Abstract
Background
Gastric carcinoma is the second leading cause of cancer-associated death worldwide. The high mortality associated with this disease is in part due to limited knowledge about gastric carcinogenesis and a lack of available therapeutic and prevention strategies. MUC1 is a high molecular weight transmembrane mucin protein expressed at the apical surface of most glandular epithelial cells and a major component of the mucus layer above gastric mucosa. Overexpression of MUC1 is found in approximately 95% of human adenocarcinomas, where it is associated with oncogenic activity. The role of MUC1 in gastric cancer progression remains to be clarified.
Methodology
We downregulated MUC1 expression in a gastric carcinoma cell line by RNA interference and studied the effects on cellular proliferation (MTT assay), apoptosis (TUNEL assay), migration (migration assay), invasion (invasion assay) and aggregation (aggregation assay). Global gene expression was evaluated by microarray analysis to identify alterations that are regulated by MUC1 expression. In vivo assays were also performed in mice, in order to study the tumorigenicity of cells with and without MUC1 downregulation in MKN45 gastric carcinoma cell line.
Results
Downregulation of MUC1 expression increased proliferation and apoptosis as compared to controls, whereas cell-cell aggregation was decreased. No significant differences were found in terms of migration and invasion between the downregulated clones and the controls. Expression of TCN1, KLK6, ADAM29, LGAL4, TSPAN8 and SHPS-1 was found to be significantly different between MUC1 downregulated clones and the control cells. In vivo assays have shown that mice injected with MUC1 downregulated cells develop smaller tumours when compared to mice injected with the control cells.
Conclusions
These results indicate that MUC1 downregulation alters the phenotype and tumorigenicity of MKN45 gastric carcinoma cells and also the expression of several molecules that can be involved in tumorigenic events. Therefore, MUC1 should be further studied to better clarify its potential as a novel therapeutic target for gastric cancer.
Introduction
Gastric cancer is one of the most common and life-threatening cancers worldwide (for review see [1]).The poor prognosis of this disease reflects our poor understanding of its etiological factors and pathogenesis and the lack of effective treatments.
MUC1 is a high molecular weight transmembrane protein that is expressed at the apical surface of most glandular epithelial cells [2]. MUC1 is overexpressed in almost 95% of cancer cells [3], a molecular pathological feature that is associated with carcinogenesis and poor prognosis [4], [5], [6], [7], [8]. Moreover, aberrant glycosylation and loss of apical expression of MUC1 have been reported for gastric carcinomas [9], [10], [11].
MUC1 protein consists of a highly variable extracellular domain composed of a variable number of tandem repeats (VNTR), and a highly conserved cytoplasmic domain (CD), which are both essential for MUC1-driven oncogenic activities [12], [13]. The MUC1 extracellular domain can be extensively glycosylated [14] and was shown to interact with several extracellular ligands, including ICAM-1[15] and galectin-3[16]. These interactions influence cell adhesion [16], motility and migration [17], [18], metastasis [19] and cell-cell aggregation [20], which contribute to the maintenance of a normal cell phenotype, and upon disregulation contribute to tumor progression. The MUC1 cytoplasmic domain (MUC1-CD) engages in signal transduction through several residues that can be phosphorylated by receptor tyrosine kinases (and other kinases), which in turn regulate MUC1-CD affinity to other mediators of signal transduction and transcriptional regulation [for review see [21] and [22]. MUC1-CD associates with molecules such as β-catenin, c-Src, Grb2/Sos, p53, GSK-3β, EGFR and PKC-δ [for review see [22], Lyn [23] , Lck and Zap 70 [24] , ER-α [25], NFKβ [26], c-Abl [27], ATM [28] and CAML [29], that regulate processes of cell survival, proliferation, apoptosis, adhesion, migration and cell-cell aggregation. These functions of MUC1 are known to contribute to tumor progression and poor survival of cancer patients [for review see [6], [30]. Nonetheless, the relevance of MUC1 in gastric cancer progression has not been previously investigated.
In the report presented here we used retrovirus-mediated transfection of short-hairpin RNAs (shRNA) to induce a stable downregulation of MUC1 in the gastric carcinoma- derived cell line MKN45. The effects of MUC1 downregulation were studied in vitro with respect to cell proliferation, apoptosis, migration, invasion and cell-cell aggregation. MUC1 downregulated cells were more proliferative and apoptotic than the controls and exhibited lower degrees of cell-cell aggregation. No significant differences were found in terms of cell migration and invasion. Global gene expression analysis, evaluated by oligonucleotide microarrays, identified several genes influenced by MUC1 downregulation that may contribute to the observed phenotypic alterations. In vivo studies have shown that MUC1 downregulation impacts tumor development.
Materials and Methods
Cell culture
A human cell line derived from diffuse-type gastric carcinoma – MKN45 (poorly differentiated adenocarcinoma, Japan Health Sciences Foundation [31]) was grown in RPMI 1640 containing GlutamaxTMI and 25mM HEPES, supplemented with 10% fetal bovine serum (FBS) and 50 µg/ml gentamicin (Invitrogen). The packaging cell line PhoenixGP [32] was maintained in Dulbecco's Modified Eagle Medium containing GlutamaxTMI, 4,500 mg/l D-Glucose and Sodium Pyruvate, supplemented with 10% FBS and 1% (v/v) penicillin/streptomycin. Stable MUC1 downregulated clones derived from MKN45 cells were grown in standard growth medium supplemented with 5 µg/ml puromycin (Sigma). After evaluation of MUC1 levels at different time points in culture, all the assays were performed considering the time of cell culture in which the downregulation was higher, at 96 hours of cell culture. Cells were grown at 37°C with 5% CO2 in humidified atmosphere.
