Abstract
Nocturnin is a member of the CCR4 deadenylase family, and its expression is under circadian control with peak levels at night. Because it can remove poly(A) tails from mRNAs, it is presumed to play a role in post-transcriptional control of circadian gene expression, but its target mRNAs are not known. Here we demonstrate that Nocturnin expression is acutely induced by the endotoxin lipopolysaccharide (LPS). Mouse embryo fibroblasts (MEFs) lacking Nocturnin exhibit normal patterns of acute induction of TNFα and iNOS mRNAs during the first three hours following LPS treatment, but by 24 hours, while TNFα mRNA levels are indistinguishable from WT cells, iNOS message is significantly reduced 20-fold. Accordingly, analysis of the stability of the mRNAs showed that loss of Nocturnin causes a significant decrease in the half-life of the iNOS mRNA (t1/2 = 3.3 hours in Nocturnin knockout MEFs vs. 12.4 hours in wild type MEFs), while having no effect on the TNFα message. Furthermore, mice lacking Nocturnin lose the normal nighttime peak of hepatic iNOS mRNA, and have improved survival following LPS injection. These data suggest that Nocturnin has a novel stabilizing activity that plays an important role in the circadian response to inflammatory signals.
Introduction
Living organisms require dynamic changes in protein composition to correspond to changes in the environment. Some of these changes are unpredictable – such as exposure to pathogens or toxins, while others are predictable – such as the daily cyclic changes in light levels and nutrient availability. Many types of acute signaling systems are in place to mediate the biological responses to the former, while the circadian system regulates the response to the latter, driving daily temporal changes in protein type and quantity to allow the organism to coordinate physiological events with time of day. In addition, interactions between the systems also occur since some acute responses can alter the circadian system, and the circadian system can cause changes in the magnitude and types of acute responses at different times of day. Although transcriptional changes are important for both types of response systems, it has become increasingly clear that post-transcriptional mechanisms play a critical role in dictating the protein complement of the cell. Indeed, tight control of RNA stability and/or translatability provides a way to rapidly modulate responses and quickly terminate protein production.
Nocturnin is a deadenylase, a 3′ exoribonuclease with specificity for tracts of poly(A), that is under both circadian and acute regulation. Identified originally as a rhythmic mRNA in Xenopus retina [1], [2], [3], it is expressed with high amplitude rhythms in many tissues in mice with peak levels at night [4], [5]. In addition to this circadian expression, it is also acutely inducible by a number of stimuli, exhibiting immediate early gene responses [6]. Mice lacking this gene have a metabolic phenotype, resistance to diet-induced obesity, and changes in rhythmic expression of metabolic-related genes in several different tissues/cell types [7]. Therefore, based on Nocturnin's known activity and expression pattern, our working hypothesis is that Nocturnin is involved in the post-transcriptional regulation of both rhythmic and acutely inducible mRNAs. However the specific target mRNAs upon which Nocturnin acts are not known.
Deadenylation, or poly(A) tail removal, is a rate-limiting and highly regulated step in the mRNA decay pathway, and also can regulate translatability of the message [8], [9], [10]. Nocturnin is a member of the CCR4 family of deadenylases, of which there are 5 in mice and humans [11], [12]. A single Ccr4p protein exists in yeast and interacts with Caf1p/Pop2p proteins as part of the Ccr4-Not complex – the major deadenylase in yeast [13], [14], [15]. In mammals, the members of this family all contain a conserved Mg++-dependent nuclease domain similar to that found in yeast Ccr4p and typified by the apurinic/apyrimidinic endonuclease (APE) family of proteins [11], but only two of the members of this family (called “Ccr4a” and “Ccr4b” in humans) have N-terminal extensions that contain leucine-rich repeats like those necessary for the Caf1p/Pop2p interaction in yeast. The other three members, including Nocturnin (also known as “hCcr4c” in humans) are shorter, lacking these N-terminal domains. One of these short Ccr4s in humans (hCcr4d) has been shown to be part of an “unconventional” deadenylase complex found in nuclear Cajal bodies [12] and it has been proposed by these authors that the other short Ccr4s may also have diverged to take on specialized functions. Although recombinant mouse Nocturnin has been shown to possess bona fide deadenylase activity [6], the in vitro activity is not robust compared to other deadenylases, and the human Nocturnin has been reported to lack activity, at least within the conditions of the in vitro assays and using generic substrates [12].
Because Nocturnin is an immediate early gene, and its expression is up-regulated by many acute stimuli [4], [6], we examined the promoter region of the Nocturnin gene to identify the in vivo signals responsible for Nocturnin's acute induction. This led us to the discovery of multiple nuclear factor-kappa B (NF-κB) sites, indicating Nocturnin might be induced in response to inflammatory signals. Since many mRNAs involved in the inflammatory response are highly regulated and under post-transcriptional control, we used a candidate approach to explore whether Nocturnin expression altered the stabilization of these transcripts. This led to the surprising discovery that the presence of Nocturnin stabilizes the pro-inflammatory transcript, iNOS, rather than destabilizing it. This results in significant in vivo changes in iNOS expression profiles that may impact how the circadian clock modulates the inflammatory response of the animal.
