60 transcription factor. Importantly, the VvMYB30 gene product, which shares 47% identity to VvMYB60, did not complement the atmyb60-1 mutation. This result is in agreement with functional studies which indicate that, despite the high degree of homology between the AtMYB30 and AtMYB60 aminoacidic sequences, these two proteins play two distinct functional roles. In Arabidopsis, AtMYB30 mediates brassinosteroid-induced gene expression [15] and pathogen-induced hypersensitive response [16], whereas AtMYB60 positively regulates light-induced stomatal opening and modulates water loss under drought [10].
As a whole, our findings indicate a role for VvMYB60 in the regulation of guard cell activity and transpiration rate in grapevine. Stomatal conductance is a key trait in grapevine, as it directly determines the isohydric/anisohydric behavior displayed by different genotypes. These differences are due to stomatal control over evaporative demand rather than stomatal density in vegetative tissues [37]. Cultivar-specific differences have also been described for the effects of water deficit on ABA metabolism and signaling [38]. Anisohydric cultivars such as Pinot Noir possess insufficient stomatal regulation and show high transpiration rates and stomatal conductance, whereas isohydric cultivars as Shiraz, display much lower values [39]. In this perspective, it will be interesting to survey variations in naturally occurring VvMYB60 alleles and to establish their contribution to differences in stomatal activity in different Vitis species and cultivars.
Conclusions
VvMYB60 could represent a valuable target for downstream biotechnological applications. Although grapevine is a highly productive water stress-adapted plant, the availability of molecular targets for engineering or breeding of new cultivars with enhanced stomatal responses represents an attractive approach to increase water use efficiency and, possibly, to reduce pathogen penetration through the stomatal pore [40,41].
Methods
Phylogeny reconstruction and bootstrap analysis
Alignments were performed using the BLOSUM matrix (Gap opening and extension penalties of 10 and 5, respectively) of the ClustalW algorithm-based AlignX® module from Mega4 Software [42]. A rooted tree was constructed using the Neighbour Joining Method (NJ) in MEGA4 and confirmed with MEGA3. Tree nodes were evaluated by bootstrap analysis for 100 replicates (pairwise deletion, uniform rates and Poisson correction options). Publicly available sequences were collected from Genbank via NCBI (http://www.ncbi.nlm.nih.gov/). The corresponding cDNAs of the complete coding sequences of VvMYB30 and VvMYB60 were amplified from PN40024 genomic DNA and leaf cDNA, respectively, using the following primer combinations: VvMYB30F1-VvMYB30R3 and Vv60L2F4-Vv60L2R4 (see below for primer sequences).
Field sampling of grape organs and nucleic acid extraction
Grapevine organs (Vitis vinifera L. cv. Cabernet Sauvignon) were collected from a commercial vineyard in the Maipo Valley, Chile, and frozen in liquid nitrogen for RNA extraction. For grape berry skin and seed sampling, a total of nine grape clusters were collected from three plants every two weeks during fruit development, beginning two weeks after fruit set and ending at eight weeks after veraison. Total RNA was isolated from all grapevine tissues as described [43]. For cDNA synthesis, one μg of total RNA was reverse transcribed with random hexamer primers using StrataScript® reverse transcriptase (Stratagene) according to the manufacturer's instructions.
ABA and salt treatment experiments
For ABA treatments, young leaves of Vitis vinifera cv. Cabernet Sauvignon were cut from two month old plantlets cultivated in vitro and placed in petri dishes supplemented with 50 μM, 100 μM ABA (+/- cis, trans ABA; SIGMA), dissolved in 100% ethanol, or with an equal amount of 100% ethanol (mock solution). Samples were maintained in a growth chamber at 20°C in the light (120 μmol m-2 sec-1 of measured light irradiance), and every two hours three leaves from each treatment were collected for RNA extraction.
For salt treatments, nodal explants of Vitis vinifera cv. Cabernet Sauvignon were placed vertically on sterile MS medium supplemented with 3 mM NaCl (standard), 100 mM NaCl or 200 mM NaCl and left for a month in a growth chamber (20°C; 16 h photoperiod). At the end of the experiment, complete plantlets were collected for RNA extraction.