MUC1 downregulation strategy
MUC1 downregulated cells were produced using a retroviral expression system with short hairpin RNAs. Briefly, a 21-nucleotide sequence of the MUC1 gene, with no homology to other DNA sequences detected in a BLAST search, was chosen according to standard RNAi rules [33]. The scramble control was designed and tested for homology in a BLAST search as well. Sense and antisense oligos (Proligo) were ligated and inserted in the pSUPER.retro.puro vector (Oligoengine). The oligos used were the following: MUC1 Exon 2 (sense: GATCCCCACCTCCAGTTTAATTCCTCTTCAAGAGAGAGGAATT AAACTGGAGGTTTTTTA; antisense: AGCTTAAAAAACCTCCAGTTTAATTCCTCTC TCTTGAAGAGGAATTAAACTGGAGGTGGG; the MUC1-cDNA target region is underlined) and scramble control (sense: GATCCCCATCACCTTCGTACTCCTTA TTCAAGAGATAAGGAGTACGAAGGTGATTTTTTA, antisense: AGCTTAAAAAATC ACCTTCGTACTCCTTATCTCTTGAATAAGGAGTACGAAGGTGATGGG; the “unpaired”-cDNA target region is underlined). The MUC1 specific target or the scrambled control constructs were transfected into PhoenixGP packaging cell line by calcium-phosphate mediated transfection and transfected cells were selected using puromycin. Stable transfectants were seeded in a 6-well plate (1x106cells/well) and incubated for 24 hours at 32°C. The media containing the virus was collected, filtered through a 0.45 µm filter to remove remnant cells, and used to infect MKN45 cells, during 24 hours at 37°C. The viral supernatant was then replaced by the standard growth medium and cells were incubated 48 hours at 37°C. Efficiently transduced cells were selected and grown in standard media supplemented with puromycin. Two independent MUC1 downregulated clones (C1 and C2) were isolated and expanded for three times using cloning rings.
Immunofluorescence
MKN45 cells at 96h in culture were harvested, seeded in 12-well slides (Cell Line) and air-dried overnight at room temperature. Cells were then fixed in ice cold acetone for 5 minutes, washed twice with PBS and blocked with normal rabbit serum (DAKO) diluted 1∶5 in 10% bovine serum albumin (BSA) for 30 minutes. Serum was then replaced by the MUC1 monoclonal antibody HMFG1 (NovoCastra) diluted 1∶50 in 5% BSA, and incubated overnight at 4°C. After three washes with PBS, cells were incubated with a rabbit anti-mouse FITC labeled antibody (DAKO) diluted 1∶70 in 5% BSA for 30 minutes in the dark at room-temperature. Cells were washed 3 times with PBS and mounted in vectashield (Vectorlabs). Images were acquired in a Leica DMIRE2 fluorescent microscope. Results are representative of three independent experiments.
Protein extraction and Western blot
MKN45 cells were cultured in 60-mm dishes to 80–90%-confluence at 96h in culture. After washing twice with PBS, lysis buffer (10mM Tris pH 7.4, 150mM NaCl, 0.1% (p/v) SDS, 1mM PMSF, 1% (v/v) Triton X-100) was added and cells were scraped. Lysates were incubated on ice for 1 hour and centrifuged for 2 minutes at 4°C to collect the supernatants. Protein content was assessed by the bicinchoninic acid method (Pierce), as described in the manufacturer's instruction manual. Protein extracts were analyzed by a 4–10% SDS-PAGE (Invitrogen), transferred to a nitrocellulose membrane (Amersham Biosciences), and blotted overnight at 4°C with anti MUC1-Ab5 monoclonal antibody (1∶300, ThermoScientific), anti-beta-actin polyclonal antibody (1∶8,000, Sigma), anti ERK1/2(1∶1,000, Cell Signaling Technology) and anti β-catenin (1∶1,000, BD Transduction Laboratories) in 5% non-fat milk in TBS-0.1%Tween20 (Sigma). Membranes were washed 3 times with TBS-0.1%Tween20 and the primary antibodies were revealed using goat anti-mouse peroxidase-conjugated antibody (1∶1,000, DAKO) in 5% non-fat milk in TBS-0.1%Tween20, followed by ECL detection kit (BioRad). Results are representative of three independent experiments.
RNA extraction and Real-Time PCR
Total RNA was isolated from MKN45 cells at 96h in culture using TriReagentTM (Sigma), according to the manufacturer's instructions. 5 µg of RNA were primed with random hexamers (Invitrogen) and reverse transcribed with Superscript II (Invitrogen) in a final volume of 20 µl. 2 µl of a 1∶10 dilution of cDNA were amplified with 300nM of each primer and SYBRGreen (Applyed BioSystems) in a final volume of 20 µl, using the fluorescence reader ABI Prism 7000. Each sample was run in duplicate. The primers used were the following: MUC1 (sense: CTCCTTTCTTCCTGCTGCTG, antisense: CTGGAGAGTACGCTGCTGGT) and 18S (sense: CGCCGCTAGAGGTGAAATTC, antisense: CATTCTTGGCAAATGCTTTCG), and their specificity was confirmed using the software BLASTn on-line and by melt curve analysis. For each sample, the level of 18S RNA was measured and used for normalization of target genes abundance. Relative mRNA levels were then calculated using the comparative Ct method [34]. Data is expressed as a ratio of the results obtained with each clone and the scramble control, from three independent experiments. Statistical analysis was performed using the Mann-Whitney test.
MTT proliferation assay
Cells were plated in triplicate in 96-well plates at 5,000 cells per well and incubated at normal conditions for 96h for MKN45 cells. At each time point, the medium was removed and cells incubated with 20 µl of MTT solution (5mg/ml, Sigma) for 3 hours at normal conditions. MTT was removed and 200 µl of DMSO were added to each well to dissolve formazan. Finally, formazan optical density was measured using a microplate reader at a wavelength of 540nm. The relative growth was defined as the following formula: Relative Growth = (A540nm at Tn / A540nm at T024h). Data is expressed as a ratio of the results obtained with each clone and the scramble control, from three independent experiments. Statistical analysis was performed using the Mann-Whitney test.
Terminal Transferase dUTP Nick End Labeling (TUNEL) assay
Post-confluent cells at 96h in culture were harvested and fixed with 4% paraformaldehyde in PBS for 15 minutes. Fixed cells were seeded in 12-well slides (Cell Line) and air-dried overnight at room temperature. Following washing with PBS, cells were permeabilized with ice-cold freshly-made PBSTrCit solution (PBS + 0.1%TritonX + 0.1% Sodium Citrate) for 2 minutes on ice. Cells were washed again twice, and incubated with TUNEL reaction mix (enzyme solution, label solution and dilution buffer, 1∶9∶10, In Situ Death Detection Kit, Fluorescein, Roche) for 1 hour at 37°C. Two additional washing steps were performed and slides were mounted in Vectashield with DAPI (Vectorlabs). Results were analyzed under a Leica DMIRE2 fluorescent microscope and data is expressed as a ratio of the results obtained with each clone and the scramble control, from three independent experiments. Statistical analysis was performed using the Mann-Whitney test.