Results
Nocturnin is induced by LPS
Nocturnin has been shown to be rapidly induced in response to various acute signals in culture, but in vivo signals that induce Nocturnin are not well characterized [6]. We analyzed the genomic regions 10 kb upstream from the murine and human Nocturnin translation start sites to identify putative enhancer sites that may regulate Nocturnin's acute induction. This analysis revealed many sequences with high similarity to canonical NF-κB transcription binding sites (Figure 1A). Since NF-κB is a central regulator of inflammatory responses, we investigated whether lipopolysaccharide (LPS), could induce Nocturnin expression. Indeed, we observed a 5–6 fold induction of the Nocturnin mRNA and a 3-fold increase in Nocturnin protein levels in mouse embryonic fibroblasts (MEFs) within 3 hours following LPS treatment (Figure 1B–C).
Figure 1. Nocturnin is induced by LPS in MEFs.
(A) Putative NF-κB enhancer sites located in conserved regions in the Nocturnin gene and flanking sequence. Human and mouse genomic sequences spanning 10 Kb upstream of the Nocturnin gene were aligned and analyzed by Matlnspector [48]. Shown are relative positions of predicted conserved NF-κB binding sites (black boxes) near the mouse Nocturnin gene. +1 denotes transcription start site and negative and positive numbers denote nucleotide position within the mouse sequence relative to that site. (B,C) MEFs treated with LPS for indicated times were collected for total RNA isolation and protein extraction. iNOS mRNA levels were determined by quantitative RT-PCR and normalized to Gapdh mRNA level. Data represent mean (±SEM) of three independent MEF lines. (B). NOC protein levels were determined by Western blotting and normalized to Tubulin. The fold changes of NOC protein at different time points are indicated (C).
Nocturnin is necessary for the normal accumulation of iNOS protein following LPS stimulation
The observation that Nocturnin is induced by LPS led us to investigate whether Nocturnin plays a role in regulating the LPS-induced inflammatory response. To do this, we examined the induction profiles of TNFα and iNOS, two known LPS-responsive mRNAs, in wild-type (WT) and Noc KO MEFs. Both of the mRNAs showed significant induction by LPS after 3 hours in both the WT and Noc KO MEFs, and this induction was not significantly different between genotypes (Figure 2). By 24 hours after LPS treatment, the mRNA levels of TNFα decreased in both WT and Noc KO cells, returning nearly to basal levels (Figure 2A). In contrast, although the iNOS mRNA continues to rise in the WT cells over 24 hours, it was nearly undetectable in the Noc KO cells (Figure 2B). The Noc KO cells also had undetectable levels of the iNOS protein 24 hours following LPS, in contrast to the high levels observed in the WT cells at that time point (Figure 3A). To verify that the loss of iNOS induction in the KO MEFs was due solely to the lack of Nocturnin, we expressed heterologous Nocturnin protein in the Noc KO cells. Nocturnin expression was sufficient to rescue iNOS protein induction by LPS, and did so in a dose-dependent manner (Figure 3B). This indicates that Nocturnin is necessary for the normal accumulation of iNOS protein following LPS stimulation in MEFs.
Figure 2. iNOS mRNA is induced by LPS, but does not accumulate in Noc KO MEFs.
MEFs treated with LPS for the indicated times were collected for total RNA isolation. mRNA levels of TNFα (A) and iNOS (B) were determined by quantitative RT-PCR and normalized against β2 microglobin. Data represent mean (±SEM) of three independent MEF lines and comparison between genotypes at each time point was done by two-way ANOVA followed by Bonferroni post tests (Prism). N.S. = no significant difference between genotypes (p>0.05).
Figure 3. Accumulation of iNOS protein following LPS requires Nocturnin.
(A) iNOS protein is reduced in Noc KO MEFs following LPS treatment. WT and Noc KO MEFs treated with LPS for twenty-four hours were collected for protein extraction. Proteins were analyzed by Western blotting with α-iNOS antibody (a representative blot is shown in top panel) and protein levels were determined (normalized to β-tubulin levels) and plotted as mean (±SEM) of three independent MEF lines (bottom panel). (B) Expression of Nocturnin in Noc KO MEFs rescues the LPS induction of iNOS protein. Noc KO MEFs were electroporated with either empty FLAG vector (control) or a vector expressing a FLAG-Nocturnin fusion protein (NOC-FLAG). Forty-eight hours after electroporation, MEFs were treated for 24 hours with LPS and were then collected for protein extraction. Proteins were analyzed by Western blotting with α-iNOS and α-FLAG antibodies. Shown are extracts expressing two different levels of NOC-FLAG (compare lanes 3 and 4, top blot), which correlate with the level of rescued iNOS protein (compare lanes 3 and 4, bottom blot). The arrowhead marks a non-specific band (this band is not iNOS, as it is still present in samples from iNOS knockout mice, data not shown). The data shown are from two independently-derived WT and two independently-derived KO MEF lines (one per lane). This experiment was also repeated in a third independent MEF line for each genotype with similar results (data not shown).