Quantitative comparison of gene expression in grape tissues
Relative transcript quantification of isolated genes was achieved by real time RT-PCR, using the Brilliant® SYBR® Green QPCR Master Reagent Kit (Stratagene) and the Mx3000P detection system (Stratagene) as described in the manufacturer's manual. Primers qPCR_VvMYB30fw (5'-CTCAAGTCCCTCTCACAATG-3') and qPCR_VvMYB30rev (5'-TGTCAATTAGGTCTTCTTGTTC-3'), qPCR_VvMYB60fw (5'-TTGAGTACGAAAACCTGAATGAT-3') and qPCR_VvMYB60rev (5'-GGAGGGTTGTGCTTCTTCTGAT-3') were used for amplification and qPCR quantification of VvMYB30 (81 bp) and VvMYB60 (121 bp), respectively. Amplification of the UBIQUITIN (99 bp) or GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE (G3PDH) genes was used for normalization [44], depending on their expression variations for each experimental condition. PCR conditions, standard quantification curves for each gene and relative gene expression calculations were conducted according to Matus et al. [45].
Plasmid constructs, generation and analysis of Arabidopsis transgenic lines
To generate the pVvMYB30:GUS construct, a region of 2,173 bp upstream of the translational start codon was amplified from grape genomic DNA (Pinot noir, PN40024), using primers VvMYB30F3 (5'-AAGCTTCTGACGCAGTTTTCAACCATC-3'), containing a HindIII site, and VvMYB30R4 (5'-TCTAGAGGTGGCCTCCCCTTGGCT-3'), containing an XbaI site. The HindIII-XbaI fragment was cloned upstream of the uidA coding sequence in the pBI101.3 binary vector (Stratagene). Similarly, the 2,239 bp putative pVvMYB60:GUS promoter was amplified with primers Vv60F3 (5'-AAGCTTATGAGAGGTCGTATAAGTA-3'), containing a HindIII site, and Vv60R3 (5'-TCTAGAGGCCTTCCTATGGCTT-3'), containing an XbaI site, and the PCR fragment was cloned in the pBI101.3 vector. The VvMYB30 full length cDNA, was obtained by amplification of PN40024 leaf cDNA with the primers VvMYB30F1 (5'-GGATCCATGGGGAGGCCACCTTG-3'), containing a BamHI site and VvMYB30R3 (5'-GATATCTAGAAGAGCTGAGCAGTGTCCT-3'), containing an EcoRV site. The full length VvMYB60 cDNA was amplified with the primer Vv60L2F4 (5'-GGATCCATGGGAAGGCCTCCTTGCTGTG-3'), containing a BamHI site and the primer Vv60L2R4 (5'-GAGCTCTCAGAATATTGGAGAGAGTTGATCC-3'), containing a SacI site. Amplified cDNAs were sequenced and then cloned in a modified version of the pPZP221 binary vector, containing the 1.3 kb AtMYB60 promoter and the nos terminator (Galbiati, unpublished). All constructs were introduced in Arabidopsis (Col-0) by Agrobacterium-mediated transformation as described [22]. Transformed lines were selected on antibiotic containing media, and the presence of the transgene was confirmed by PCR. For analysis of transgene expression, total RNA was isolated with the RNeasy Mini Kit (Qiagen) and reverse-transcribed with the RT Superscript II kit (Invitrogen). Semi-quantitative RT-PCRs were performed for 25 cycles with primer pairs: Vv30F2 (5'-GGATCCATGGGAAGGCCTCCTTGCT-3') and Vv30R2 (5'-AAGTCTGACAGTGATGAGAGGAGC-3'); Vv60F2 (5'-CTCCTTGCTGTGATAAAGTTGGTAT-3') and Vv60R2 (5'-ATTCAGGTTTTCGTACTCAAGAATG-3'). The control AtACTIN2 gene (At3g18780) [46] was amplified using primers AtACT2F (5'-GTGTTGGACTCTGGAGATGGTGTG-3') and AtACT2R (5'-GCCAAAGCAGTGATCTCTTTGCTC-3'). Homozygous T3 lines were selected for GUS staining and functional complementation analyses.