Migration assay
MKN45 cells were cultured in 60-mm dishes for a full-confluence at 96h in culture. The epithelial cells monolayer was then washed with PBS and wounded with a 10 µl micropipette tip. Non-adherent cells were removed by washing twice with PBS. Images of cells at the edge of the wound were acquired automatically at 20x magnification in a Leica DMIRE2 fluorescence microscope with a Leica DFC Twain camera for 144 frames at 10-minute intervals (corresponding to 24 hours) controlled by Leica FW4000 software. Frames from 0, 6, 12, 18 and 24 hours were used to quantify the percentage of migration: a grid of 50x30 squares was used to fulfill the wound space and the percentage of migration was calculated by the number of squares occupied by cells at each time point. Data is expressed as a ratio of the results obtained with each clone and the scramble control, from three independent experiments. Statistical analysis was performed using the Mann-Whitney test.
Matrigel invasion assay
Cell invasion was studied by using BD Biocoat™ Matrigel™ invasion chambers with 8-µm size pores (BD Biosciences), according to the manufacturer's instructions. MKN45 cells at 96h in culture were harvested and seeded in duplicate at 250,000 cells per insert (sized for 24-well plates) in 1% FBS containing medium, and 20% FBS containing medium was added to the bottom of the growth well, as an attractant. Cells were allowed to invade for 22 hours (37°C, 5% CO2 atmosphere). The non-invading cells were then swabbed from the top of the inserts and the invading cells on the lower surface were fixed with 100% methanol and stained with DAPI for 15 minutes in the dark. The membranes were removed and cells were counted under a Leica DMIRE2 fluorescence microscope. Data are expressed as a ratio of the results obtained with each clone and the scramble control, from three independent experiments. Statistical analysis was performed using the Mann-Whitney test.
Cell-cell Aggregation assay
MKN45 cells at 96h in culture were harvested and seeded in duplicate at 250,000 cells per well in 24-well plates. Plates were placed at 37°C with constant stirring (150rpm) for 1 and 2 hours. Cells were fixed with 100 µl of 25% glutaraldehyde at time zero and at the end of the incubation. Aggregates were photographed under a light microscope and isolated cells were counted (cells in duplicates were counted as isolated cells). The aggregation index was defined as the following formula: Aggregation index = 1- (number of isolated cells at Tn / number of isolated cells at T0). Data are expressed as a ratio of the results obtained with each clone and the scramble control, from three independent experiments. Statistical analysis was performed using the Mann-Whitney test.
Gene expression analysis
The expression of 12,135 genes in MUC1 downregulated clones and the respective scramble control were evaluated following the same protocol as before [35]. Briefly, following RNA extraction (as described previously), cDNA was obtained by reverse transcription, during which labeled nucleotides were incorporated: MKN45-C1 and MKN45-C2 cDNAs were labeled with Cy3 (green emission) and MKN45-SC control with Cy5 (red emission). After hybridization, the mixture was hybridized with the array overnight and then the array was digitalized with the ScanArray4000 (Perkin-Elmer) system and fluorescence analyzed by the QuantArray software package (Perkin-Elmer).
Normalization and background subtraction were performed and ratios for downregulated clones /Scrambled Control and Scrambled Control/downregulated clones were calculated using Microsoft Excel software. Gene expression with a ratio higher than 2 was considered statistically significant.
All data is MIAME compliant and that the raw from the microarray experiments were uploaded onto the Gene Expression Omnibus Database http://www.ncbi.nlm.nih.gov/geo (Geo accession numbers: GSM717858 and GSM717859).
In Vivo tumor growth Assays
Six-week-old female N:NIH(s)II:nu/nu nude mice were obtained previously from the Medical School, University of Cape Town in 1991 and maintained and housed at IPATIMUP Animal House at the Medical Faculty of the University of Porto, in a pathogen-free environment under controlled conditions of light and humidity. Males and females, aged 6–8 weeks, were used for in vivo experiments. Animal experiments were carried out in accordance with the Guidelines for the Care and Use of Laboratory Animals, directive 86/609/EEC. Mice were subcutaneously injected in the dorsal flanks using a 25-gauge needle with 1x105 of MKN45-SC (2 male and 2 female mice) or MKN45-C2 cells (3 male and 3 female mice). Mice were weighed, and tumor width and length were measured with calipers every week. Mice were euthanized 21 days after cell injection, at the time when the first tumor reached maximum allowable volume. For statistical analysis, the Mann Whitney test-StatView Software version 5.0 (SAS Institute, Cary, NC) was used. A P value of less than 0.05 was considered as statistically significant.
Results
MUC1 downregulation by shRNA
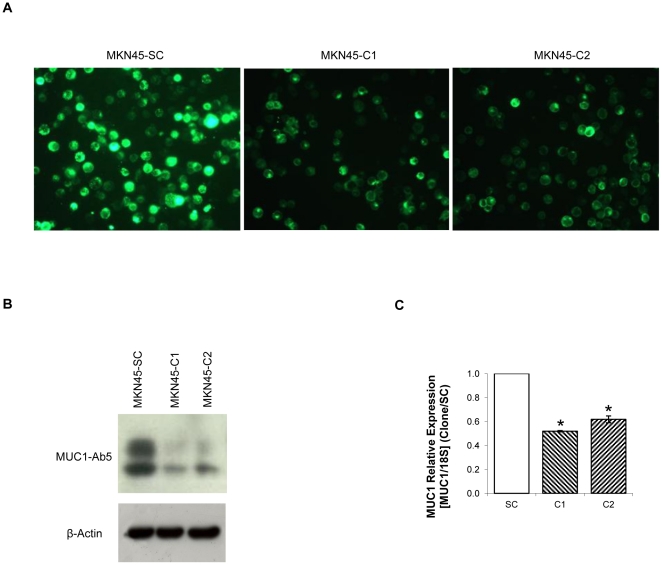
We established two independent MUC1 downregulated clones, MKN45-C1 and MKN45-C2 and one scramble control, MKN45-SC. MUC1 downregulation was verified by immunofluorescence (Figure 1A), Western Blot (Figure 1B) and Real-Time PCR (Figure 1C).
