Abstract
Many neuroblastoma cell lines can be induced to differentiate into a mature neuronal cell type with retinoic acid and other compounds, providing an important model system for elucidating signalling pathways involved in this highly complex process. Recently, it has become apparent that miRNAs, which act as regulators of gene expression at a post-transcriptional level, are differentially expressed in differentiating cells and play important roles governing many aspects of this process. This includes the down-regulation of DNA methytransferases that cause the de-methylation and transcriptional activation of numerous protein coding gene sequences. The purpose of this article is to review involvement of miRNAs and DNA methylation alterations in the process of neuroblastoma cell differentiation. A thorough understanding of miRNA and genetic pathways regulating neuroblastoma cell differentiation potentially could lead to targeted therapies for this disease.
Keywords: ATRA, MYCN, neuroblastoma, microRNA, NCOR2, differentiation, NOS1, DNA methylation
1. Introduction
1.1 Neuroblastoma as a clinically and genetically heterogeneous disease
Neuroblastomas originate from precursor cells of the sympathetic nervous system, and although these tumors are the leading cause of childhood cancer deaths, clinical outcome is highly heterogeneous, ranging from spontaneous regression to rapid progression in spite of intensive multimodal chemotherapy. Several clinical parameters are predictive of outcome, including age at diagnosis, disease stage, and histopathological characteristics of the tumors [1]. In addition, high risk neuroblastoma tumors are characterized by a number of genomic aberrations, including amplification of the MYCN oncogenic transcription factor, loss of heterozygosity of chromosome 1p or 11q regions, or gain of 1q or 17q regions, as summarized in a review by Stallings [2]. MYCN amplification and loss of 11q material seldom occur within the same tumor and therefore actually represent two clinically unfavorable genetic subtypes of this disease. The presence of any segmental chromosomal imbalances alone is also predictive of suboptimal patient survival [3]. In contrast, tumors that contain mostly whole chromosome gains or losses and that lack segmental imbalances are associated with significantly better patient survival, particularly when tumors have specific patterns of metastases that define stage 4S disease in infants. A number of signatures based on the patterns of expression of specific combinations of protein coding genes [4, 5] or microRNAs [6-8] are also predictive of patient survival.
1.2. Importance of differentiation in neuroblastoma biology and therapy
The degree of differentiation of neuroblastoma tumors is particularly important in assessment of prognosis. Shimada et al., (2001) reported overall patient survival to be 30% in undifferentiated tumors versus 61% in poorly differentiated tumors and 81.4% in differentiating tumors [9]. In some instances, tumors in infants with stage 4s disease can differentiate into a benign ganglioneuroma, further underscoring the importance of differentiation in this highly heterogeneous disease.
Given that more differentiated tumors are clinically less aggressive, it is perhaps not surprising that differentiation therapy involving 13-cis-retinoic acid, a derivative of vitamin A (retinol), following high dose chemotherapy improves event free survival [10]. A related compound, all trans-retinoic acid (ATRA) causes a number of neuroblastoma cell lines to undergo dramatic increases in neurite length during the process of neural cell differentiation [11]. Retinoic acid (RA) binds to heterodimers of retinoic acid receptors (RAR) or the retinoid X receptor (RXR), which in turn bind to retinoic acid response elements (RARE) located in the 5′ upstream regions of target genes. RA can modulate the expression of many protein coding genes and non-coding RNA sequences through causing the displacement of repressor complexes with activator complexes [12]. Although naturally occurring, RA levels are tightly controlled during normal embryonic development [13].
The process of neuroblastoma cell differentiation is clearly a complex, poorly understood process involving many genes and genetic pathways. Given the importance of differentiation therapy, a more thorough understanding of signalling pathways that trigger neuroblastoma cell differentiation is warranted and is of potential therapeutic benefit. Recently, a number of studies have demonstrated that both microRNAs (miRNAs) and genome–wide DNA methylation alterations are also involved with regulating this process. Here, we will review the role of miRNAs and DNA methylation changes in regulating neuroblastoma cell differentiation.
