Abstract
In this study, we investigated whether DHA, a nutritionally important n-3 unsaturated fatty acid, modulated the sensitivity of brain tumor cells to the anticancer drug, etoposide (VP16). Medulloblastoma (MB) cell lines, Daoy and D283, and glioblastoma (GBM) cell lines, U138 and U87, were exposed to DHA or VP16 alone or in combination. The effects on cell proliferation and the induction of apoptosis were determined by using MTS and Hoechest 33342/PI double staining. U87 and U138 cells were found to be insensitive to the addition of DHA and VP16, whereas the two MB cell lines showed high sensitivity. DHA or VP16 alone showed little effect on cell proliferation or death in either the MB or GBM cell lines, but pretreatment with DHA enhanced the responsiveness to VP16 in the MB cell lines. To understand the mechanisms of combined DHA and VP16 on MB cells, pathway specific oligo array analyses were performed to dissect possible signaling pathways involved. The addition of DHA and VP16, in comparison to VP16 added alone, resulted in marked suppression in the expression of several genes involved in DNA damage repair, cell proliferation, survival, invasion, and angiogenesis, including PRKDC, Survivin, PIK3R1, MAPK14, NFκB1, NFκBIA, BCL2, CD44, and MAT1. These results suggest (1) that the effects of DHA and VP16 in brain tumor cells are mediated in part by the down regulation of events involved in DNA repair and the PI3K/MAPK signaling pathways and (2) that brain tumors genotypically mimicked by MB cells may benefit from therapies combining DHA with VP16.
Keywords: Apoptosis, docosahexaenoic acid, etoposide, glioblastoma, medulloblastoma
INTRODUCTION
Glioblastomas (GBM) and medulloblastomas (MB) are commonly diagnosed brain tumors that show highly variable rates of morbidity and mortality. GBM patients typically have a median survival of only 15 months, and the 2-year survival rate is only 27% [1, 2]. In contrast, MB patients frequently show a 5-year survival rate of 70–80% using existing cytotoxic therapies [3]. Since conventional treatment regimens for both GBM and MB have significant short-term side-effects and ill-defined long-term complications, it is imperative to develop novel adjunctive therapies to counteract the unfavorable toxicities and to enhance the efficacy of current chemotherapies.
Docosahexaenoic acid (DHA) is a popular dietary supplement with marked general human health benefits, specifically in the maintenance of normal brain function [4]. Recent studies indicate that DHA has autoimmune preventive activity [5, 6] and anti-carcinogenic properties in vitro and in vivo [7-9]. Moreover, in vitro studies show that when DHA was administered in combination with well-studied anticancer drugs (e.g., doxorubicin, taxane, 5-fluorouracil, and celecoxib), exhibits additive or even supra-active effects in breast cancer [10-12], colorectal cancer [13], colon cancer, and prostate cancer cells [14, 15]. However, the mechanism(s) by which DHA augments the anti-tumor agents have not been completely elucidated. Conceivably, DHA might induce synergistic efficacy in colorectal carcinoma and neuroblastoma cells by targeting the PI3K/MAPK or ERK signaling pathways [16, 17]. Exposure to DHA combined with 5-fluorouracil increases the expression of BAX in gastric carcinoma cells [18]. The combination of DHA and docetaxel suppresses the expression of genes in NFκB pathway in prostate cancer cells [19], and activation of p53 in DHA-treated prostate and colon cancer cells [20, 21]. In this study, we investigated the effect of DHA on the sensitivity of MB and GBM cells to etoposide (VP16), an anticancer drug currently used to treat brain tumors [22]. We also studied the mechanism by which the cells respond to the combination of DHA with VP16.
MATERIALS AND METHODS
Cell Culture and Chemicals
MB cell lines, Daoy and D283, and GBM cell lines, U87 and U138, were obtained from ATCC. Cells were maintained in minimum Essential Medium (MEM) (Cellgro) supplemented with 4 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 1% sodium pyruvate, 1% nonessential amino acids, and 10% fetal bovine serum (FBS) at 37°C with 5% CO2. Daoy and U138 cells harbor a p53 mutation but D283 and U87 cells have no mutation in p53 [23, 24]. Etoposide (VP16), DHA, and propidium iodide (PI) were obtained from Sigma. Hoechest 33342 was obtained from Invitrogen.
Determination of Inhibition of Cell Proliferation
Cells (9×103/well) maintained in complete medium were placed in 96-well plates overnight. DHA or identical volume of control (ethanol) was added to the appropriate wells at doses indicated (15 μM, 30 μM, and 60 μM). Four hours later, VP16 or control (DMSO) was added into designated wells to final concentrations of 0.2 μM, 0.8 μM, and 1.6 μM. The control and treated cells were cultured for an additional 48 hours, and the cell numbers were determined by adding 20 μl of MTS solution (Promega), and measuring the optical densities at 490 nm after 4-hour incubation. The results were expressed as the percentages of control cultures.