Figure 1. MUC1 downregulation by shRNA.
(A) MUC1 detection by immunofluorescence with HMFG-1 antibody in MKN45-C1 and MKN45-C2 and MKN45-SC control; (B) MUC1 protein detection by western-blot with MUC1-Ab5 antibody of total protein extracts from MKN45-C1 and MKN45-C2 and MKN45-SC control; (C) Quantification of MUC1 RNA in MKN45-C1 and MKN45-C2 and MKN45-SC control by real-time PCR. MUC1 expression was corrected to the house-keeping gene 18S and normalized to the data obtained with the scrambled control. *P<0.01.
There was a significant downregulation of MUC1 expression in MKN45-C1 and MKN45-C2 clones as compared to the MKN45-SC control. The expression of MUC1 at the protein level was detected with two different antibodies, one that binds the VNTR extracellular domain (HMFG-1, Figure 1A) and other that recognizes a 14–28 KDa sequence in MUC1 cytoplasmic domain (MUC1-Ab5, Figure 1B). Both showed a significant reduction in the amount of MUC1 protein in MKN45-C1 and MKN45-C2 clones when compared to the scramble control. Real-Time PCR results indicate that the MUC1 downregulation was 48% (MKN45-C1) and 38% (MKN45-C2) (Figure 1C). MUC1 RNA levels were evaluated at 48, 72 and 96h of cell culture and the highest downregulation occurred at 96h (results not shown).
Effects of MUC1 downregulation on MKN45 cells
Cell proliferation
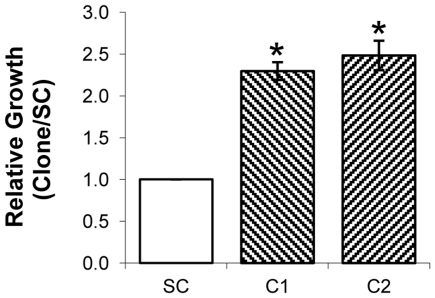
MKN45-C1 and MKN45-C2 cells showed significantly increased proliferation rates (P<0.01) when compared to the MKN45-SC control (2.29 and 2.48 vs 1), when evaluated by MTT assay (Figure 2).
Figure 2. Quantification of cell proliferation by MTT assay.
Quantification of metabolically active cells by MTT assay in MKN45-C1 and MKN45-C2 clones and MKN45-SC control at 96h in culture. Data from 24 hours was used to set time zero and results were normalized to the data obtained with the scrambled control.*P<0.01.
Cell apoptosis
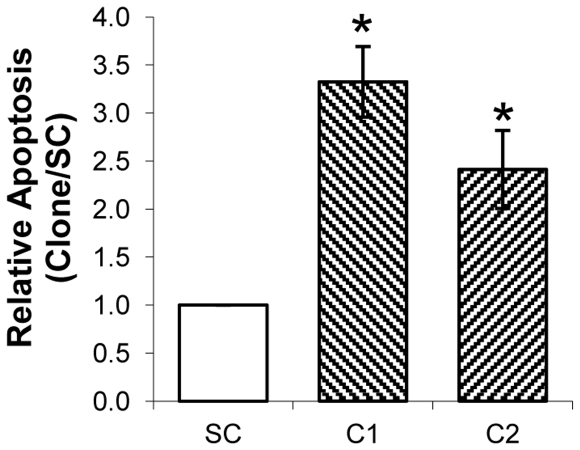
MKN45-C1 and MKN45-C2 cells showed significantly increased levels of apoptosis (P<0.01) when compared to the MKN45-SC control (3.32 and 2.41 vs 1), when evaluated by a TUNEL assay (Figure 3).
Figure 3. Quantification of apoptotic cells by TUNEL assay.
Apoptosis of MKN45-C1 and MKN45-C2 and scramble control (SC) were evaluated at 96h in culture by the TUNEL assay. Results were normalized to the data obtained with the scrambled control. *P<0.01.
Cell-cell aggregation
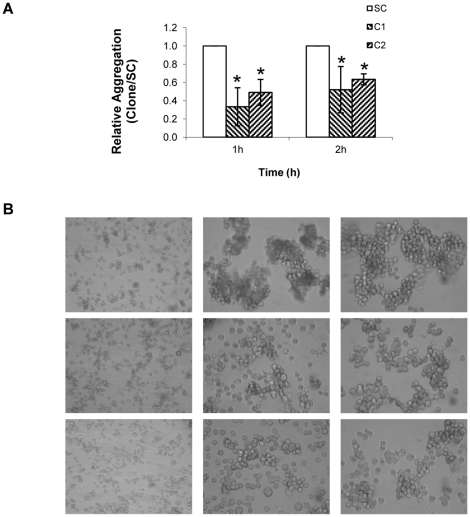
MKN45-C1 and MKN45-C2 cells showed significantly decreased cell-cell aggregation levels (P<0.01), when compared to the MKN45-SC control (0.34 and 0.49 vs 1 at 1h; 0.52 and 0.64 vs 1 at 2h), when evaluated by a cell aggregation assay (Figures 4A and 4B).
Figure 4. Quantification of cell-cell aggregation.
(A)Quantification of the cell-cell aggregation index in MKN45-C1 and MKN45-C2 and MKN45-SC control. The cell-cell aggregation index was assessed by the observed decrease in the number of isolated cells over time, and normalized to the data obtained with the scrambled control. *P<0.01; (B) Images of the aggregates formed after 1 and 2 hours of constant stirring. First column shows isolated cells at time 0h (20x magnification) and second and third columns show aggregates formed after 1h and 2h of incubation (40x magnification), in MKN45-C1 and MKN45-C2 and MKN45-SC control.
Cell migration and invasion
There were no significant differences in cell migration and invasion of MKN45-C1 and MKN45-C2 cells when compared to the MKN45-SC control, when evaluated by motility and invasion assays (results not shown).