2. MicroRNAs and neuroblastoma pathogenesis
Mature miRNAs are 19 to 22 nucleotides in length, having been processed by the RNA cleavage enzymes DROSHA and DICER from much larger RNA precursor molecules, as reviewed in a number of articles [14, 15]. These small noncoding RNAs play major post-transcriptional regulatory roles by targeting complementary sequences found on the 3′ UTR regions of mRNAs, leading to either degradation of the mRNA sequence or translational inhibition at the RNA induced silencing complex (RISC). More recently, it has become apparent that miRNAs can also target sequences found within the 5′ UTR [16] and exonic regions [17], although global proteomic studies indicate that 3′ UTR targeting has the greatest effect on the down-regulation of proteins [18]. Until recently miRNAs were considered to be primarily negative regulators of gene expression at a post-transcriptional level. However, it is apparent that specific miRNAs can also enhance mRNA translation in non-proliferating cells [19], and others are involved in directly regulating transcription through targeting complementary sequences in gene promoter regions [20]. Thus, much remains to be learned about the basic mode of action of miRNAs.
MiRNAs play major oncogenic and tumor suppressor roles in virtually all forms of cancer, affecting properties such as cell proliferation, migration, invasion, apoptosis, metastasis, angiogenesis and immune escape, as summarized in many excellent reviews [21-24]. More specifically, miRNAs play highly complex and diverse roles in the pathogenesis of neuroblastoma, as recently reviewed [25-27]. Expression profiling of miRNAs in primary tumors has led to the identification of numerous miRNAs that are associated with poor patient survival and aggressive disease course [6, 7, 28-30], while a number of functional studies have shown that some miRNAs can have either positive or negative effects on neuroblastoma cell proliferation rates and apoptosis, both in vitro and in vivo [28, 31-36]. In addition, it appears that multiple miRNAs significantly contribute to the process of ATRA induced neuroblastoma cell differentiation.
3. MiRNA expression profiling following ATRA induced neuroblastoma cell differentiation
Several studies have profiled miRNA expression in neuroblastoma cell lines following ATRA induced differentiation [28, 37-42]. Inspection of the miRNAs that were determined to be differentially expressed in response to ATRA indicated only partial overlap of miRNAs between the studies. However, these studies differed in the cell lines profiled (SK-N-BE versus SHSY-5Y), concentration of ATRA, addition of brain derived neurotrophic factor (BDNF) to the media, culture conditions, miRNA profiling platforms (qPCR, Northern blot and microarray), number of miRNAs profiled and the post-ATRA time points at which measurements were carried out. Under these circumstances, variation between studies might be expected. It is remarkable that in spite of the experimental variables, the differential expression of 37 miRNAs were validated in at least two or more studies (Table 1). Although some of the miRNAs identified in single studies will likely prove to be of great interest, the validated set of miRNAs, as detailed in Table 1, are a promising initial set for functional studies.
Table 1.
miRNAs Determined to be Differentially Expressed During ATRA Induced Differentiation of Neuroblastoma Cell Lines in at Least Two Independent Studies
| Up-regulated*: | ||||||||
|---|---|---|---|---|---|---|---|---|
| microRNA | Chen [28] | Chen [38] |
Laneve [50] |
Le [40] |
Beveridge. [37] |
Foley [39] |
Meseguer [41] |
Ragusa [42] |
| Let-7a | 1.7 | up | ||||||
| Let-7b | 1.8 | up | ~40 | 1.5 | ||||
| miR-7 | 1.7 | up | ~10 | |||||
| miR-9 | up | 1.9 | ||||||
| miR-10a | 3.98 | 10 | 6 | |||||
| miR-10b | 10 | 6 | ||||||
| miR-21 | ~10 | 2.5 | ||||||
| miR-22 | up | 2 | ||||||
| miR-24 | up | 2.3 | ||||||
| miR-26a | up | 1.7 | ||||||
| miR-27a | 1.99 | ~10 | 1.3 | |||||
| miR-30a-5p | up | 1.5 | ||||||
| miR-30b | 1.6 | 1.6 | ||||||
| miR-103/107 | 1.9 | up | ||||||
| miR-124 | up | ~ 38 | 2.05 | |||||
| miR-125a | up | 1.5 | ||||||
| miR-125b | 1.33 | up | ~ 17 | 1.6 | ||||
| miR-132 | 2.7 | 136 | 2.5 | |||||
| miR-137 | 2.2 | 1.4 | ||||||
| miR-149 | 1.6 | 1.5 | ||||||
| miR-150 | 2.1 | 1.4 | ||||||
| miR-152 | 2.5 | up | ||||||
| miR-184 | 9.0 | 5.5 | 2 | |||||
| miR-189 | ~20 | 1.5 | ||||||
| miR-197 | 1.7 | 1.7 | ||||||
| miR-199a | 1.9 | ~40 | 1.7 | |||||
| miR-199a* | 1.9 | ~40 | ||||||
| miR-200c | 2.6 | 1.4 | ||||||
| miR-210 | 1.65 | 5.5 | ||||||
| miR-214 | 1.8 | ~10 | 2.4 | |||||
| miR-330 | 2.9 | 1.6 | ||||||
| miR-331 | 1.9 | 2.17 | 1.6 | 1.5 | ||||
| miR-615 | 3 | 5 | ||||||
| Down-regulated * : | ||||||||
| microRNA | Chen [ 28 ] |
Chen
[ 38 ] |
Laneve
[ 50 ] |
Le
[ 40 ] |
Beveridge.