Arabidopsis growth, ABA treatment and analysis of GUS expression
Seeds were surface-sterilized overnight in a sealed chamber in the presence of 100 ml of commercial bleach and 3 ml of 37% HCl, and germinated in Petri dishes containing Murashige and Skoog medium, 1% w/v sucrose and 0.8% w/v agar. Plants were grown under long-day conditions (16 h light/8 h dark, at 100 μmol m-2 sec-1) at 22°C in a controlled growth chamber. For ABA treatments, plants were transferred to liquid MS medium with 3% w/v sucrose and 0.5 g/l MES, supplemented with 100 μM ABA (+/- cis, trans ABA; SIGMA), dissolved in 100% ethanol, or with an equal amount of 100% ethanol (mock solution). For detection of GUS activity, tissues were incubated for 6 hr, at 37°C, in 0.5 mg/ml X-glucuronic acid, 0.1% Triton X-100, and 0.5 mM ferrocyanidine in 100 mM phosphate buffer (pH 7). Tissues were cleared with 70% ethanol and examined using a Leica M205 FA stereoscope or a Leica DM2500 optical microscope. qPCR analysis of GUS expression was performed as described for the grape samples, using primers qPCR_GUSF1 (5'-TACGGCAAAGTGTGGGTCAATAATCA-3') and qPCR_GUSR1 (5'-CAGGTGTTCGGCGTGGTGTAGAG-3'). GUS expression was normalized using the control AtACTIN2 gene (At3g18780) [46], amplified with primers qPCR_AtACT2fw (5'-TGCTTCTCCATTTGTTTGTTTC-3') and qPCR_AtACT2rev (5'-GGCATCAATTCGATCACTCA-3').
Stomatal aperture and water loss measurements
Stomatal assays were performed on abaxial epidermis strips, incubated in 30 mM KCl, 10 mM MES-KOH, pH 6.5, at 22°C, and exposed to light (300 μmol m-2 sec-1) for 4 h. Measurements of stomatal aperture were performed using a Leica DM2500 optical microscope and the LAS Image Analysis software. For drought experiments, seeds were germinated in individual pots each containing the same amount of pre-wetted soil. Plants were regularly irrigated for 20 days. Before watering was terminated, pots were covered with tin foil to minimize evaporation from soil. Pots were weighed every other day at the same time for 18 days. At the end of the treatment, pots were dried for three days at 65°C to determine the dry weight. Water content was estimated as [(Wtn-DW)/(Wt0-DW)] × 100, where Wtn = total weight of the pot at day n; DW = dry weight of the pot and Wt0 = total weight of the pot at day 0.
Authors' contributions
MG and JTM contributed to the conception of the study, drafted the manuscript, carried out the grape genome search and cloning of MYB60 like genes and their promoter sequences. MG, PF, FR, LC and EC, produced and analysed the Arabidopsis transgenic lines described in the work. JTM carried out the phylogenetic study and, together with PC and CM, tested VvMYB60 and VvMYB30 expression in Vitis vinifera organs and experimental conditions. CT and PAJ were involved in revising the manuscript critically for important intellectual content and gave final approval of the version to be published. All authors read and approved the final manuscript.
Supplementary Material
Deduced gene structure of AtMYB60, VvMYB30 and VvMYB60. Boxes represent exons, while black lines represent introns. The location of the ATG start codon is indicated (black arrow). Gene organization and size of exons and introns were deduced by comparing the sequence of amplified genomic and cDNA fragments. Yellow and green boxes represent exon sequences coding for the R2 and R3 repeat, respectively.
Phenotypic changes in grapevine plantlets grown in the presence of growing NaCl concentration. Pictures were taken one month after the beginning of the treatment.
Activity of the grape VvMYB360 and VvMYB60 promoters in flowers and siliques from Arabidopsis lines carrying promoter:GUS fusions. (A) GUS expression in pVvMYB30:GUS flowers was localized in carpels and stigmatic tissues (arrow). (B) Most pVvMYB60:GUS flowers did not show GUS activity, with the exception of two independent lines which disclosed staining in the distal part of the anther filament (arrow). (C) pVvMYB60:GUS siliques did not show GUS expression in developing seeds (Bars = 1 mm).