Effects of MUC1 downregulation in MKN45 cells gene expression
The observed phenotypic modifications associated were likely due in part to alterations in signal transduction pathways mediated by MUC1-CD, since overexpression of MUC1 has been shown to modulate gene expression through reprograming transcription of multiple genes [36], [37]. We evaluated the net effects of downregulating MUC1 in the MKN45 gastric carcinoma cell line by performing a global analysis of gene expression by oligonucleotide microarrays (Table 1). The results revealed that a number of genes that influence proliferation, migration, invasion and motility were differentially expressed in MKN45-C1 and MKN45-C2 and the MKN45-SC control. The most significant differences were found for TCN1, KLK6 and ADAM29 (>10 fold upregulated between the MKN45-C1 and MKN45-C2 clones and the MKN45-SC control) and LGALS4, TSPAN8 and SHPS-1 (>3.5 fold downregulated between the MKN45-C1 and MKN45-C2 clones and the MKN45-SC control).
Table 1. Oligonucleotide microarray results by comparison between MKN45-C1/MKN45-C2 and MKN45-SC control cells, by order of magnitude.
| Genes upregulated >2 fold in MUC1 downregulated clones |
| Transcobalamin 1 (TCN1) |
| Kallikrein-related peptidase 6 (KLK6) |
| Desintegrin and metalloproteinase 29 (ADAM29) |
| Keratoepithelin (TGFBI) |
| MRP family of ATP transport member 2 (ABCC2) |
| Amyloid beta precursor-like protein 2 (APLP2) |
| Mitochondrial ATP synthase (ATP5I) |
| Sulfide dehydrogenase like protein (SQRDL) |
| Sarcoglycan, epsilon (SGCE) |
| Hypothetical protein (FLJ20323) |
| Galectin 1 (LGALS1) |
| Proline-histidine rich protein (PHLDA1) |
| Trypsin 2 (PRSS2) |
| Mesotrypsin (PRSS3) |
| SP2 transcription factor (SP2) |
| Ubiquitin-conjugating enzyme (UBE2L6) |
| Vitellogenic-like carboxypeptidase (CPVL) |
| Genes downregulated >2 fold in MUC1 downregulated clones |
| Galectin 4 (LGALS4) |
| Tetraspanin 8 (TSPAN8) |
| Tyrosine phosphatase SHP substrate (SHPS-1) |
| Polymerase (DNA-directed), delta 4 (POLD4) |
| H2B histone family, member J (HIST1H2BH) |
| H2B histone family, member T (HIST1H2Bk) |
| Carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) |
| Annexin IV (ANXA4) |
| Intercellular adhesion molecule 4 (ICAM4) |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 (DDX39) |
| Apolypoprotein B (APOBEC2) |
| Clusterin (CLU) |
| GDP-mannose 4,6-dehydratase (GMDS) |
| Serine/threonine kinase 38 like (STK38L) |
| CD55 (CD55) |
| Apolipoprotein B-catalytic polypeptide-like 3C (APOBEC3C) |
| Cell adhesion related-molecule (CDON) |
| Villin-1 (VIL1) |
MKN45-C1 and MKN45-C2 and the MKN45-SC control were analysed by oligonucleotide microarrays. Listed are genes with expression increased or decreased more than 2 fold in both MUC1 downregulated clones when compared to the control.
In vivo tumor growth assays
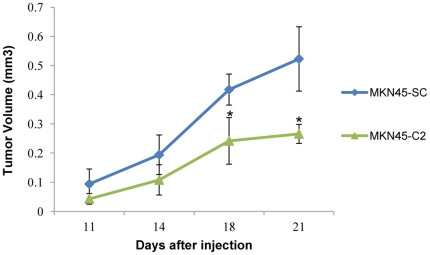
In vivo tumorigenicity assays showed that mice injected with MUC1-downregulated cells (MKN45-C2) developed smaller and slower-growing tumors, when compared to mice injected with the MKN45-SC control cells (Figure 5).
Figure 5. Study of the tumorigenicity of MKN45 gastric carcinoma cells in vivo.
Tumor growth curves. 1×105 cells were subcutaneously injected in mice at day 0. The curves show tumor growth until day 21, the day on which all mice were sacrificed.* P<0.05, when compared to the MKN45-SC control cell line.
Discussion
In the work presented here, we evaluated the effects of MUC1 downregulation on cancer-related properties of MKN45 gastric carcinoma cells. Stable downregulation of MUC1 expression was achieved in MKN45 gastric carcinoma cell line by RNA interference. MUC1 contributes to tumor progression of adenocarcinomas and therefore its downregulation was predicted to affect the malignant properties of cancer cells, including proliferation, apoptosis, migration, invasion and cell-cell aggregation.
We found that proliferation was significantly increased in MUC1 downregulated clones MKN45-C1 and MKN45-C2 when compared to the control MKN45-SC. Similar studies with breast and pancreatic carcinoma cell lines have shown similar [38] and opposite [35], [38], [39] results. In different tumor models, MUC1 was shown to regulate cell proliferation by interacting with several proteins such as ER-α, β-catenin and EGFR [25], [40], [41]. However, for gastric carcinoma cells, such interactions have not been investigated. Results obtained by an oligonucleotide microarray analysis showed that expression of molecules affecting cell proliferation such as KLK6 and LGAL-4 [42], [43], [44] were significantly altered in MKN45-C1 and MKN45-C2 clones when compared to the MKN45-SC control. KLK6 expression was increased in MKN45-C1 and MKN45-C2 clones, whereas LGAL4 expression was decreased and these differences may explain the observed differences in proliferation. The mechanisms by which KLK6 and LGAL4 expression is altered in MKN45-C1 and MKN45-C2 when compared to MKN45-SC control remains to be elucidated.
Another important observation was that apoptosis was significantly increased in MKN45-C1 and MKN45-C2 clones when compared to the MKN45-SC control. MUC1 was previously shown to mediate a pro-apoptotic response in hamster ovary cells [45] and it was also attributed with anti-apoptotic functions in myeloma, breast and colorectal carcinoma cell lines [46], [47], [48]. However, little is known about the influence of MUC1 on cell apoptosis in gastric carcinoma cells.