[ 37 ] |
Foley
[ 39 ] |
Meseguer
[ 41 ] |
Ragusa
[ 42 ] |
| hsa-miR-7 | −1.8 | up | −2.1 | |||||
| miR-106a | −2 | −10 | ||||||
| miR-323 | down 75% | −5.1 | ||||||
| miR-424 | Down | −3.0 | ||||||
| miR-487b | −4 | −20 | ||||||
Fold Change
4. Molecular mechanisms mediating miRNA expression in response to RA
The molecular mechanisms responsible for the expressional alterations of miRNAs during ATRA induced differentiation have not been fully elucidated. In some instances, such as for miR-10a [39], the miRNAs have retinoic acid receptor elements positioned in an up-stream region and are therefore direct retinoid targets. An indirect effect of ATRA treatment is the down-regulation of MYCN [43], which directly or indirectly regulates the expression of numerous miRNAs [6-8, 44], thus MYCN could be responsible for miRNA expressional alterations following ATRA treatment. A decrease in MYCN mRNA levels can be detected within 6 hours post-ATRA treatment, occurring prior to the onset of neurite outgrowth, so that changes in MYCN levels could account for some of the changes in miRNA expression.
MYCN down-regulation might be a necessary condition for differentiation to occur in MYCN amplified cell lines, as ectopic over-expression of MYCN can render some cell lines unresponsive to retinoic acid [45]. In addition, siRNA or shRNA inhibition of MYCN in MYCN amplified cell lines can lead to a reduction in cell proliferation and/or increased neurite outgrowth [46-48]. In contrast, Edsjo et al [49] reported that high levels of ectopic over-expression in non-MYCN amplified cell lines does not prevent differentiation. Analysis of MYCN expression in embryonic tissues also would suggest that high expression is not completely incompatible with a differentiated cell state [49]. Thus, the effects of MYCN on differentiation are complex, poorly understood, and might be dependent on cell line context (e.g. MYCN amplified versus non-amplified). It is conceivable that MYCN might play a role in the differentiation process through the modulation of miRNA expression, and in this regard, a number of miRNAs have been demonstrated to effect cellular phenotypic properties when they are artificially dysregulated in neuroblastoma cell lines.
5. MiRNAs mediated regulation of neuroblastoma cell differentiation
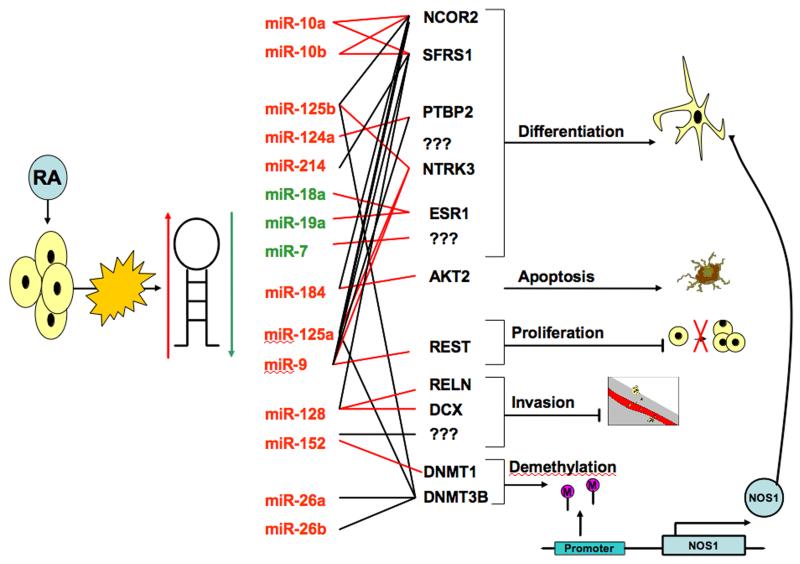
A number of functional studies have been carried out on miRNAs which were differentially expressed in response to ATRA induced differentiation, as summarized in Figure 1. Laneve et al [50] carried out one of the first functional studies, having identified miR-9, -125a and -125b as being over-expressed following ATRA treatment of SK-N-BE cells. Transfection of these miRNAs into SK-N-BE cells causes decreased cell proliferation in an additive manner. Knock-down of this set of miRNAs prior to ATRA treatment only partially restored the level of proliferation relative to untreated cells, indicating that the up-regulation of these miRNAs, while important, are not fully responsible for the differentiation phenotype. It is noteworthy that an indirect effect of the ectopic up-regulation of these three miRNAs is the down-regulation of MYCN, which is also an effect of ATRA treatment [43, 51]. None of these miRNAs are predicted to target the MYCN 3′ UTR, so presumably MYCN down-regulation is a secondary effect. Intriguingly, all three of these miRNAs target a truncated isoform of a neurotrophic tyrosine receptor kinase, NTRK3 (also known as TRKC) and siRNA mediated knock-down of this mRNA alone has similar anti-proliferative effects [50]. Neurotrophins are growth factors which regulate many important neural cell functions, including cell survival. Subsequently, miR-9-2 was determined to directly target the RE1-silencing transcription factor (REST) [52]. REST on the other hand, binds to the promoter region of this miRNA and represses transcription in coordination with the cAMP-response element binding protein (CREB), forming an auto-regulatory feedback loop [52].