Nuclear Staining of Live Cells
The cells were seeded in 6-well plates overnight. Thirty μM of DHA or an equal amount of control (ethanol) was first added to the respective wells as described in the figure legends. Four hours later, 0.8 μM of VP16 or vehicle control (DMSO) were added to the designated wells. After an additional 24 hours in culture, the cells were stained in the culture medium with 0.15 mg/ml of Hoechest 33342 and 0.2 μg/ml of PI. Images were captured using confocal microscopy (Olympus).
Gene Expression Profiling
Daoy and D283 cells, with or without pretreatment of 30 μM of DHA, were exposed to 0.8 μM of VP16 for 24 hours. Thereafter, the cells were harvested, and the total RNA was extracted using the RNeasy kit (QIAGEN, CA). The concentration of RNA was determined using a NanoDrop spectrophotometer and the quality of the RNA was evaluated by agarose gel electrophoresis. Total RNA (3 μg) from the control and treated cells was used for cDNA synthesis, followed by cRNA synthesis using TrueLabeling-AMP 2.0 kit (SABiosciences). Biotin-11-UTP-cRNA (Perkinelmer) was amplified and equal amounts of different samples were applied to Oligo GEArray (SABiosciences). Hybridization was performed according to the manufacturer’s instruction. The chemiluminescent signal was detected using Kodak film and the intensity of the signals was quantified using Alpha Innotech. The images were further analyzed using GEArray Expression Analysis Suite 2.0 (SABiosciences).
Array Data Analysis
The density of each spot and the background around each spot were determined using GEArray Expression Analysis Suite 2.0 (SABiosciences). The raw data were analyzed manually using Excel for the subtraction of the background density, and the results were further normalized using housekeeping genes. When the normalized data for the same gene were less than 0.005 in more than 3 out of 4 different conditions (control, DHA, VP16, and the combination), the data were considered as negative calls (whose expression is too low to be detected). The ratio of a gene in cells exposed to different treatments was determined using the following formula: the ratio of Sample B/Sample A = (Tb + (ABS(Ta)/Ta-1)*Ta/2)/ABS(Ta). Significance in change of expression of a gene in MB cells in response to treatment B versus treatment A required a 2-fold increase or decrease.
Quantitative RT-PCR
Total RNAs from control and treated MB cells were also used for cDNA synthesis using the QuantiTect Reverse Transcription kit (Qiagen). The primers were selected using the NCI web-based primer designing tool and were obtained from Integrated DNA Technologies. The gene expression levels were determined by real-time PCR using the QuantiTect SYBR PCR kit (Qiagen). Real time PCR was performed using a Mastercycler ep realplex (Eppendorf). The results were analyzed by Pfaffl’s method [25-28]. The ΔCt for each treatment was calculated using a formula (ΔCt = treatment Ct – control Ct ). The 2−ΔCT was then calculated to give the level of gene expression for each treatment in comparison to the control. The results were normalized using GAPDH. The primer sequences are available upon request.
Western Blot Analysis
Control and treated cells were harvested at the 24-hour time point. Lysates were prepared, analyzed by 10% SDS-PAGE, and transferred onto the nitrocellulose membrane. Immunoblots were probed with antibodies specific for CD44 (Cell Signaling), BCL2 (Santa Cruz), caspase-3 (Santa Cruz), and re-probed with β-actin (Sigma) as a loading control. The images were quantified using ImageJ with correction using β-actin.
RESULTS
Cytotoxic Effects of DHA and VP16 Individually, for Comparison with Combination of DHA and VP16
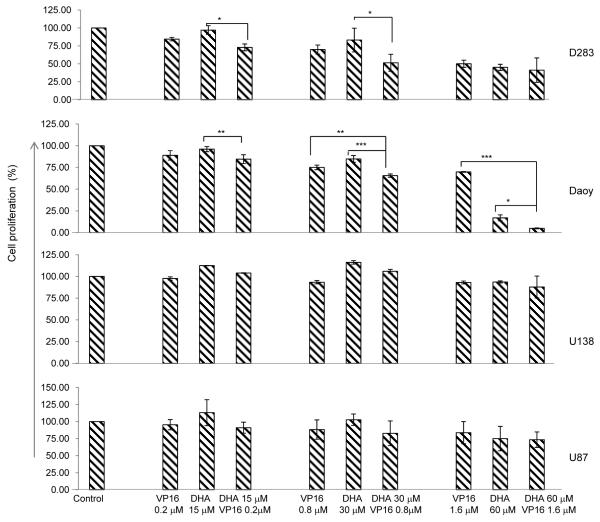
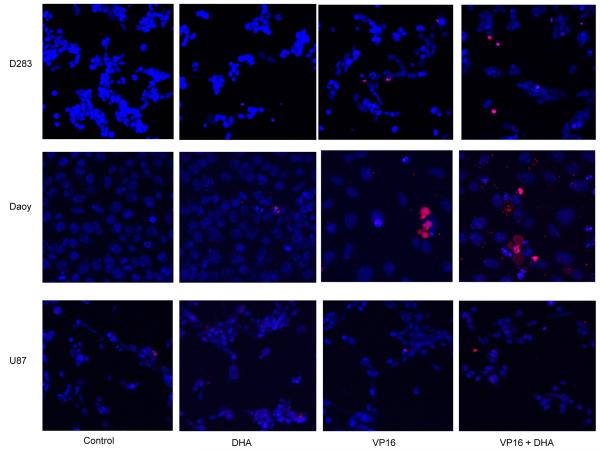
To determine if synergistic cytotoxicity might result from exposure to combined DHA and VP16, brain tumor cells were treated with DHA for 4 hours, followed by additional exposure to VP16 for 48 hours. Cell proliferation was determined by MTS. In Daoy and D283 MB cells, the addition of DHA or VP16 induced cell death in a dose-dependent manner, and exerted an additive effect when combined. In U138 and U87 GBM cells, however, no response to either DHA or VP16 treatments was observed (Fig. 1). The cytotoxic effects of DHA, VP16, and their combinations on tumor cells were validated by ascertaining the induction of apoptosis based on the staining of treated tumor cells with Hoechest 33342/PI in culture, which showed that Daoy and D283 cells were highly sensitive to the combined treatment while the U87 cells did not respond to the treatments. Interestingly no evidence of apoptosis was observed in Daoy and D283 cells treated with DHA alone (Fig. 2). These results suggest that the cytotoxicities of DHA and VP16 vary with tumor type, possibly attributed in part to their genetic backgrounds.