No significant differences were found between MUC1-C1 and C2 clones and MKN45-SC control with respect to cell migration and invasion. This is in contrast to previous findings in which MUC1 was shown to influence cell migration in breast, cervical, renal and pancreatic carcinoma cell lines [18], [49], [50], [51] and cell invasion in breast, lung, gastrointestinal, hepatic and pancreatic carcinoma cell lines [3], [52], [53].
Cell-cell aggregation was decreased in MKN45-C1 and MKN45-C2 clones when compared to the MKN45-SC control. Previous studies have shown that overexpression of different forms of MUC1 can lead to an increase or a decrease in cell-cell aggregation in a pancreatic carcinoma cell line [54], whereas others have shown that MUC1 downregulation induces an increase of cell-cell aggregation in an oral carcinoma cell line [41]. MUC1 interactions with other adhesion molecules have been shown to contribute to both adhesive [55], [56] and anti-adhesive [57], [58], [59] properties of cells. Our results showed that MUC1 plays a relevant role in MKN45 cell-cell aggregation, contributing to gastric cells adhesive properties.
Another possibility is that signaling through the MUC1-CD influences gene expression, which in turn affects the phenotypic properties of the MKN45 cell line. By oligonucleotide microarray analysis we found alterations in the transcriptional profile of cells following MUC1 downregulation when compared to control cells. These alterations are likely due to MUC1 downregulation, since MUC1 has been shown to directly conduct signals that alter the transcriptional program of tumor cells [36], [37], [60]. MUC1 cytoplasmic domain can be phosphorylated in several sites, modulating its interaction with cell signalling partners and transcription factors [21]. The phosphorylation of MUC1-CD will be dependent on the amount and availability of its signaling partners and therefore on the cell type in question. We found significant alterations in the expression levels of several genes, mainly TCN1, KLK6, ADAM29, LGALS4, TSPAN8 and SHPS-1. Some of these molecules have functions not yet fully clarified yet others are known to be associated with cell proliferation and migration, including KLK6, LGAL4 and SHPS-1 [42], [43], [59], [61], [62], invasion, including KLK6 [59] and motility, including LGAL4 [43]. MUC1 may be facilitating the transcription of these genes and therefore be contributing to the observed phenotypic alterations observed.
In vivo assays confirmed that cells with decreased levels of MUC1 form smaller and slower-growing tumors than the control cells. This result emphasizes that MUC1 contributes to gastric tumor progression in the context of the multicellular environment of tumor growth in vivo.
MUC1 overexpression has been associated with the neoplastic progression of several tumors, including the acquisition of invasive and metastatic properties. Phenotypic studies in cell models other than gastric cancer have suggested that MUC1 influences events such as proliferation, apoptosis, migration, invasion, adhesion and cell-cell aggregation. Previous studies of MUC1 in breast carcinogenesis models show mixed results for different breast cancer cell lines [38], which reinforces the relevance of the molecular context on the MUC1-mediated cancer progression. The effects of MUC1 in gastric carcinogenesis will thus be dependent on MUC1 and the molecules interacting with MUC1, which will significantly differ between cell lines. Evaluation of different gastric cell lines will complement the data regarding the impact of MUC1 gastric carcinogenesis.
The work presented here shows for the first time that MUC1 expression influences proliferation, apoptosis and cell-cell aggregation of MKN45 gastric carcinoma cells. The results are consistent with the view that MUC1 modulates different signaling pathways in a manner that is dependent on the expression and activity of other regulatory mechanisms and molecules, which are influenced by the cellular and biological context of the cell type that is overexpressing MUC1.
Acknowledgments
We thank Ana Rita Barbosa and Nuno Mendes for technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Fundação para a Ciência e a Tecnologia (FCT), Portugal, Projects POCTI/SAU-IMI/56895/2004 and PTDC/SAU-OBD/65616/2006 and fellowships SFRH/BD/21693/2005 and SFRH/BD/36961/2007 and Fundação Luso-Americana (FLAD). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Power DG, Kelsen DP, Shah MA. Advanced gastric cancer--slow but steady progress. Cancer Treat Rev. 2010;36:384–392. doi: 10.1016/j.ctrv.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Patton S, Gendler SJ, Spicer AP. The epithelial mucin, MUC1, of milk, mammary gland and other tissues. Biochim Biophys Acta. 1995;1241:407–423. doi: 10.1016/0304-4157(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 3.Yonezawa S, Sato E. Expression of mucin antigens in human cancers and its relationship with malignancy potential. Pathol Int. 1997;47:813–830. doi: 10.1111/j.1440-1827.1997.tb03713.x. [DOI] [PubMed] [Google Scholar]
- 4.Klinge CM, Radde BN, Imbert-Fernandez Y, Teng Y, Ivanova MM, et al. Targeting the intracellular MUC1 C-terminal domain inhibits proliferation and estrogen receptor transcriptional activity in lung adenocarcinoma cells. Mol Cancer Ther. 2011. [DOI] [PMC free article] [PubMed]
- 5.Ye Q, Yan Z, Liao X, Li Y, Yang J, et al. MUC1 induces metastasis in esophageal squamous cell carcinoma by upregulating matrix metalloproteinase 13. Lab Invest. 2011;91:778–787. doi: 10.1038/labinvest.2011.12. [DOI] [PubMed] [Google Scholar]
- 6.Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29:2893–2904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resende C, Thiel A, Machado JC, Ristimaki A. Gastric cancer: basic aspects. Helicobacter. 2011;16(Suppl 1):38–44. doi: 10.1111/j.1523-5378.2011.00879.x. [DOI] [PubMed] [Google Scholar]