Figure 1.
Summary of functional studies carried out on miRNAs that are either up-regulated (red) or down-regulated (green) in neuroblastoma cell lines treated with ATRA. Experimentally validated mRNA targets are connected by red lines to each miRNA, while non-validated computationally predicted targets are designated by black lines. The biological effects of resulting from artificial over or under-expression of each miRNA is designated on the right side of the panel.
It is unclear from the study of Laneve et al [50] whether over-expression of miR-125b in SK-N-BE cells results in neuronal differentiation, or only a decrease in the cell proliferation rate, as cell morphology changes were not discussed. A more recent study by Le et al [40] provided evidence that ectopic over-expression of miR-125b, as well as miR-124a, cause an increase in neurite outgrowth in the SH-SY5Y neuroblastoma cell line. Induced differentiation of the non-cancer derived RVM cell line by removal of growth factors also results in the up-regulation of miR-125b, while ectopic up-regulation of this miRNA in these cells promotes neurite outgrowth [40]. Further experiments with miR-125b revealed that this miRNA resulted in over-expression of neuronal markers MAP2AB and GABBR1 and decreased expression of the neural progenitor marker MSI1. A very elegant analysis based on mRNA expression profiling was provided of the down-stream genetic pathways that are regulated by miR-125b. mRNA expression profiling of SH-SY5Y cells ectopically over-expressing this miRNA identified 164 down-regulated genes with predicted target sites. Le et al [40] developed a model whereby miR-125b directly post-transcriptionally represses ten genes that are key effectors of neuronal differentiation, launching a complex cascade of transcriptional changes of down-stream effectors for neuronal differentiation. Some of the miRNAs that are up-regulated in response to ATRA appear to overlap targets in the differentiation pathway, such as miR-184, which directly targets AKT2 [32], a down-stream effector in the pathway.
As previously mentioned, Le et al [40] determined that ectopic over-expression of miR-124a causes human SH-SY5Y cells to under differentiation. Prior to this observation, Makeyev et al [53] determined that ectopic up-regulation of miR-124 at physiological levels causes the mouse CAD neuroblastoma cell line to undergo differentiation. They further showed that the polypyrimidine tract binding protein 1 (PTBP1), which is a regulator of mRNA splicing, is directly targeted by miR-124 and that a decrease in PTBP1 protein is required for differentiation of CAD cells. PTBP1 represses a related family member, PTBP2, by causing exon skipping that generates a nonsense codon, subjecting PTBP2 to nonsense-mediated decay. Thus, miR-124 mediated inhibition of PTBP1 leads to correctly spliced PTBP2 mRNA that is translated into PTBP2 protein, which in turn causes a shift towards a neuronal pattern of pre-mRNA splicing of genes and a differentiated phenotype.