Occurrence of [A/T]AAAG motifs in the 300 bp regulatory region located upstream of the translational start codon of the AtMYB60, VvMYB30, VvMYB60 and VvSIRK genes. [A/T]AAAG nucleotides on the + strand are highlighted in yellow, whereas [A/T]AAAG nucleotides on the - strand are highlighted in pale blue. The predicted TATA box is in italic and highlighted in green, the ATG codon is highlighted in dark blue. Sequences encompassing clusters of [A/T]AAAG motifs (see text for definition) are in bold and underlined.
Generation and selection of the transgenic lines used for the complementation of the atmyb60-1 Arabidopsis mutant (atmyb60-C60 and atmyb60-C30). (A) and (B), schematic representation of the constructs used in the complementation test (not to scale). (C) and (D), RT-PCR analysis of transgene expression (VvMYB60 and VvMYB30) in three independent homozygous T3 transformed atmyb60-1 lines. (), lane 1 = atmyb60-C60-1; lane 2 = atmyb60-C60-2; lane 3 = atmyb60-C60-3; lane 4 = atmyb60-1; lane 5 = dH2O. (D), lane 1 = dH2O; lane 2 = atmyb60-C30-1; lane 3 = atmyb60-C30-2; lane 4 = atmyb60-C30-3; lane 5 = atmyb60-1. The Arabidopsis AtACTIN2 gene (At3g18780) was used as a control.
Contributor Information
Massimo Galbiati, Email: massimo.galbiati@unimi.it.
José Tomás Matus, Email: tomas.matus@cragenomica.es.
Priscilla Francia, Email: Priscilla.Francia@unimi.it.
Fabio Rusconi, Email: fabio.rusconi@studenti.unimi.it.
Paola Cañón, Email: pcanong@gmail.com.
Consuelo Medina, Email: cmedina@bio.puc.cl.
Lucio Conti, Email: lucio.conti@unimi.it.
Eleonora Cominelli, Email: eleonora.cominelli@unimi.it.
Chiara Tonelli, Email: chiara.tonelli@unimi.it.
Patricio Arce-Johnson, Email: parce@bio.puc.cl.
Acknowledgements
We thank Michael Handford (Universidad de Chile) for critically reviewing the manuscript. This work was partially supported by FONDECYT 1100709, the Chilean Fruit Consortium, CORFO-Innova 07Genoma01, Millennium Nucleus for Plant Functional Genomics (P06-009-F), by the Italian "Progetto AGER, bando Viticoltura da Vino" (SERRES, 2010-2105) and by Fondazione Umberto Veronesi per il Progresso delle Scienze, Milano, Italy.
References
- Chaves M, Zarrouk O, Francisco R, Costa J, Santos T, Regalado A, Rodrigues M, Lopes C. Grapevine under deficit irrigation: hints from physiological and molecular data. Ann Bot. 2010;105(5):661–676. doi: 10.1093/aob/mcq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Dokoozlian N, Krstic M. Cultural practice and environmental impacts on the flavonoid composition of grapes and wine: A review of recent research. Am J Enol Vitic. 2006;57(3):257–268. [Google Scholar]
- Lovisolo C, Hartung W, Schubert A. Whole-plant hydraulic conductance and root-to-shoot flow of abscisic acid are independently affected by water stress in grapevines. Funct Plant Biol. 2002;29(5):1349–1356. doi: 10.1071/FP02079. [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Chaves MM, Wendler R, David MM, Quick P, Leegood R, Stitt M, Pereira JS. Osmotic adjustment in water stressed grapevine leaves in relation to carbon assimilation. Aust J Plant Physiol. 1993;20(3):309–321. doi: 10.1071/PP9930309. [DOI] [Google Scholar]
- Flexas J, Barón M, Bota J, Ducruet J, Gallé A, Galmés J, Jiménez M, Pou A, Ribas-Carbó M, Sajnani C. et al. Photosynthesis limitations during water stress acclimation and recovery in the drought-adapted Vitis hybrid Richter-110 (V. berlandieri × V. rupestris) J Exp Bot. 2009;60(8):2361–2377. doi: 10.1093/jxb/erp069. [DOI] [PubMed] [Google Scholar]
- Escalona J, Flexas J, Medrano H. Stomatal and non-stomatal limitations of photosynthesis under water stress in field-grown grapevines. Aust J Plant Physiol. 1999;26(5):421–433. doi: 10.1071/PP99019. [DOI] [Google Scholar]