- 8.Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, et al. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Zhang L, Hu G. Potential application of alternatively glycosylated serum MUC1 and MUC5AC in gastric cancer diagnosis. Biologicals. 2009;37:18–25. doi: 10.1016/j.biologicals.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki Y, Tang W, Xu H, Nakata M, Mafune K, et al. Sustained aberrant localization of KL-6 mucin and beta-catenin at the invasion front of human gastric cancer cells. Anticancer Res. 2011;31:535–542. [PubMed] [Google Scholar]
- 11.Benjamin JB, Jayanthi V, Devaraj H. MUC1 expression and its association with other aetiological factors and localization to mitochondria in preneoplastic and neoplastic gastric tissues. Clin Chim Acta. 2010;411:2067–2072. doi: 10.1016/j.cca.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Kohlgraf KG, Gawron AJ, Higashi M, Meza JL, Burdick MD, et al. Contribution of the MUC1 tandem repeat and cytoplasmic tail to invasive and metastatic properties of a pancreatic cancer cell line. Cancer Res. 2003;63:5011–5020. [PubMed] [Google Scholar]
- 13.Hattrup CL BJ, Kotlarczyk KL, Madsen CS, Hentz JG, et al. The MUC1 Cytoplasmic Tail and Tandem Repeat Domains Contribute to Mammary Oncogenesis in FVB Mice. Breast Cancer. 2008;1:57–63. doi: 10.4137/bcbcr.s655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendler S, Taylor-Papadimitriou J, Duhig T, Rothbard J, Burchell J. A highly immunogenic region of a human polymorphic epithelial mucin expressed by carcinomas is made up of tandem repeats. J Biol Chem. 1988;263:12820–12823. [PubMed] [Google Scholar]
- 15.Hayashi T, Takahashi T, Motoya S, Ishida T, Itoh F, et al. MUC1 mucin core protein binds to the domain 1 of ICAM-1. Digestion. 2001;63(Suppl 1):87–92. doi: 10.1159/000051917. [DOI] [PubMed] [Google Scholar]
- 16.Yu LG, Andrews N, Zhao Q, McKean D, Williams JF, et al. Galectin-3 interaction with Thomsen-Friedenreich disaccharide on cancer-associated MUC1 causes increased cancer cell endothelial adhesion. J Biol Chem. 2007;282:773–781. doi: 10.1074/jbc.M606862200. [DOI] [PubMed] [Google Scholar]
- 17.Shen Q, Rahn JJ, Zhang J, Gunasekera N, Sun X, et al. MUC1 initiates Src-CrkL-Rac1/Cdc42-mediated actin cytoskeletal protrusive motility after ligating intercellular adhesion molecule-1. Mol Cancer Res. 2008;6:555–567. doi: 10.1158/1541-7786.MCR-07-2033. [DOI] [PubMed] [Google Scholar]
- 18.Rahn JJ, Chow JW, Horne GJ, Mah BK, Emerman JT, et al. MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM-1. Clin Exp Metastasis. 2005;22:475–483. doi: 10.1007/s10585-005-3098-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Q, Guo X, Nash GB, Stone PC, Hilkens J, et al. Circulating galectin-3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Res. 2009;69:6799–6806. doi: 10.1158/0008-5472.CAN-09-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Q, Barclay M, Hilkens J, Guo X, Barrow H, et al. Interaction between circulating galectin-3 and cancer-associated MUC1 enhances tumour cell homotypic aggregation and prevents anoikis. Mol Cancer. 2010;9:154. doi: 10.1186/1476-4598-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Carraway KL, 3rd, Funes M, Workman HC, Sweeney C. Contribution of membrane mucins to tumor progression through modulation of cellular growth signaling pathways. Curr Top Dev Biol. 2007;78:1–22. doi: 10.1016/S0070-2153(06)78001-2. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Chen W, Ren J, Yu WH, Li Q, et al. DF3/MUC1 signaling in multiple myeloma cells is regulated by interleukin-7. Cancer Biol Ther. 2003;2:187–193. doi: 10.4161/cbt.2.2.282. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Ren J, Kufe D. Interaction of human MUC1 and beta-catenin is regulated by Lck and ZAP-70 in activated Jurkat T cells. Biochem Biophys Res Commun. 2004;315:471–476. doi: 10.1016/j.bbrc.2004.01.075. [DOI] [PubMed] [Google Scholar]
- 25.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad R, Raina D, Joshi MD, Kawano T, Ren J, et al. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raina D, Ahmad R, Kumar S, Ren J, Yoshida K, et al. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. EMBO J. 2006;25:3774–3783. doi: 10.1038/sj.emboj.7601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang L, Liao X, Beckett M, Li Y, Khanna KK, et al. MUC1-C Oncoprotein Interacts Directly with ATM and Promotes the DNA Damage Response to Ionizing Radiation. Genes Cancer. 2010;1:239–250. doi: 10.1177/1947601910368059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guang W, Kim KC, Lillehoj EP. MUC1 mucin interacts with calcium-modulating cyclophilin ligand. Int J Biochem Cell Biol. 2009;41:1354–1360. doi: 10.1016/j.biocel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor-Papadimitriou J, Burchell J, Miles DW, Dalziel M. MUC1 and cancer. Biochim Biophys Acta. 1999;1455:301–313. doi: 10.1016/s0925-4439(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 31.Motoyama T, Hojo H, Watanabe H. Comparison of seven cell lines derived from human gastric carcinomas. Acta Pathol Jpn. 1986;36:65–83. doi: 10.1111/j.1440-1827.1986.tb01461.x. [DOI] [PubMed] [Google Scholar]
- 32.Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 33.Tuschl T. RNA interference and small interfering RNAs. Chembiochem. 2001;2:239–245. doi: 10.1002/1439-7633(20010401)2:4<239::AID-CBIC239>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 34.Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30:503–512. doi: 10.1016/s0301-472x(02)00806-8. [DOI] [PubMed] [Google Scholar]
- 35.Tsutsumida H, Swanson BJ, Singh PK, Caffrey TC, Kitajima S, et al. RNA interference suppression of MUC1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin Cancer Res. 2006;12:2976–2987. doi: 10.1158/1078-0432.CCR-05-1197. [DOI] [PubMed] [Google Scholar]
- 36.Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, et al. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67:5201–5210. doi: 10.1158/0008-5472.CAN-06-4647. [DOI] [PubMed] [Google Scholar]