Artificial modulation of a number of additional miRNAs appears to also cause neuroblastoma cells to undergo differentiation. For example, ectopic over-expression of miR-214, and conversely, inhibition of the endogenous miR-7, causes an increase in neurite outgrowth in SH-SY5Y cells. However, the essential mRNA targets of these miRNAs have not been identified [38]. The oncogenic miR-17-5p-92 polycistronic cluster, which is up-regulated in poor prognosis neuroblastoma tumors [6-8, 54], is down-regulated following ATRA-induced differentiation of neuroblastoma cells [37]. In a very interesting study, Loven et al [55] demonstrated that miR-18a and -19a, which are members of this polycistronic cluster, repress the estrogen receptor-α (ES1) transcription factor. ES1 is involved with the process of differentiation and these authors demonstrated that ectopically introduced ES1 in SK-N-BE cells induces neuronal differentiation, while inhibition of endogenous miR-18a results in neurite outgrowth.
MiR-128 is another miRNA which is up-regulated in response to ATRA, resulting in decreased neuroblastoma cell motility and invasiveness through inhibiting the Reelin and DCX genes when ectopically over-expressed [56]. The function of miR-128 has some redundancy with miR-9, -125a and -125b, as all four of these miRNAs target the truncated isoform of NTRK3 [57]. Interestingly, the full length and truncated isoforms of NTRK3 have non-overlapping 3′ UTRs and are regulated by different sets of miRNAs [57]. Guidi et al [57] demonstrated that ectopic up-regulation of miR-128 causes SH-SY5Y cells to become smaller, with shortened neurites, and to have increased cell proliferation. siRNA mediated inhibition of the truncated NTRK3 recapitulated the phenotype obtained by miR-128, indicating that NTRK3 silencing was primarily responsible for the observed effects of this miRNA. mRNA expression microarray analysis of cells over-expressing miR-128 indicated that anti-apopotic genes, most notably BCL2, tended to be up-regulated as an indirect effect of miR-128 up-regulation. The finding that miR-128 enhances cell proliferation is not necessarily contradictory to the earlier conclusion by Evangelisti et al [56] that miR-128 causes decreased cell motility and invasiveness, as these phenotypic traits can be independently regulated [58].
Recent studies have shown that miR-10a and -10b are extremely potent inducers of neuroblastoma cell differentiation. As demonstrated by Meseguer et al [41] and our own group [39], anti-miR knockdown of miR-10a/b prior to ATRA treatment of SHSY-5Y or SK-N-BE results in a partial inhibition of neurite outgrowth. Although Meseguer et al [41] found that ectopic over-expression of these miRNAs only marginally increased the expression of several markers of differentiation, we determined that over-expression of either miR-10a and -10b caused highly significant increased neurite outgrowth in SH-SY5Y, SK-N-BE and LAN5 cells, along with highly significant increases in the neuronal markers GAP43 and β-3-tubulin at mRNA and protein level. In addition, over-expression of either miRNA resulted in highly significant reduction in MYCN mRNA and protein in all three cell lines, similar to that which is observed following ATRA induced differentiation [39].
Through mRNA expression profiling, we also demonstrated that miR-10a and -10b over-expression inhibits a large number of genes with predicted target sites and launches a cascade of secondary and tertiary alterations to the transcriptome. In the study by Meseguer et al [41], the SR-family splicing factor (SFRS1) was determined to be a direct target of miR-10a/b and that miRNA inhibition of this splicing factor can alter levels of mRNA isoforms, although this genes does not affect differentiation. In our study, the nuclear receptor co-repressor 2 (NCOR2, also known as SMRT), was identified as the primary target of miR-10a and -10b that is responsible for causing the differentiation phenotype, as siRNA mediated inhibition of this gene completely recapitulated a differentiated phenotype [39]. Moreover, co-transfection of an NCOR2 expression construct lacking a 3′UTR target sequence with miR-10a synthetic mimics completely blocked differentiation [39].
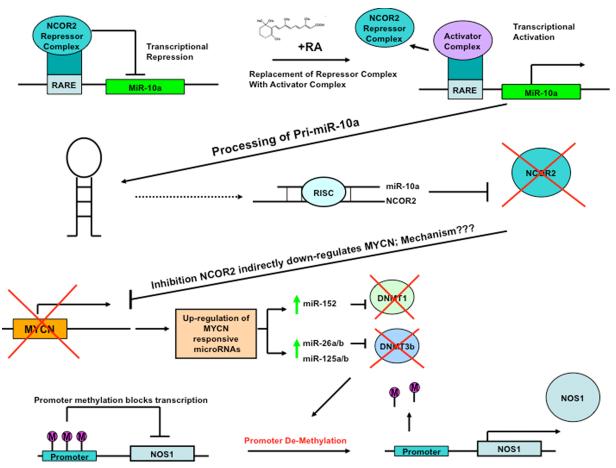
A model of some of the steps involved with ATRA induced differentiation is summarized in Figure 2, with miR-10a over-expression being an early event. MiR-10a localizes within a HOX gene cluster on chromosome 17q, and available evidence indicates that this miRNA is a direct retinoic acid target, having a retinoic acid response element [59]. ATRA causes replacement of the NCOR2 associated repressor complex with an activator complex, leading to transcriptional activation of miR-10a, which in turn keeps NCOR2 mRNA suppressed through direct targeting. This auto-regulatory feedback loop, as originally described by Foley et al, [39], explains why a differentiated state is maintained in the absence of ATRA. There seems to be considerable redundancy in the roles played by miRNAs in the process of differentiation, as miR-10a/10b are predicted to target some of the same targets as miR-125b (for example, ITCH, GAB2 and MKNK2), while NCOR2 is also a predicted target of miR-125b, as originally discussed by Foley et al[39]. For future studies, it would be of great interest to ascertain the direct targets of NCOR2 that are responsible for the induction of differentiation.