Fig. (1). The effects of DHA or VP16 alone and their combination on cell proliferation in brain tumor cells.
Cells in complete medium were placed in 96-well plates overnight before treatment. After 48 hours, the number of viable cells in each well was determined using MTS. The optical densities were measured at 490 nm. The results were calculated as the percentage of control cultures and presented as mean ± SD. The statistical differences were determined using paired student’s t-test and performed using SPSS. * p<0.05, **p<0.01, and ***p<0.001.
Fig. (2). The effects of cytotoxicity of DHA or VP16 alone and their combination in brain tumor cells.
The cells were seeded in 6-well plates for treatments. After 24 hours, the cells were stained in culture with Hoechest 33342/PI. Images were captured using confocal microscopy. Hoechest 33342 labelled the nueclei of cells in blue and PI labelled the apoptotic cells in red.
DHA Modulating Multiple Signaling Pathways in MB Cells
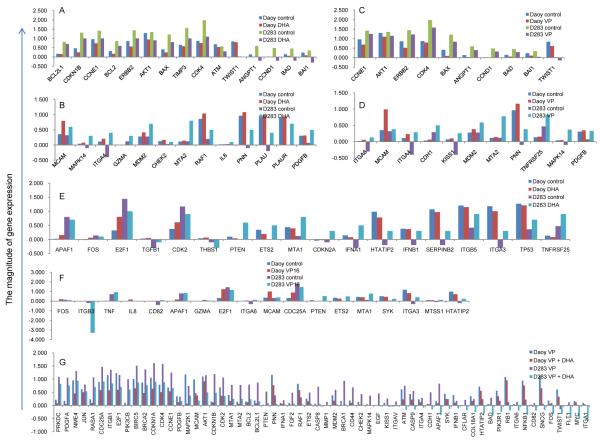
To elucidate the mechanism by which DHA potentiated the effects of VP16 in the MB cell lines, Human Cancer Pathway Finder Oligo GEArray was used to analyze the gene profiles in Daoy and D283 cells exposed to 0.8 μM of VP16 for 24 hours with or without pretreatment by 30 μM DHA. The data showed that DHA modulated the expression of several genes in MB cells. Exposure to DHA alone resulted in down regulation of fifteen genes involved in growth signaling, cell cycle control, DNA damage repair, and anti-apoptotic pathway, in both Daoy and D283 cells (Fig. 3A). Using the same criteria, thirteen genes including MAPK14, were found to be up regulated (Fig. 3B). In VP16-treated Daoy and D283 cells, ten genes were down regulated including those involved in the suppression of apoptosis such as BAX, BAD, and BAI1 (Fig. 3C); while eleven genes, including MAPK14, MCAM and MDM2, were up regulated (Fig. 3D). Additional genes found to be differentially regulated in response to DHA or VP16 included PTEN and Tp53 involved in cell proliferation and cell death pathways (Figs. 3E and 3F). These results reinforce the notion that cells with different genetic backgrounds respond differently to the same chemical treatments.
Fig. (3). Regulation of gene expressions in MB cells treated with DHA and VP16.
The Y axis represents the normalized intensity for the genes chosen for analysis using Human Cancer Pathway Finder Oligo GEArray. The ratio of the changes was calculated using the formula described in the material and methods section. A negative number indicates low to no expression for the gene in the cells. A) up regulated genes in response to DHA compared to control; B) down regulated genes in response to DHA compared to control; C) up regulated genes in response to VP16 compared to control; D) down regulated genes in response to VP16 compared to control; E) differentially regulated genes in response to DHA compared to the control; F) differentially regulated genes in response to VP16; G) down regulated genes in response to DHA + VP16 compared to VP16 alone.