- Schroeder J, Kwak J, GJ GA. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410(6826):327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Tonelli C. Transcription factors controlling stomatal movements and drought tolerance. Transcription. 2010;1(1):1–5. doi: 10.4161/trns.1.1.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wang L, Sack F, Grotewold E. Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J. 2010;64(5):731–739. doi: 10.1111/j.1365-313X.2010.04364.x. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15(13):1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S, Reid J. et al. A High Quality Draft Consensus Sequence of the Genome of a Heterozygous Grapevine Variety. PloS ONE. 2007;2(12):e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008;8:(83). doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C. et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16(2):263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Wang F, Lian H, Kang C, Yang H. Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol Plant. 2010;3(1):246–259. doi: 10.1093/mp/ssp097. [DOI] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58(2):275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, Huard C, Blee E, Mongrand S, Domergue F, Roby D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20(3):752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P, Park C. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010;186(2):471–483. doi: 10.1111/j.1469-8137.2010.03183.x. [DOI] [PubMed] [Google Scholar]
- Seo P, Xiang F, Qiao M, Park J, Lee Y, Kim S, Lee Y, Park W, Park C. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151(1):275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281(49):37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Ding Z, Li S, An X, Liu X, Qin H, Wang D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics. 2009;36(1):17–29. doi: 10.1016/S1673-8527(09)60003-5. [DOI] [PubMed] [Google Scholar]
- Liang Y, Dubos C, Dodd I, Holroyd G, Hetherington A, Campbell M. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15(13):1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Plesch G, Ehrhardt T, Mueller-Roeber B. Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J. 2001;28(4):455–464. doi: 10.1046/j.1365-313x.2001.01166.x. [DOI] [PubMed] [Google Scholar]
- Galbiati M, Simoni L, Pavesi G, Cominelli E, Francia P, Vavasseur A, Nelson T, Bevan M, Tonelli C. Gene trap lines identify Arabidopsis genes expressed in stomatal guard cells. Plant J. 2008;53(5):750–762. doi: 10.1111/j.1365-313X.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- Pratelli R, Lacombe B, Torregrosa L, Gaymard F, Romieu C, Thibaud JB, Sentenac H. A grapevine gene encoding a guard cell K(+) channel displays developmental regulation in the grapevine berry. Plant Physiol. 2002;128(2):564–577. doi: 10.1104/pp.010529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q. et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Waterhouse A. Wine phenolics. Ann N Y Acad Sci. 2002;957:21–36. doi: 10.1111/j.1749-6632.2002.tb02903.x. [DOI] [PubMed] [Google Scholar]
- Blanke M, Pring R, Baker E. Structure and elemental composition of grape berry stomata. J Plant Physiol. 1999;154:477–481. [Google Scholar]
- Gambetta G, Matthews M, Shaghasi T, McElrone A, Castellarin S. Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta. 2010;232(1):219–234. doi: 10.1007/s00425-010-1165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Kwon Y, Kim J, Noh H, Hong S, Lee H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol Biol. 2011;77(1-2):91–103. doi: 10.1007/s11103-011-9796-7. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson G, Provart N. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissectionvof the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9(3):355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim H, Shin J, Chung C, Ohlrogge J, Suh M. Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5'-UTR intron. Mol Genet Genomics. 2006;276(4):351–368. doi: 10.1007/s00438-006-0148-2. [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti M, Jang C, Holtan H, Repetti P, Galbraith D, Girke T, Bailey-Serres J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. PNAS. 2009;106(44):18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell. 2006;18(11):2958–2970. doi: 10.1105/tpc.106.045229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogiers S, Gree D, Hutton R, Landsberg J. Does night-time transpiration contribute to anisohydric behaviour in a Vitis vinifera cultivar? J Exp Bot. 2009;60(13):3751–3763. doi: 10.1093/jxb/erp217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Quilici D, Decendit A, Grimplet J, Wheatley M, Schlauch K, Mérillon J, Cushman J, Cramer G. Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genomics. 2009;8:212. doi: 10.1186/1471-2164-10-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz H. Differences in hydraulic architecture account for near-isohydric and anisohydric behaviour of two field-grown Vitis vinifera L. cultivars during drought. Plant Cell Environ. 2003;26(8):1393–1405. doi: 10.1046/j.1365-3040.2003.01064.x. [DOI] [Google Scholar]