Figure 2.
Model depicting some of the miRNA and DNA demethylation driven events important for initiating and maintaining neuroblastoma cells in a differentiated state following treatment with ATRA.
From all of the above cited studies it is obvious that miRNAs contribute in a major and highly complex way to the process of neuroblastoma cell differentiation. It is likely that many additional miRNAs that are either up or down-regulated in response to ATRA also contribute to the complex ATRA induced differentiated phenotype. As discussed in the next section, miR-152, which is up-regulated in response to ATRA in multiple neuroblastoma cell lines, has a profound impact on cell phenotype and on the DNA methylation status of many genes through direct targeting and down-regulation of a DNA methyltransferase (DNMT1) [60].
6. The role of DNA methylation changes in neuroblastoma differentiation
To date, limited information regarding the role of DNA methylation alterations in ATRA induced cellular differentiation has been reported. Previously, Love et al. [61] demonstrated hypermethylation and resulting transcriptional repression of the hTERT gene in response to ATRA induced differentiation of HL60 leukemia cells. The authors demonstrated that the typical phenotypic traits associated with ATRA induced differentiation such as reduced cell proliferation and induced apoptosis in HL60 cells were only observed after inhibition of the telomerase activity, thus highlighting the important role of DNA methylation in granulocytic differentiation. In contrast, Rowling et al. [62] reported an overall reduction in DNA methylation levels in rats treated with ATRA.
Recently, we have identified genome-wide DNA methylation alterations which occur during the process of ATRA induced neuroblastoma differentiation [60]. Using the ATRA sensitive neuroblastoma cell line SK-N-BE, a genome-wide methylation analysis of pre- and post-ATRA differentiated states revealed approximately 402 and 88 gene promoters which were hypo- and hypermethylated, respectively. Integration with mRNA expression data revealed 82 hypomethylated genes were over-expressed greater (>2-fold) and 13 hypermethylated genes were under expressed (>2-fold), indicating that many of the DNA methylation alterations had functional consequences. DNA demethylation has been previously shown to have a role in normal development [63, 64] as well as disease progression [65]. The reduced levels of DNA methylation observed in the ATRA treated neuroblastoma cell lines is also consistent with what is detected in more differentiated tumor subtypes, ganglioneuroblastoma (GNB) and ganglioneuroma (GN) versus immature neuoblastoma [66]. On average, fewer hypermethylated loci were detected in the more differentiated GNB/GN tumors than in the more clinically aggressive neuroblastomas [67].
An example of one of the genes that was demethylated and over expressed in several neuroblastoma cell lines treated with ATRA was NOS1 [60]. The NOS1 gene on chromosome 12 catalyzes the generation of nitric oxide and has been implicated in neural development [68] as well as neuroblastoma proliferation and differentiation [69]. Ciani et al. [69] reported that over expression of NOS1 in SK-N-BE cells, resulting in an increase in nitric oxide, enhances neurite extension and decreases cell proliferation, thus highlighting the crucial role of NOS1 in the differentiation process.