Exposure to Combined DHA and VP16 Targets DNA Damage Repair Genes and Metastatic Related Gene Expression in MB Cells
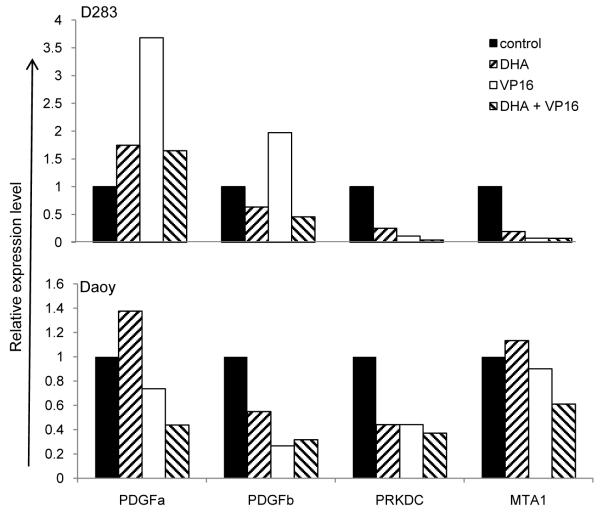
Compared to exposure to VP16 alone, Daoy and D283 cells treated with both DHA and VP16 showed that over sixty genes were significantly down regulated in both cell lines (Fig. 3G). The most pronounced suppression occurred in genes involved in DNA damage repair (e.g. PRKDC, BRCA1), cell death (e.g. BIRC5 [Survivin], APAF1), cell adhesion (e.g. CD44, integrins), cell proliferation (e.g. PDGF), and metastasis (e.g. MAT1 and MAT2) (Fig. 3G). This notion was supported by the analysis of the array data, and further validated in the expression levels of PRKDC, MAT1, PDGFA and PDGFB by real-time PCR analysis using the total RNA prepared from Daoy and D283 cells with or witout 30 μM of DHA pretreatment and additionally exposed to 0.8 μM of VP16 for 24 hours (Fig. 4). It is noteworthy that PRKDC, a nuclear serine/threonine protein kinase gene, is required for DNA repair in response to DNA damage [29]. Thus, our results raise the possibility that combined DHA and VP16 treatment downregulates the DNA repair pathway, which may contribute to MB cell death.
Fig. (4). Confirmation of down regulated genes involved in cell proliferation, DNA repair, and metastasis in MB cells in response to DHA + VP16 compared to VP16 alone by real time PCR.
Total RNAs from Daoy and D283 Cells exposed to DHA, VP16 and their combination were used for cDNA synthesis, and it was then subject to be used for real time PCR using SYBR. The results were calculated using Pfaffl’s method [25-28], normalized using GAPDH and presented as relative expression level. The results shown were one representative of three independent experiments.
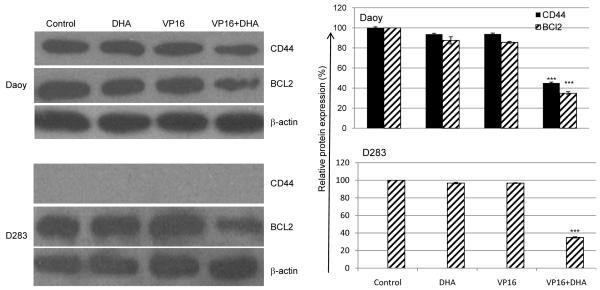
Additional evidence that the combination of DHA and VP16 affected the expression of genes not responsive to either agent alone can be found in the protein expression of CD44, a cell surface adhesion molecule demonstrated to play an important role in tumor metastasis in lymphoma, breast cancer, and colon cancer [30-35]. Although CD44 protein was not detected in D283 cells, the expression levels of the CD44 and BCL2 in Daoy cells were unchanged by DHA or VP16 alone but were significantly reduced by the combination of DHA and VP16 (Fig. 5).
Fig. (5). Down regulation of CD44 and BCL2 in MB cells in response to DHA + VP16 compared to VP16 alone.
A). Total cellular proteins were prepared using lysis buffer containing 2% SDS, 100 mM DTT, 60 mM Tris (pH 6.8), 10% glycerol and separated on a 10% SDS-polyacrylamide gel, and transferred onto nitrocellulose membrane. Immunoblots were probed with antibodies specific for CD44, BCL2, caspase-3. The blots were then re-probed with β-actin as a loading control. B). Quantification of the results from A using ImageJ, expressed as a ratio of the expression level of target genes against β-actin. The results were calculated as mean ± SD. The statistical differences between the combination treatment vs. control or DHA or VP16 alone were determined using paired student’s t-test and performed using SPSS. ***p<0.001.