Nocturnin stabilizes iNOS mRNA
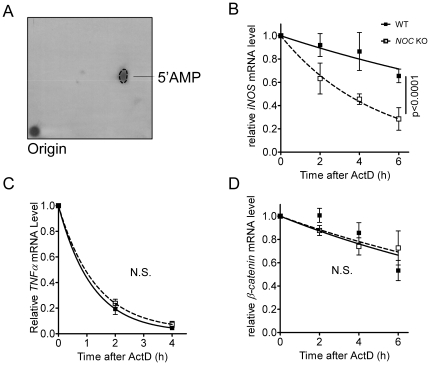
Nocturnin has deadenylase activity in in vitro experiments, removing 5′adenosine monophosphates from the 3′ end of polyadenylated substrates [Figure 4A; [1], [6]]. Removal of the poly (A) tail from transcripts typically alters the stability of the mRNAs, causing them to degrade more rapidly. To determine whether Nocturnin influences the iNOS mRNA half-life, we treated WT and Noc KO MEFs with LPS for 3 hours to induce iNOS mRNA and then blocked transcription with the addition of Actinomycin D and collected samples at different intervals to measure mRNA stability. In these experiments, we saw a clear and surprising effect of Nocturnin on the half-life of iNOS mRNA (Figure 4B). The iNOS mRNA was significantly less stable (t1/2 = 3.3 hours vs. 12.4 hours) in the MEFs lacking Nocturnin, suggesting that the presence of Nocturnin has a stabilizing effect on iNOS message. In the Noc KO MEFs we did not observe any change in the half-lives of TNFα mRNA (Figure 4C) or β-catenin (Figure 4D), a message unrelated to the inflammatory response, indicating that the Nocturnin-dependent stabilization of iNOS is transcript specific and is not a general effect.
Figure 4. Nocturnin alters iNOS mRNA half-life.
(A) Nocturnin-mediated deadenylation releases 5′AMP. Recombinant GST-NOC protein was incubated with an RNA substrate containing a 100 nt poly(A) tail labeled with [α-32P] -ATP at 37°C for 30 min. The radiolableled product of the reaction was determined by two-dimensional thin layer chromatography analysis as described in the Materials and Methods. The dotted line denotes the position of the cold 5′AMP marker. (B–D) The stability of the iNOS mRNA (B), but not TNFα (C) or β-catenin (D) is significantly reduced in the MEFs lacking Nocturnin. MEFs were treated with LPS for 3 hours before the experiment. At time 0, LPS was removed and replaced with media containing actinomycin D and cells were collected for RNA isolation at the times indicated. mRNA levels were determined by quantitative RT-PCR and normalized against Gapdh mRNA. In each case, the mRNA level at time 0 was set as 100% and data were plotted as mean (±SEM) of 4–6 independent MEF cell lines for each genotype.
Nocturnin does not localize in mRNA-storage cytoplasmic subdomains or polysomes
Considering that the deadenylase CCR4a/b can localize to processing bodies (P-bodies; [16], [17]), that a number of translationally silenced mRNAs can be stored in a deanylated form in P-bodies or stress granules —two cytoplasmic subdomains that are important for RNA storage, decay, and translational silencing [18], [19], and that the stabilized iNOS mRNA may be stored in those foci, we analyzed whether Nocturnin localizes to some of these structures. However, Nocturnin does not colocalize with markers of stress granules or P-bodies (Figure 5A). Additionally, we hypothesized that Nocturnin may deadenylate the iNOS mRNA in polysomes, inducing changes in its translational status, but polysome analysis demonstrated that Nocturnin is not present in polysomes (Figure 5B). This implies that Nocturnin may be functioning outside of the known mRNA processing pathways.
Figure 5. Subcellular localization of Nocturnin protein.