- 37.Behrens ME, Grandgenett PM, Bailey JM, Singh PK, Yi CH, et al. The reactive tumor microenvironment: MUC1 signaling directly reprograms transcription of CTGF. Oncogene. 2010;29:5667–5677. doi: 10.1038/onc.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hattrup CL, Gendler SJ. MUC1 alters oncogenic events and transcription in human breast cancer cells. Breast Cancer Res. 2006;8:R37. doi: 10.1186/bcr1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan Z, Liu X, Wong S, Machan JT, Chung MA. MUC1 Knockdown With RNA Interference Inhibits Pancreatic Cancer Growth. J Surg Res. 2009;157:e39–46. doi: 10.1016/j.jss.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Lillehoj EP, Lu W, Kiser T, Goldblum SE, Kim KC. MUC1 inhibits cell proliferation by a beta-catenin-dependent mechanism. Biochim Biophys Acta. 2007;1773:1028–1038. doi: 10.1016/j.bbamcr.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Wang L, Nunes DP, Troxler RF, Offner GD. Suppression of MUC1 synthesis downregulates expression of the epidermal growth factor receptor. Cancer Biol Ther. 2005;4:968–973. doi: 10.4161/cbt.4.9.1913. [DOI] [PubMed] [Google Scholar]
- 42.Nathalie HV, Chris P, Serge G, Catherine C, Benjamin B, et al. High kallikrein-related peptidase 6 in non-small cell lung cancer cells: an indicator of tumour proliferation and poor prognosis. J Cell Mol Med. 2009;13:4014–4022. doi: 10.1111/j.1582-4934.2009.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satelli A, Rao PS, Thirumala S, Rao US. Galectin-4 functions as a tumor suppressor of human colorectal cancer. Int J Cancer. 2010. [DOI] [PMC free article] [PubMed]
- 44.Satelli A, Rao PS, Thirumala S, Rao US. Galectin-4 functions as a tumor suppressor of human colorectal cancer. Int J Cancer. 2011;129:799–809. doi: 10.1002/ijc.25750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaturvedi R, Srivastava RK, Hisatsune A, Shankar S, Lillehoj EP, et al. Augmentation of Fas ligand-induced apoptosis by MUC1 mucin. Int J Oncol. 2005;26:1169–1176. [PubMed] [Google Scholar]
- 46.Kawano T, Ahmad R, Nogi H, Agata N, Anderson K, et al. MUC1 oncoprotein promotes growth and survival of human multiple myeloma cells. Int J Oncol. 2008;33:153–159. [PMC free article] [PubMed] [Google Scholar]
- 47.Agata N, Ahmad R, Kawano T, Raina D, Kharbanda S, et al. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68:6136–6144. doi: 10.1158/0008-5472.CAN-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren J, Agata N, Chen D, Li Y, Yu WH, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Li M, Wang Y, Zhang Y, Jin S, et al. RNA interference targeting CML66, a novel tumor antigen, inhibits proliferation, invasion and metastasis of HeLa cells. Cancer Lett. 2008;269:127–138. doi: 10.1016/j.canlet.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 50.Aubert S, Fauquette V, Hemon B, Lepoivre R, Briez N, et al. MUC1, a new hypoxia inducible factor target gene, is an actor in clear renal cell carcinoma tumor progression. Cancer Res. 2009;69:5707–5715. doi: 10.1158/0008-5472.CAN-08-4905. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Z, Wong S, Borrelli A, Chung MA. Down-regulation of MUC1 in cancer cells inhibits cell migration by promoting E-cadherin/catenin complex formation. Biochem Biophys Res Commun. 2007;362:740–746. doi: 10.1016/j.bbrc.2007.08.074. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder JA, Adriance MC, Thompson MC, Camenisch TD, Gendler SJ. MUC1 alters beta-catenin-dependent tumor formation and promotes cellular invasion. Oncogene. 2003;22:1324–1332. doi: 10.1038/sj.onc.1206291. [DOI] [PubMed] [Google Scholar]
- 53.Gao J, McConnell MJ, Yu B, Li J, Balko JM, et al. MUC1 is a downstream target of STAT3 and regulates lung cancer cell survival and invasion. Int J Oncol. 2009;35:337–345. [PMC free article] [PubMed] [Google Scholar]
- 54.McDermott KM, Crocker PR, Harris A, Burdick MD, Hinoda Y, et al. Overexpression of MUC1 reconfigures the binding properties of tumor cells. Int J Cancer. 2001;94:783–791. doi: 10.1002/ijc.1554. [DOI] [PubMed] [Google Scholar]
- 55.Regimbald LH, Pilarski LM, Longenecker BM, Reddish MA, Zimmermann G, et al. The breast mucin MUCI as a novel adhesion ligand for endothelial intercellular adhesion molecule 1 in breast cancer. Cancer Res. 1996;56:4244–4249. [PubMed] [Google Scholar]
- 56.Nath D, Hartnell A, Happerfield L, Miles DW, Burchell J, et al. Macrophage-tumour cell interactions: identification of MUC1 on breast cancer cells as a potential counter-receptor for the macrophage-restricted receptor, sialoadhesin. Immunology. 1999;98:213–219. doi: 10.1046/j.1365-2567.1999.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wesseling J, van der Valk SW, Vos HL, Sonnenberg A, Hilkens J. Episialin (MUC1) overexpression inhibits integrin-mediated cell adhesion to extracellular matrix components. J Cell Biol. 1995;129:255–265. doi: 10.1083/jcb.129.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wesseling J, van der Valk SW, Hilkens J. A mechanism for inhibition of E-cadherin-mediated cell-cell adhesion by the membrane-associated mucin episialin/MUC1. Mol Biol Cell. 1996;7:565–577. doi: 10.1091/mbc.7.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klucky B, Mueller R, Vogt I, Teurich S, Hartenstein B, et al. Kallikrein 6 induces E-cadherin shedding and promotes cell proliferation, migration, and invasion. Cancer Res. 2007;67:8198–8206. doi: 10.1158/0008-5472.CAN-07-0607. [DOI] [PubMed] [Google Scholar]
- 60.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, et al. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985–26995. doi: 10.1074/jbc.M805036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henkhaus RS GE, Ignatenko NA. Kallikrein 6 is a mediator of K-RAS-dependent migration of colon carcinoma cells. Biological Chemistry. 2008;389(6):757–764. doi: 10.1515/BC.2008.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling Y, Maile LA, Lieskovska J, Badley-Clarke J, Clemmons DR. Role of SHPS-1 in the regulation of insulin-like growth factor I-stimulated Shc and mitogen-activated protein kinase activation in vascular smooth muscle cells. Mol Biol Cell. 2005;16:3353–3364. doi: 10.1091/mbc.E04-10-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]