DISCUSSION
In this study, we tested if DHA alters the effects of anticancer drug VP16 in brain tumor cell lines and performed studies to gain insight into the mechanism of induced synergy and cytotoxicity in cells exposed to the combination of DHA and VP16. Several interesting observations have resulted from these experiments. First, DHA at ≥ 30 μM significantly increased cell death and restricted cell proliferation in MB cells; moreover, DHA combined with VP16 induced synergistic activities on cell proliferation and cell death, in striking contrast to GBM cells which failed to respond to either DHA or VP16. Second, insights on how DHA enhanced VP16 cytotoxicity in MB cells were gleaned by the pathway finder array analysis aiming to dissect the gene profiles of Daoy and D283 cells treated with DHA, VP16 or both. The data provided evidence in support of the notion that DHA modulated the expression of genes involved in multiple pathways including DNA damage repair, cell cycle, angiogenesis, cell adhesion, and metastasis. Further, common genes, notably PRKDC, Survivin, MAPK14, PIK3R1, NFκB1 NFκBIA, BCL2, CD44, and MAT1, were down regulated in response to DHA or VP16 (Fig. 3) in both Daoy and D283 cells, which is consistent with the inhibition response pattern made evident by the determination of cell proliferation and cell death (Figs. 1 & 2).
The unlimited cell proliferation of tumor cells is fueled by the activation of growth signaling pathways. Previous studies have reported that anticancer activities of DHA in human colorectal carcinoma cells involve the modulation of the PI3K/p38 MAPK [16]. In MB cells, robust expression of PDGFs and PDGFRs may contribute to tumor development [28, 36-40] possibly by the activation of the PI3K /MAPK pathway [28, 40]. Notably, our current study showed that the combination of DHA and VP16 down regulated the expression of PDGFa and PDGFb in MB cells while the addition of VP16 alone did not. These observations are consistent with our previous observation that PDGF/PDGFR signaling plays an important role in MB cell proliferation and cell death [41]. Conceivably, the down regulation of PDGF ligands could disrupt the autocrine signaling loop necessary for the activation of the PI3K/MAPK pathway and the subsequent survival of MB cells.
The concerted suppression of apoptosis in synchrony with the inhibition of cell proliferation is vital to tumor cell survival. One mechanism to diminish the efficacy of induced cell death during chemotherapy is the enhancement of DNA repair activity. In this regard, the down regulation of the expression of DNA-PKcs (encoded by PRKDC), an important DNA damage repair protein, could contribute to the induction of cell death in MB cells in response to the combined DHA and VP16 treatment but not VP16 alone. DNA-PKcs is a nuclear serine/threonine protein kinase involved in the DNA repair signaling pathway for the DNA non-homologous-end-joining, and it contributes to the maintenance of genomic stability and prevention of cancer. Several studies have demonstrated that GBM cells lack DNA-PKcs activity which may lead to persistence of oxidative induced cell death [42-44]. Moreover, the cell death pathway could be activated via BCL2/BAD/caspase [45] in a p53-dependent or p53-independent manner. BCL2 is believed to be an anti-apoptotic molecule that blocks caspase activation [46]. The observed decrease in the expression of BCL2 in MB cells without change in the protein level of caspase-3 (data not shown) but the concomitant down regulation of caspases 8 and 9 (Fig. 3G) suggest that, in MB cells, the combined DHA and VP16 treatment likely activates a caspase-independent cell death pathway. Since p53 is a well-known inducer of apoptosis whose function is tightly coupled to the expression of MDM2 [47] and because our array data showed that the expression of MDM2 was decreased in MB cells treated with DHA and VP16 compared to VP16 alone, it is possible that the co-administration of DHA and VP16 impinges on the regulation of MDM2/p53 interplay in D283 cells harboring the wild type p53. However, as the same effects were also observed in the mutant p53-containing Daoy cells, therefore, the combined treatment of DHA and VP16 in MB cells suggests that modulation of the MDM2/p53 pathway may not be the primary mechanism of apoptosis. Alternatively, down regulation of Survivin, another critical regulator of mitosis and apoptosis in cancer cells [48], could play a role in the apoptotic effect of DHA and VP16 treated MB cells.
Finally, because metastasis plays a pivotal role as the driver of tumor progression, and because CD44 plays an important role in metastasis in numerous tumor types [33, 34, 49], we measured the expression of CD44. We found that the combination of DHA and VP16 treated cells had markedly a lower CD44 expression compared to cells treated with VP16 alone or untreated control cells. Of note, CD44 expression was detected in both MB cell lines at the mRNA level using array analysis (Fig. 3), while CD44 protein was only detected in Daoy but not in D283 cells (Fig. 5). A possible explanation may relate to the probes of CD44 covering more CD44 variants while the pan-CD44 antibody displays a more restricted detection capability. Alternatively, CD44 expression at the protein level in D283 may be too low for detection by Western blotting. These different possibilities are currently under consideration for additional investigation in our laboratory.
CONCLUSION
In conclusion, our study demonstrated that DHA modulates the cytotoxic effect of VP16 in brain tumor cells, in a cell type dependent manner. Our data indicate that the combination of DHA with VP16 may be more efficacious for treating patients with MB but not GBM. We also demonstrated that the combination of DHA with VP16 in MB cells altered multiple signaling pathways. Among them, the down regulating DNA repair and limiting growth factors/PI3K/MAPK pathways are likely the major contributors to the cytotoxic effects.