(A) Nocturnin protein does not co-localize with stress granules (SGs) or P-bodies (PBs). NIH3T3 cells were transfected with NOC-FLAG only (arsenite-induced SG and PB experiments) or pE-eGFP-G3BP1 and NOC-FLAG (G3BP1-induced SG experiments) constructs. Specific antibodies were used to detect NOC-FLAG and GE-heds (marker for P-bodies). Part of the image was amplified on the right to more clearly show the lack of overlap. Shown are representative examples from 3 independent experiments. (B) Nocturnin protein does not co-localize with polysomes. Polysome analysis was conducted by sucrose gradients. EDTA or cycloheximide was utilized to disassociate or freeze the polysomes, respectively. Fractions were collected and separated by SDS-PAGE and analyzed by Western blotting. Tubulin and ribosomal P antigen were blotted as markers of non-polysome fractions and ribosome/polysome fractions, respectively. This fractionation was performed on two independent occasions with indistinguishable results.
Nocturnin regulates iNOS rhythmicity
We next examined whether Nocturnin controls iNOS levels in vivo by measuring the levels of iNOS mRNA in WT and Noc KO mouse livers, a tissue which expresses iNOS and in which Nocturnin shows a very high amplitude rhythm of expression [Figure 6A; [5]]. Analysis of the circadian profile of basal iNOS mRNA revealed high amplitude rhythms in the liver with peak levels occurring during the late night, when Nocturnin protein is highest (Figure 6B). In Noc KO mice, however, this rhythm is lost, demonstrating that Nocturnin protein is necessary for the nighttime accumulation of the iNOS mRNA (Figure 6C).
Figure 6. Nocturnin regulates rhythmic expression of iNOS in mouse liver.
(A,B) Nocturnin mRNA and protein are rhythmic in mouse liver. Mice were maintained on a 12∶12 light-dark cycle, and liver samples were collected at different times of day as indicated. “ZT” refers to Zeitgeber Time (hrs), where ZT0 is defined as light onset and ZT12 is defined as dark onset. (A) Nocturnin mRNA levels were determined by quantitative RT PCR and normalized against β-2 microglobin. (B) Nocturnin protein levels were determined by measuring the chemiluminesence density from three independent western blots (n = 3 mice) and plotted as mean±SEM. (C) iNOS mRNA level is rhythmic in WT, but not in Noc KO, mouse livers. iNOS mRNA levels were measured in the same samples described in (A) by quantitative RT-PCR and normalized to β-2 microglobin. Data are presented as mean (±SEM) of 3–6 mice for each time point. For all the curves, data from ZT0 was re-plotted as ZT24 to improve visualization of the 24 hour cycle. Rhythmicity of curves was determined by CircWave v1.4 software (http:/hutlab.nl; R. A. Hut, University of Groningen).
Nocturnin is involved in the in vivo response to LPS challenge
LPS-induced endotoxic shock, is widely used as a model of the inflammatory responses [20], [21], [22], [23], [24] and therefore, to further explore whether Nocturnin is involved in regulating an inflammatory response in vivo, we examined the survival rate of WT and Nocturnin KO mice following injection with LPS. Blind subjective scoring of “sickness,” 24 hours following the injection, demonstrated that the WT mice were significantly more adversely affected by the LPS injection than were the KO mice (Figure 7A). This genotype-specific difference continued for several days, with decreased “survival” in the WT mice (see Methods: Figure 7B).
Figure 7. Noc KO mice are more resistant to an LPS challenge.
(A) WT mice were more adversely affected than Noc KO mice 24 hours after LPS injection. Mice were injected with 10 mg/kg of body weight of LPS at ZT4, and “sickness” of mice was assessed using subjective scoring criteria (Materials and Methods). The score 24 hours after injection were plotted as mean (±SEM) of 6 (WT) and 12 (Noc KO) mice. (B) WT mice had a lower survival rate than Noc KO mice following LPS injection. Mice described in (A) were monitored for 5 days. All mice still alive on the fifth day post-injection had a sickness score less than 3 (recovered).
Discussion
We have shown that the stability of iNOS mRNA is regulated by the circadian deadenylase Nocturnin, and we have demonstrated that Nocturnin is necessary for the circadian rhythm of iNOS mRNA levels in the liver. Our data suggest that the iNOS rhythmicity is a result of a stabilization of the iNOS mRNA by Nocturnin during the night when Nocturnin levels are high. Regulation of iNOS at the level of mRNA stability has been described in many contexts including hepatocytes and Kupffer cells of the liver [25], [26]. The 3′UTR of the iNOS transcript contains several AU-rich elements (AREs) and several ARE-binding proteins have been shown to bind to these elements, including HuR, AU-binding factor 1/heterogeneous nuclear ribonucleoprotein D (hnRNP D), and KSRP [27], [28], [29]. Additionally, several other RNA binding proteins such as hnRNP I/polypyrimidine tract-binding protein (PTB) and hnRNP L also have been shown to interact with other regions of the iNOS 3′UTR and contribute to its proper post-transcriptional control [27], [30], [31]. In hepatocytes, iNOS mRNA levels are also regulated by a fascinating mechanism in which a natural antisense transcript of the iNOS gene interacts with the iNOS message, forming a direct binding platform for the HuR protein that stabilizes the message [32].