- Allegre M, Heloir MC, Trouvelot S, Daire X, Pugin A, Wendehenne D, Adrian M. Are grapevine stomata involved in the elicitor-induced protection against downy mildew? Mol Plant Microbe Interact. 2009;22(8):977–986. doi: 10.1094/MPMI-22-8-0977. [DOI] [PubMed] [Google Scholar]
- Pou A, Flexas J, Alsina Mdel M, Bota J, Carambula C, de Herralde F, Galmes J, Lovisolo C, Jimenez M, Ribas-Carbo M. et al. Adjustments of water use efficiency by stomatal regulation during drought and recovery in the drought-adapted Vitis hybrid Richter-110 (V. berlandieri × V. rupestris) Physiol Plant. 2008;134(2):313–323. doi: 10.1111/j.1399-3054.2008.01138.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;10(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Reid K, Olsson N, Schlosser J, Pengù F, Lund S. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6:27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Harvey J, Robinson S. Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.) Aust J Grape Wine Res. 2003;9(2):110–121. doi: 10.1111/j.1755-0238.2003.tb00261.x. [DOI] [Google Scholar]
- Matus JT, Loyola R, Vega A, Pena-Neira A, Bordeu E, Arce-Johnson P, Alcalde JA. Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J Exp Bot. 2009;60(3):853–867. doi: 10.1093/jxb/ern336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Yokota E, Wada E, Shimmen T, Okada K. An Arabidopsis ACT2 Dominant-Negative Mutation, which Disturbs F-actin Polymerization, Reveals its Distinctive Function in Root Development. Plant Cell Physiol. 2003;44(11):1131–1140. doi: 10.1093/pcp/pcg158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Deduced gene structure of AtMYB60, VvMYB30 and VvMYB60. Boxes represent exons, while black lines represent introns. The location of the ATG start codon is indicated (black arrow). Gene organization and size of exons and introns were deduced by comparing the sequence of amplified genomic and cDNA fragments. Yellow and green boxes represent exon sequences coding for the R2 and R3 repeat, respectively.
Phenotypic changes in grapevine plantlets grown in the presence of growing NaCl concentration. Pictures were taken one month after the beginning of the treatment.
Activity of the grape VvMYB360 and VvMYB60 promoters in flowers and siliques from Arabidopsis lines carrying promoter:GUS fusions. (A) GUS expression in pVvMYB30:GUS flowers was localized in carpels and stigmatic tissues (arrow). (B) Most pVvMYB60:GUS flowers did not show GUS activity, with the exception of two independent lines which disclosed staining in the distal part of the anther filament (arrow). (C) pVvMYB60:GUS siliques did not show GUS expression in developing seeds (Bars = 1 mm).
Occurrence of [A/T]AAAG motifs in the 300 bp regulatory region located upstream of the translational start codon of the AtMYB60, VvMYB30, VvMYB60 and VvSIRK genes. [A/T]AAAG nucleotides on the + strand are highlighted in yellow, whereas [A/T]AAAG nucleotides on the - strand are highlighted in pale blue. The predicted TATA box is in italic and highlighted in green, the ATG codon is highlighted in dark blue. Sequences encompassing clusters of [A/T]AAAG motifs (see text for definition) are in bold and underlined.
Generation and selection of the transgenic lines used for the complementation of the atmyb60-1 Arabidopsis mutant (atmyb60-C60 and atmyb60-C30). (A) and (B), schematic representation of the constructs used in the complementation test (not to scale). (C) and (D), RT-PCR analysis of transgene expression (VvMYB60 and VvMYB30) in three independent homozygous T3 transformed atmyb60-1 lines. (), lane 1 = atmyb60-C60-1; lane 2 = atmyb60-C60-2; lane 3 = atmyb60-C60-3; lane 4 = atmyb60-1; lane 5 = dH2O. (D), lane 1 = dH2O; lane 2 = atmyb60-C30-1; lane 3 = atmyb60-C30-2; lane 4 = atmyb60-C30-3; lane 5 = atmyb60-1. The Arabidopsis AtACTIN2 gene (At3g18780) was used as a control.