ACKNOWLEDGEMENTS
We thank Dr. Chenwen Sun (North Dakota State University) and Dr. Bin Guo (North Dakota State University) for their discussions during this study. We wish to extend our thanks to Mary Pull (North Dakota State University) for her thoughtful reading of the manuscript. This project was supported by NCI/NIH 1R15CA140833 (SQ), NCRR/NIH 5P20 RR015566 (SQ), A Reason To Ride research fund (ETW), ND EPSCoR (EW), North Dakota State University faculty funds (EW), and Pilot Project Grant (EW) from the Centers of Biomedical Research Excellence (COBRE) grant NIH P20 RR020151 from the National Center for Research Resources (NCRR). NCRR is a component of the National Institutes of Health (NIH). The contents of this report are solely the responsibility of the authors and do not necessarily reflect the official views of the NIH or NCRR.
ABBREVIATIONS
- PRKDC
Protein kinase, DNA-activated, catalytic polypeptide
- DNA-PKcs
DNA-dependent protein kinase, catalytic subunit (encoded by PRKDC)
- PDGF
Platelet-derived growth factor
- PIK3R1
Phosphoinositide-3-kinase, regulatory subunit 1
- MAPK14
Mitogen activated protein kinase 14
- NFκB1
Nuclear factor of kappa light polypeptide gene enhancer in B cells 1
- NFκBIA
Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha
- BCL2
B-cell CLL/lymphoma 2
- MAT1
Metastasis associate1
- MDM2
Murine double minute 2
- MTS
3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H- tetrazolium, inner salt
- DMSO
Dimethyl sulfoxide
REFERENCES
- [1].Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- [2].Krakstad C, Chekenya M. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer. 2010;9:135. doi: 10.1186/1476-4598-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–31. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- [4].Lukiw WJ, Bazan NG. Docosahexaenoic acid and the aging brain. J Nutr. 2008;138(12):2510–4. doi: 10.3945/jn.108.096016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Russo GL. Dietary n-6 and n-3 polyunsaturated fatty acids: from biochemistry to clinical implications in cardiovascular prevention. Biochem Pharmacol. 2009;77(6):937–46. doi: 10.1016/j.bcp.2008.10.020. [DOI] [PubMed] [Google Scholar]
- [6].Ferguson LR, Philpott M. Cancer prevention by dietary bioactive components that target the immune response. Curr Cancer Drug Targets. 2007;7(5):459–64. doi: 10.2174/156800907781386605. [DOI] [PubMed] [Google Scholar]
- [7].Hardman WE. Dietary canola oil suppressed growth of implanted MDA-MB 231 human breast tumors in nude mice. Nutr Cancer. 2007;57(2):177–83. doi: 10.1080/01635580701277445. [DOI] [PubMed] [Google Scholar]
- [8].Chamras H, Ardashian A, Heber D, Glaspy JA. Fatty acid modulation of MCF-7 human breast cancer cell proliferation, apoptosis and differentiation. J Nutr Biochem. 2002;13(12):711–6. doi: 10.1016/s0955-2863(02)00230-9. [DOI] [PubMed] [Google Scholar]
- [9].Mandal CC, Ghosh-Choudhury T, Yoneda T, Choudhury GG, Ghosh-Choudhury N. Fish oil prevents breast cancer cell metastasis to bone. Biochem Biophys Res Commun. 2010;402(4):602–7. doi: 10.1016/j.bbrc.2010.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vibet S, Goupille C, Bougnoux P, Steghens JP, Gore J, Maheo K. Sensitization by docosahexaenoic acid (DHA) of breast cancer c ells to anthracyclines through loss of glutathione peroxidase (GPx1) response. Free Radic Biol Med. 2008;44(7):1483–91. doi: 10.1016/j.freeradbiomed.2008.01.009. [DOI] [PubMed] [Google Scholar]
- [11].Maheo K, Vibet S, Steghens JP, Dartigeas C, Lehman M, Bougnoux P, et al. Differential sensitization of cancer cells to doxorubicin by DHA: a role for lipoperoxidation. Free Radic Biol Med. 2005;39(6):742–51. doi: 10.1016/j.freeradbiomed.2005.04.023. [DOI] [PubMed] [Google Scholar]
- [12].Menendez JA, Lupu R, Colomer R. Exogenous supplementation with omega-3 polyunsaturated fatty acid docosahexaenoic acid (DHA ; 22:6n-3) synergistically enhances taxane cytotoxicity and downregulates Her-2/neu (c-erbB-2) oncogene expression in human breast cancer cells. Eur J Cancer Prev. 2005;14(3):263–70. doi: 10.1097/00008469-200506000-00011. [DOI] [PubMed] [Google Scholar]
- [13].Calviello G, Di Nicuolo F, Serini S, Piccioni E, Boninsegna A, Maggiano N, et al. Docosahexaenoic acid enhances the susceptibility of human colorectal cancer cells to 5-fluorouracil. Cancer Chemother Pharmacol. 2005;55(1):12–20. doi: 10.1007/s00280-004-0846-6. [DOI] [PubMed] [Google Scholar]