To determine the mechanism underlying the overall reduced levels of DNA methylation following ATRA induced differentiation, mRNA and protein levels for the three DNA methyltransferases (DNMT1, DNMT3A and DNMT3B) were determined post-ATRA treatment [60]. DNMTs are responsible for de novo methylation and maintenance of methylation patterns in the cell [70]. A statistically significant decrease in expression of both DNMT1 and DNMT3A was identified post ATRA treatment in several neuroblastoma cell lines. To determine a possible mechanism for the observed decrease in DNMT expression, a miRNA expression screen was performed pre- and post-ATRA which identified 17 up-regulated miRNAs. One of these up-regulated miRNAs, miR-152, was determined to directly target DNMT1, which maintains methylation patterns during cell division with a preference for hemi-methylated DNA [71]. MiR-152 targeting of DNMT1 was also recently demonstrated by Huang et al. [72] in hepatic cell lines, resulting in decreased methylation levels. In neuroblastoma, miR-152 is under-expressed in MYCN amplified tumors [6], so it is possible that the reduction in MYCN that occurs in ATRA treated cells is responsible for the miR-152 up-regulation. Ectopic over-expression of miR-152 in SK-N-BE cells did not cause the cells to undergo differentiation, but did have a negative effect on cell invasion without affecting proliferation rate. Ragusa et al [42], on the other hand, reported that inhibition of endogenous miR-152 in SH-SY5Y cells resulted in decreased cell invasion without affecting cell proliferation. This result, taken together with our result, might indicate that miR-152 targets genes that both promote and antagonize cell invasion, and that disturbing the balances between such genes is what causes a phenotypic effect.
Interestingly, other miRNAs up-regulated in response to ATRA, such as miR-125a/b and miR-26a/b, are predicted to target DNMT3B, and thus might also contribute to changes in the DNA methylome during differentiation. Our model for a miRNA driven differentiation process triggered by ATRA, which includes genome-wide DNA methylation alterations, is summarized in Figure 1. In this model, changes in gene expression occurring as a consequence of DNA demethylation are relatively late events. In summary, the shift in DNA methylation patterns observed during the process of ATRA induced neuroblastoma in vitro differentiation reflects the flexibility and dynamism of the epigenome and represents an area of cross-talk between two distinctly different modes of epigenetic regulation, namely miRNA and DNA methylation.
7. Concluding remarks and areas for further research
Figure 2 summarizes some of the events that have been determined to be of importance in ATRA induced differentiation of MYCN amplified neuroblastoma cell lines. As a direct retinoid target, up-regulation of miR-10a is predicted to be a very early event in differentiation. The displacement of the NCOR2 co-repressor complex with an activator complex results in miR-10a up-regulation and subsequent constitutive post-transcriptional down-regulation of NCOR2 through direct miR-10a targeting. The down-regulation of NCOR2 then causes a cascade of direct and indirect alterations to the transcriptome, including the down-regulation of MYCN. The identification of the direct targets of the NCOR2 co-repressor critical for inducing differentiation would be of great interest. Likewise, the exact mechanism leading to the inhibition of MYCN as a secondary consequence of NCOR2 inhibition would be of interest. It is tempting to speculate that MYCN is down-regulated post-transcriptionally, as MYCN has predicted target sites for some of the miRNAs that are up-regulated in response to ATRA. Later events in the model include the targeting of DNA methytransferases by miRNAs such as miR-152, which are up-regulated after being released from the repressive effects of MYCN. The down-regulation of the DNA methytransferases ultimately leads to the demethylation and re-expression of genes known to be important in differentiation, such as NOS1. Clearly, alterations in miRNAs expression and DNA methylation play important and complex roles in the process of neuroblastoma cell differentiation. Although neuroblastoma has proven to be an excellent model for the elucidation of signaling pathways involved with the process of neural cell differentiation [45], it is important that we also further test the models developed from these lines in cells of non-cancerous origin.
Acknowledgments
Funding This work was supported in part by grants from This work was supported in part by Science Foundation Ireland (07/IN.1/B1776), Children’s Medical and Research Foundation, Cancer Research Ireland and the U.S. National Institutes of Health (5R01CA127496).