Why does the loss of Nocturnin shorten the half-life of the iNOS mRNA? Deadenylation is generally thought to be an important regulatory step for targeting a message for degradation [8], [9], and yet loss of the Nocturnin deadenylase causes the iNOS mRNA to degrade more rapidly, calling into question Nocturnin's in vivo role. However, our data do not discriminate between direct and indirect effects of Nocturnin on the iNOS message, and it is possible that Nocturnin is destabilizing some other message that encodes an iNOS destabilizer. However, the recent observation that many RNAs can exist in stable, but short-tailed forms [33] suggests that the role of deadenylation may be more complex than previously thought. Nevertheless, we have shown that Nocturnin does not localize to known RNA storage compartments such as stress granules or P-bodies, and is not present in polysomes. Also, loss of Nocturnin in MEFs results in loss of both iNOS mRNA and iNOS protein, suggesting that Nocturnin does not simply convert the iNOS message into a stable short-tailed but translationally silent form.
It is possible that Nocturnin competes with other more robust deadenylases for certain target mRNAs. In in vitro assays, Nocturnin has a less robust activity than other deadenylases such as PARN [6], although there are many caveats in interpreting such assays with generic substrates and isolated enzymes. Perhaps a “slow” or inefficient deadenylase protects messages from rapid deadenylation and subsequent decay. Repeated attempts to measure the poly(A) tail lengths of the iNOS mRNA in our Noc KO cells were unsuccessful due to the extremely low levels of iNOS message. But support for such an idea comes from analysis of the CCR4 family members in humans where the “yeast-like” members with the extended leucine-rich N-terminal domains (CCR4a and CCR4b) have robust deadenylase activity while the other members, including Nocturnin, do not. One of the members of this latter class, Ccr4d, was shown to participate in a deadenylation complex in nuclear Cajal bodies [12]. Perhaps Nocturnin has diverged to take on a role that stabilizes mRNAs, either through its deadenylase activity or some other unknown activity.
Recent reports have shown that the circadian clock modulates the innate immune system in physiologically important ways [34], [35]. Both the clock and the immune system are under significant post-transcriptional control [36], [37], [38], [39] although the inflammatory system has been much more extensively studied in this regard. Many mRNAs that encode inflammatory response proteins have short half-lives and this instability is necessary to avoid accumulation of potentially harmful protein products in times when they are not needed [39]. Our data strongly suggest that Nocturnin plays an important role in stabilization of the hepatic iNOS message both following LPS stimulation and during the night within the circadian cycle.
Why are the basal levels of iNOS mRNA rhythmic in the liver? Perhaps this is related to a circadian change in susceptibility to inflammatory signals. It has been reported that mice exhibit significantly higher lethality to LPS and TNFα administration during their resting phase (day) and that this rhythm is under circadian control [40], [41]. Our demonstration that mice lacking Nocturnin exhibit higher survivability following LPS injection suggests that Nocturnin is playing a role in this daily change in sensitivity. Whether this resistance to LPS is due to the changes we observe on iNOS stability is not yet known and further studies will be required to more carefully examine other aspects of the innate immune response in the Noc KO mice, and to determine whether other inflammatory signals may also be Nocturnin targets.
Materials and Methods
Plasmids
To generate the Noc-FLAG construct, annealed oligos encoding three copies of FLAG tag were inserted between XhoI and SalI sites of a pCMV-Tag4A vector (Stratagene). The mouse Nocturnin cDNA was then subcloned into the EcoRI and XhoI sites. Mouse Noc (GenBank Accession Number AF199491) was cloned in a pDEST15 vector (Invitrogen) for Escherichia coli expression with an N-terminal GST-tag. pE-eGFP-G3BP1 plasmid was a generous gift of Dr. Nancy Kedersha and Dr. Paul Anderson.
Antibodies and Western blot
The custom α-Nocturnin antibody was raised in rabbits (using services provided by Sigma Life Sciences) against a recombinant mouse Nocturnin protein (NP_033964) starting from the amino acid 65 (methionine). The Ribosomal P antigen antibody was obtained from ImmunoVision, the α-FLAG antibody, the α-Tubulin antibody and the α-GE-heds antibody were obtained from Sigma and the α-iNOS antibody was purchased from Santa Cruz Biotechnology. All western blot signals were quantified by the Storm 820 Phosphorimager (GE Healthcare Life Sciences).
Animals
WT and Noc KO (C57BL/6J background) mice were maintained under a 12∶12 h light-dark cycle with free access to food and water. For circadian profiles of protein and mRNA, liver tissue was collected at 4 hour intervals over 24 hours, dark time points were collected using infrared goggles.