- [14].Narayanan NK, Narayanan BA, Reddy BS. A combination of docosahexaenoic acid and celecoxib prevents prostate cancer cell growth in vitro and is associated with modulation of nuclear factor-kappaB, and steroid hormone receptors. Int J Oncol. 2005;26(3):785–92. [PubMed] [Google Scholar]
- [15].Swamy MV, Cooma I, Patlolla JM, Simi B, Reddy BS, Rao CV. Modulation of cyclooxygenase-2 activities by the combined action of celecoxib and decosahexaenoic acid: novel strategies for colon cancer prevention and treatment. Mol Cancer Ther. 2004;3(2):215–21. [PubMed] [Google Scholar]
- [16].Toit-Kohn JL, Louw L, Engelbrecht AM. Docosahexaenoic acid induces apoptosis in colorectal carcinoma cells by modulating the PI3 kinase and p38 MAPK pathways. J Nutr Biochem. 2009;20(2):106–14. doi: 10.1016/j.jnutbio.2007.12.005. [DOI] [PubMed] [Google Scholar]
- [17].Wu H, Ichikawa S, Tani C, et al. Docosahexaenoic acid induces ERK1/2 activation and neuritogenesis via intracellular reactive oxygen species production in human neuroblastoma SH-SY5Y cells. Biochim Biophys Acta. 2009;1791(1):8–16. doi: 10.1016/j.bbalip.2008.10.004. [DOI] [PubMed] [Google Scholar]
- [18].Zhuo Z, Zhang L, Mu Q, et al. The effect of combination treatment with docosahexaenoic acid and 5-fluorouracil on the mRNA expression of apoptosis-related genes, including the novel gene BCL2L12, in gastric cancer cells. In Vitro Cell Dev Biol Anim. 2009;45(1-2):69–74. doi: 10.1007/s11626-008-9154-5. [DOI] [PubMed] [Google Scholar]
- [19].Shaikh IA, Brown I, Schofield AC, Wahle KW, Heys SD. Docosahexaenoic acid enhances the efficacy of docetaxel in prostate cancer cells by modulation of apoptosis: the role of genes associated with the NF-kappaB pathway. Prostate. 2008;68(15):1635–46. doi: 10.1002/pros.20830. [DOI] [PubMed] [Google Scholar]
- [20].Narayanan NK, Narayanan BA, Bosland M, Condon MS, Nargi D. Docosahexaenoic acid in combination with celecoxib modulates HSP70 and p53 proteins in prostate cancer cells. Int J Cancer. 2006;119(7):1586–98. doi: 10.1002/ijc.22031. [DOI] [PubMed] [Google Scholar]
- [21].Kato T, Kolenic N, Pardini RS. Docosahexaenoic acid (DHA), a primary tumor suppressive omega-3 fatty acid, inhibits growth of colorectal cancer independent of p53 mutational status. Nutr Cancer. 2007;58(2):178–87. doi: 10.1080/01635580701328362. [DOI] [PubMed] [Google Scholar]
- [22].Gandola L, Massimino M, Cefalo G, et al. Hyperfractionated accelerated radiotherapy in the Milan strategy for metastatic medulloblastoma. J Clin Oncol. 2009;27(4):566–71. doi: 10.1200/JCO.2008.18.4176. [DOI] [PubMed] [Google Scholar]
- [23].Saylors RL, 3rd, Sidransky D, Friedman HS, et al. Infrequent p53 gene mutations in medulloblastomas. Cancer Res. 1991;51(17):4721–3. [PubMed] [Google Scholar]
- [24].Vogelbaum MA, Tong JX, Perugu R, Gutmann DH, Rich KM. Overexpression of bax in human glioma cell lines. J Neurosurg. 1999;91(3):483–9. doi: 10.3171/jns.1999.91.3.0483. [DOI] [PubMed] [Google Scholar]
- [25].Tian Z, Palmer N, Schmid P, et al. A practical platform for blood biomarker study by using global gene expression profiling of peripheral whole blood. PLoS One. 2009;4(4):e5157. doi: 10.1371/journal.pone.0005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu E, Palmer N, Tian Z, et al. Comprehensive dissection of PDGF-PDGFR signaling pathways in PDGFR genetically defined cells. PLoS One. 2008;3(11):e3794. doi: 10.1371/journal.pone.0003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang H, Bajraszewski N, Wu E, et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117(3):730–8. doi: 10.1172/JCI28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34(3):285–97. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Harada N, Mizoi T, Kinouchi M, et al. Introduction of antisense CD44S CDNA down-regulates expression of overall CD44 isoforms and inhibits tumor growth and metastasis in highly metastatic colon carcinoma cells. Int J Cancer. 2001;91(1):67–75. doi: 10.1002/1097-0215(20010101)91:1<67::aid-ijc1011>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- [31].Marhaba R, Zoller M. CD44 in cancer progression: adhesion, migration and growth regulation. J Mol Histol. 2004;35(3):211–31. doi: 10.1023/b:hijo.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- [32].Wang HS, Hung Y, Su CH, et al. CD44 cross-linking induces integrin-mediated adhesion and transendothelial migration in breast cancer cell line by up-regulation of LFA-1 (alpha L beta2) and VLA-4 (alpha4beta1) Exp Cell Res. 2005;304(1):116–26. doi: 10.1016/j.yexcr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- [33].Miletti-Gonzalez KE, Chen S, Muthukumaran N, et al. The CD44 receptor interacts with P-glycoprotein to promote cell migration and invasion in cancer. Cancer Res. 2005;65(15):6660–7. doi: 10.1158/0008-5472.CAN-04-3478. [DOI] [PubMed] [Google Scholar]
- [34].Subramaniam V, Vincent IR, Gardner H, Chan E, Dhamko H, Jothy S. CD44 regulates cell migration in human colon cancer cells via Lyn kinase and AKT phosphorylation. Exp Mol Pathol. 2007;83(2):207–15. doi: 10.1016/j.yexmp.2007.04.008. [DOI] [PubMed] [Google Scholar]
- [35].Wang SJ, Wreesmann VB, Bourguignon LY. Association of CD44 V3-containing isofor ms with tumor cell growth, migration, matrix metalloproteinase expression, and lymph node metastasis in head and neck cancer. Head Neck. 2007;29(6):550–8. doi: 10.1002/hed.20544. [DOI] [PubMed] [Google Scholar]
- [36].Gilbertson RJ, Clifford SC. PDGFRB is overexpressed in metastatic medulloblastoma. Nat Genet. 2003;35(3):197–8. doi: 10.1038/ng1103-197. [DOI] [PubMed] [Google Scholar]
- [37].Gilbertson RJ, Langdon JA, Hollander A, et al. Mutational analysis of PDGFR-RAS/MAPK pathway activation in childhood medulloblastoma. Eur J Cancer. 2006;42(5):646–9. doi: 10.1016/j.ejca.2005.11.023. [DOI] [PubMed] [Google Scholar]
- [38].Yuan L, Santi M, Rushing EJ, Cornelison R, MacDonald TJ. ERK activation of p21 activated kinase-1 (Pak1) is critical for medulloblastoma cell migration. Clin Exp Metastasis. 2010;27(7):481–91. doi: 10.1007/s10585-010-9337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Abouantoun TJ, Macdonald TJ. Imatinib blocks migration and invasion of medulloblastoma cells by concurrently inhibiting activation of platelet-derived growth factor receptor and transactivation of epidermal growth factor receptor. Mol Cancer Ther. 2009;8(5):1137–47. doi: 10.1158/1535-7163.MCT-08-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].MacDonald TJ, Brown KM, LaFleur B, et al. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet. 2001;29(2):143–52. doi: 10.1038/ng731. [DOI] [PubMed] [Google Scholar]
- [41].Wang FMAP, Doucette M, Bhat K, Li X, Law S, Kohane S, Wu E. PDGFR alpha and PDGFR beta differentially regulate cell proliferation and migration/invasion in medulloblastoma cells. 101st AACR Annual Meeting; 2010 April 17-21. [Google Scholar]
- [42].Peddi P, Loftin CW, Dickey JS, et al. DNA-PKcs deficiency leads to persistence of oxidatively induced clustered DNA lesions in human tumor cells. Free Radic Biol Med. 2010;48(10):1435–43. doi: 10.1016/j.freeradbiomed.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen GG, Sin FL, Leung BC, Ng HK, Poon WS. Glioblastoma cells deficient in DNA-dependent protein kinase are resistant to cell death. J Cell Physiol. 2005;203(1):127–32. doi: 10.1002/jcp.20230. [DOI] [PubMed] [Google Scholar]
- [44].Chen GG, Sin FL, Leung BC, Ng HK, Poon WS. Differential role of hydrogen peroxide and staurosporine in induction of cell death in glioblastoma cells lacking DNA-dependent protein kinase. Apoptosis. 2005;10(1):185–92. doi: 10.1007/s10495-005-6073-8. [DOI] [PubMed] [Google Scholar]
- [45].Yang E, Korsmeyer SJ. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood. 1996;88(2):386–401. [PubMed] [Google Scholar]
- [46].Burz C, Berindan-Neagoe I, Balacescu O, Irimie A. Apoptosis in cancer: key molecular signaling pathways and therapy targets. Acta Oncol. 2009;48(6):811–21. doi: 10.1080/02841860902974175. [DOI] [PubMed] [Google Scholar]
- [47].Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–87. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- [48].Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14(16):5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- [49].Wiranowska M, Ladd S, Smith SR, Gottschall PE. CD44 adhesion molecule and neuro-glial proteoglycan NG2 as invasive markers of glioma. Brain Cell Biol. 2006;35(2-3):159–72. doi: 10.1007/s11068-007-9009-0. [DOI] [PubMed] [Google Scholar]