LPS treatment
Mice were injected intraperitoneally with 10 mg/kg body weight of LPS (Sigma) at ZT4. The “sickness” score and survival rate of mice were monitored for 5 days after LPS injection, and the experiment was terminated when all the animals alive had a score less than 3. The “sickness” scores were tallied with the observation time point (24 hours after LPS injection) according to the criteria listed below. All scoring was done “blind” with genotypes unknown to the scorer.
All LPS experiments were carried out under the supervision of veterinary staff,, and with the approval of the IACUC committee at University of Virginia. The number of animals in the procedure was kept to the smallest number necessary to obtain statistical confidence in the replicability of the results (WT 6 mice, KO 12 mice). We chose a commonly used dose (10 mg/kg body weight; [20], [21], [22]) for eliciting the endotoxic response. To minimize the pain and distress that animals might suffer, as well as for assessing the severity of the inflammatory response, a humane endpoint was determined by a small pilot study following the guideline of the IACUC committee in University of Virginia. The mice were monitored twice daily and softened food pellets were placed on the cage floor for easy access. A “sickness” score (see below) was given to each mouse during each monitoring session, and all scoring was done “blind” with genotypes unknown to the scorer. To prevent unnecessary suffering, any animal with a score of 8 or higher, and any mice with 2 or more maximum scores (in any category) were euthanized. These euthanized animals are included in the “dead” category in the survivability curve. The “sickness” score and survival rate of mice were monitored for 5 days after LPS injection, and the experiment was terminated when all the animals alive had a score less than 3. The “sickness” scores were tallied with the observation time point (24 hours after LPS injection) according to the criteria listed below.
Sickness Scoring:
Body Weight Changes: 0 = normal; 1 = <10% weight loss; 2 = 10–15% weight loss; 3 = >20% weight loss.
Physical Condition, Haircoat: 0 = normal, well-groomed; 1 = rough haircoat; 2 = rough coat, hair loss, ungroomed.
Physical Condition, Eyes and Nose: 0 = normal; 1 = eyes closed or squinted, no discharge; 2 = eyes closed or squinted, discharge or porphyrin staining
Behavior, Activity: 0 = normal; 1 = decreased activity, locomotion after slight stimulation; 2 = inactive, less alert, locomotion after moderate stimulation; 3 = self-mutilation, either very restless or immobile, or no locomotion after moderate stimulation.
Behavior, Posture: 0 = normal; 1 = sitting in hunched up position; 2 = hunched posture/head on cage floor; 3 = lying prone on cage floor.
MEFs primary cell culture
Primary cultures of MEF cells from WT and Noc KO mice were made as described in [42] and maintained in MEF media (DMEM supplemented with 10% FBS, MEM nonessential amino acids, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol, 20 mM HEPES, pH7.3, and antibiotic-antimycotic mix solution). For LPS experiments, asynchronous MEFs were cultured in MEF media without FBS and phenol red for 18 hours before LPS treatment and then replaced with fresh media containing 5 µg/ml of LPS for indicated times. For mRNA decay experiments, MEF media (without FBS and phenol red) containing 1 µg/ml of actinomycin D was added upon the removal of the LPS media. For protein analysis, cells were lysed in T-PER buffer (Pierce) with protease inhibitor cocktail (Sigma) 24 hrs after LPS addition. Rescue experiments were conducted by electroporating Noc KO MEFs with NOC-FLAG or FLAG only expression vectors using a MEF1 nucleofector kit (Lonza). Forty-eight hours after electroporation MEFs were treated for 24 hours with LPS as described above and collected for protein extraction. Proteins were analyzed by western blotting with α-iNOS and α-FLAG antibodies.
Quantitative reverse transcriptase PCR
Total RNA was isolated using Trizol reagent according to the manufacturer's protocol (Invitrogen) and reverse transcribed using random primers. The quantitative RT-PCR reaction was conducted with either SYBR Green (Invitrogen) or Taqman gene expression assay (ABI). The relative expression of indicated RNAs was normalized to an internal control using the ΔCt method (Gapdh or β2-microglobin) or ΔΔCt as described by the ABI protocol respectively. Primers for iNOS, β-Catenin, β2-microglobin, and Gapdh are obtained from Harvard Primer Bank and the sequences are: Noc 5′-ACCAGCCAGACATACTGTGC-3′, 5′-CTTGGGGAAAAACGTGCCT-3′; iNOS 5′-GTTCTCAGCCCAACAATACAAGA-3′, 5′-GTGGACGGGTCGATGTCAC-3′; β-Catenin 5′-AGTTTCCTATGGGAACAGTCG-3′, 5′-AGGTTCACTAGAACATAACACT-3′; β2-microglobin 5′-TTCTGGTGCTTGTCTCACTGA-3′, 5′-CAGTATGTTCGGCTTCCCATTC-3′; Gapdh 5′-AGGTCGGTGTGAACGGATTTG-3′, 5′-TGTAGACCATGTAGTTGAGGTCA-3′. Taqman primer sets were purchased from Applied Biosystems: TNFα-Mm00443258_m1; iNOS- Mm00440485_m1; β2-microglobin- Mm00437762_m1.
In vitro deadenylase assay and two-dimensional thin layer chromatography
The deadenylase assay was carried out as described previously [1], [6], [43], [44]. The RNA substrate was synthesized from the G52 plasmid containing the last 86 nt of the β-globin 3′UTR followed by a 100 nt poly(A) tail. Radiolabeled RNA substrate was synthesized by transcribing BamHI linearized DNA constructs with SP6 polymerase (GIBCO-BRL) in the presence of [α-32P]-ATP (800 Ci/mmol, Perkin Elmer) and 625 µM of a 5′ GpppG cap analog (Amersham Pharmacia Biotech). Resulting RNA products were separated on 6% polyacrylamide/7 M urea gels and eluted in TNES buffer (0.1 M Tris-HCl pH 7.5, 0.3 M NaCl, 0.01 M EDTA, 2% w/v SDS) overnight at room temperature. GST-Noc (final concentration 0.026 µg/µl) was incubated with purified [α-32P]-ATP-labeled RNA (final concentration 0.015 µM-0.18 µM) at 37°C for 30 min. 1 µl of the stopped reaction was spotted on a PEI cellulose plate, and developed in 15∶1∶10 isobutyric acid∶NH4OH∶H2O as the first dimension solvent, 0.75 M KH2PO4 as the second dimension solvent. Cold 5′AMP were analyzed on the same plate as a marker, and the position of it was determined under 254 nm UV light.
Polysome analysis
Polysome analysis was conducted as described previously [45]. For the EDTA group, NIH3T3 cells were harvested in polysome extraction buffer (10 mM Tris-HCl, pH 7.4; 1% TritonX-100; 15 mM MgCl2; 0.3 M NaCl; 1 mg/mL Heparin) with 30 mM EDTA. For the cycloheximide group, cells were incubated in medium containing 0.1 mg/mL cycloheximide for 3 min before harvesting and were collected in 1 ml of polysome extraction buffer with 0.1 mg/mL cycloheximide. Sample was centrifuged at 12,000×g at 4°C for 10 min and the supernatant was loaded on a 10–50% consistency sucrose gradient and centrifuged at 35,000 rpms at 4°C for 190 min in an SW41 rotor. The gradient and fraction collection were done in a Gradient Station (BioComp). Tubulin and ribosomal P antigen were blotted as markers of non-polysome fractions and polysome fractions respectively.
Immunocytochemistry
For Nocturnin subcellular localization, NIH3T3 cells were transfected with NOC-FLAG only (arsenite–induced P-bodies) or pE-eGFP-G3BP1 and NOC-FLAG (G3BP1–induced stress granules) constructs using Fugene6 transfection reagent (Roche). The α-FLAG antibody (Sigma) and α-GE-heds (PB marker; Santa Cruz) antibodies were used for staining. Immunodetection of stress granules and P-bodies was conducted according to Kedersha and Anderson [46].
Statistics
For Nocturnin mRNA induction, one-way ANOVA was used to determine the induction. For iNOS mRNA two-way ANOVA followed by Bonferroni post-tests were used to determine the induction. For the mRNA half-life data, the Global Curve Comparison (Prism) was used. For iNOS protein induction and “sickness” score comparison, we used two-tailed student's T-test. For circadian profiles, rhythmicity of curves was determined by CircWave v1.4 software [http:/hutlab.nl; R. A. Hut, University of Groningen; [47]] For survival curve comparison, we used Log-rank (Mantel-Cox) test (Prism).
Acknowledgments
We are very thankful to Abraham Chiyedath for his skilled technical assistance, Dr. Nancy Kedersha and Dr. Paul Anderson for expression clones and Dr. Todd Stukenberg and John Tooley for use of the Gradient Station for the polysome analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Institutes of Health grant R01 GM076626 (to CG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baggs JE, Green CB. Nocturnin, a Deadenylase in Xenopus laevis Retina. A Mechanism for Posttranscriptional Control of Circadian-Related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 2.Green CB, Besharse JB. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc Natl Acad Sci USA. 1996;93:14884–14888. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green CB, Besharse JC. Use of a high stringency differential display screen for identification of retinal mRNAs that are regulated by a circadian clock. Mol Brain Res. 1996;37:157–165. doi: 10.1016/0169-328x(95)00307-e. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert MR, Douris N, Tongjai S, Green CB. Nocturnin expression is induced by fasting in the white adipose tissue of restricted fed mice. PLoS One. 2011;6:e17051. doi: 10.1371/journal.pone.0017051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, et al. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001;1:9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, et al. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao D, Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 10.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 11.Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, et al. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner E, Clement SL, Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies. Mol Cell Biol. 2007;27:1686–1695. doi: 10.1128/MCB.01483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 16.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, et al. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu C, York B, Wang S, Feng Q, Xu J, et al. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franco-Molina MA, Mendoza-Gamboa E, Castillo-Leon L, Tamez-Guerra RS, Rodriguez-Padilla C. Bovine dialyzable leukocyte extract protects against LPS-induced, murine endotoxic shock. Int Immunopharmacol. 2004;4:1577–1586. doi: 10.1016/j.intimp.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Kuo SM, Tan CH, Dragan M, Wilson JX. Endotoxin increases ascorbate recycling and concentration in mouse liver. J Nutr. 2005;135:2411–2416. doi: 10.1093/jn/135.10.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Netea MG, Kullberg BJ, Joosten LA, Sprong T, Verschueren I, et al. Lethal Escherichia coli and Salmonella typhimurium endotoxemia is mediated through different pathways. Eur J Immunol. 2001;31:2529–2538. doi: 10.1002/1521-4141(200109)31:9<2529::aid-immu2529>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 24.Tracz MJ, Juncos JP, Grande JP, Croatt AJ, Ackerman AW, et al. Renal hemodynamic, inflammatory, and apoptotic responses to lipopolysaccharide in HO-1−/− mice. Am J Pathol. 2007;170:1820–1830. doi: 10.2353/ajpath.2007.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geller DA, de Vera ME, Russell DA, Shapiro RA, Nussler AK, et al. A central role for IL-1 beta in the in vitro and in vivo regulation of hepatic inducible nitric oxide synthase. IL-1 beta induces hepatic nitric oxide synthesis. J Immunol. 1995;155:4890–4898. [PubMed] [Google Scholar]
- 26.Kitade H, Sakitani K, Inoue K, Masu Y, Kawada N, et al. Interleukin 1 beta markedly stimulates nitric oxide formation in the absence of other cytokines or lipopolysaccharide in primary cultured rat hepatocytes but not in Kupffer cells. Hepatology. 1996;23:797–802. doi: 10.1053/jhep.1996.v23.pm0008666334. [DOI] [PubMed] [Google Scholar]
- 27.Kleinert H, Schwarz PM, Forstermann U. Regulation of the expression of inducible nitric oxide synthase. Biol Chem. 2003;384:1343–1364. doi: 10.1515/BC.2003.152. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Pascual F, Hausding M, Ihrig-Biedert I, Furneaux H, Levy AP, et al. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J Biol Chem. 2000;275:26040–26049. doi: 10.1074/jbc.M910460199. [DOI] [PubMed] [Google Scholar]
- 29.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, et al. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soderberg M, Raffalli-Mathieu F, Lang MA. Inflammation modulates the interaction of heterogeneous nuclear ribonucleoprotein (hnRNP) I/polypyrimidine tract binding protein and hnRNP L with the 3′untranslated region of the murine inducible nitric-oxide synthase mRNA. Mol Pharmacol. 2002;62:423–431. doi: 10.1124/mol.62.2.423. [DOI] [PubMed] [Google Scholar]
- 31.Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, et al. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J Biol Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- 32.Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, et al. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–697. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 33.Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, et al. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35:e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haus E, Smolensky MH. Biologic rhythms in the immune system. Chronobiol Int. 1999;16:581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- 35.Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- 37.Keene JD. Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol. 2007;72:157–165. doi: 10.1101/sqb.2007.72.013. [DOI] [PubMed] [Google Scholar]
- 38.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoecklin G, Anderson P. Posttranscriptional mechanisms regulating the inflammatory response. Adv Immunol. 2006;89:1–37. doi: 10.1016/S0065-2776(05)89001-7. [DOI] [PubMed] [Google Scholar]
- 40.Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med. 1960;103:142–144. doi: 10.3181/00379727-103-25439. [DOI] [PubMed] [Google Scholar]
- 41.Hrushesky WJ, Langevin T, Kim YJ, Wood PA. Circadian dynamics of tumor necrosis factor alpha (cachectin) lethality. J Exp Med. 1994;180:1059–1065. doi: 10.1084/jem.180.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J. Preparation, Culture, and Immortilization of Mouse Embryonic Fibroblasts. Curr Protoc Mol Biol. 2005:28.21.21–28.21.28. doi: 10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 43.Copeland PR, Wormington M. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA. 2001;7:875–886. doi: 10.1017/s1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baggs JE, Green CB. Functional analysis of nocturnin: a circadian clock-regulated gene identified by differential display. Methods Mol Biol. 2006;317:243–254. doi: 10.1385/1-59259-968-0:243. [DOI] [PubMed] [Google Scholar]
- 45.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. Rna. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 47.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- 48.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]