Footnotes
Conflict of Interests The authors declare that they have no conflicting interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- [2].Stallings RL. Are chromosomal imbalances important in cancer? Trends Genet. 2007;23:278–83. doi: 10.1016/j.tig.2007.03.009. [DOI] [PubMed] [Google Scholar]
- [3].Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–33. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- [4].Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–8. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- [5].Vermeulen J, De Preter K, Laureys G, Speleman F, Vandesompele J. 59-gene prognostic signature sub-stratifies high-risk neuroblastoma patients. Lancet Oncol. 2009;10:1030. doi: 10.1016/S1470-2045(09)70325-0. [DOI] [PubMed] [Google Scholar]
- [6].Bray I, Bryan K, Prenter S, Buckley PG, Foley NH, Murphy DM, et al. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS One. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mestdagh P, Fredlund E, Pattyn F, Schulte JH, Muth D, Vermeulen J, et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2010;29:1394–404. doi: 10.1038/onc.2009.429. [DOI] [PubMed] [Google Scholar]
- [8].Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, Eilers UC, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- [9].Shimada H, Umehara S, Monobe Y, Hachitanda Y, Nakagawa A, Goto S, et al. International neuroblastoma pathology classification for prognostic evaluation of patients with peripheral neuroblastic tumors: a report from the Children’s Cancer Group. Cancer. 2001;92:2451–61. doi: 10.1002/1097-0142(20011101)92:9<2451::aid-cncr1595>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- [10].Matthay KK, Villablanca JG, Seeger RC, Stram DO, Harris RE, Ramsay NK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N Engl J Med. 1999;341:1165–73. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- [11].Abemayor E, Sidell N. Human neuroblastoma cell lines as models for the in vitro study of neoplastic and neuronal cell differentiation. Environ Health Perspect. 1989;80:3–15. doi: 10.1289/ehp.89803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- [13].Noy N. Between death and survival: retinoic acid in regulation of apoptosis. Annu Rev Nutr. 2010;30:201–17. doi: 10.1146/annurev.nutr.28.061807.155509. [DOI] [PubMed] [Google Scholar]
- [14].He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- [15].Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- [16].Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- [17].Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–7. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- [20].Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2009;42:1316–29. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- [22].Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- [23].Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–11. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Calin GA. MicroRNAs and cancer: what we know and what we still have to learn. Genome Med. 2009;1:78. doi: 10.1186/gm78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stallings RL. MicroRNA involvement in the pathogenesis of neuroblastoma: potential for microRNA mediated therapeutics. Curr Pharm Des. 2009;15:456–62. doi: 10.2174/138161209787315837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stallings RL, Foley NH, Bryan K, Buckley PG, Bray I. Therapeutic targeting of miRNAs in neuroblastoma. Expert Opin Ther Targets. 2010;14:951–62. doi: 10.1517/14728222.2010.510136. [DOI] [PubMed] [Google Scholar]
- [27].Eggert A, Schulte JH. A small kiss of death for cancer. Nat Med. 2010;16:1079–81. doi: 10.1038/nm1010-1079. [DOI] [PubMed] [Google Scholar]
- [28].Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–83. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- [29].Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–50. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schulte JH, Schowe B, Mestdagh P, Kaderali L, Kalaghatgi P, Schlierf S, et al. Accurate prediction of neuroblastoma outcome based on miRNA expression profiles. Int J Cancer. 2010;127:2374–85. doi: 10.1002/ijc.25436. [DOI] [PubMed] [Google Scholar]
- [31].Bray I, Tivnan A, Bryan K, Foley NH, Watters KM, Tracey L, et al. MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma. Cancer Lett. 2011;303:56–64. doi: 10.1016/j.canlet.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Foley NH, Bray IM, Tivnan A, Bryan K, Murphy DM, Buckley PG, et al. MicroRNA-184 inhibits neuroblastoma cell survival through targeting the serine/threonine kinase AKT2. Mol Cancer. 2010;9:83. doi: 10.1186/1476-4598-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tivnan A, Foley NH, Tracey L, Davidoff AM, Stallings RL. MicroRNA-184-mediated inhibition of tumour growth in an orthotopic murine model of neuroblastoma. Anticancer Res. 2010;30:4391–5. [PMC free article] [PubMed] [Google Scholar]
- [35].Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–22. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- [37].Beveridge NJ, Tooney PA, Carroll AP, Tran N, Cairns MJ. Down-regulation of miR-17 family expression in response to retinoic acid induced neuronal differentiation. Cell Signal. 2009;21:1837–45. doi: 10.1016/j.cellsig.2009.07.019. [DOI] [PubMed] [Google Scholar]
- [38].Chen H, Shalom-Feuerstein R, Riley J, Zhang SD, Tucci P, Agostini M, et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394:921–7. doi: 10.1016/j.bbrc.2010.03.076. [DOI] [PubMed] [Google Scholar]
- [39].Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, et al. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18:1089–98. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, et al. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Meseguer S, Mudduluru G, Escamilla JM, Allgayer H, Barettino D. Micro-RNAs-10a and -10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF) J Biol Chem. 2010 doi: 10.1074/jbc.M110.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ragusa M, Majorana A, Banelli B, Barbagallo D, Statello L, Casciano I, et al. MIR152, MIR200B, and MIR338, human positional and functional neuroblastoma candidates, are involved in neuroblast differentiation and apoptosis. J Mol Med. 2010;88:1041–53. doi: 10.1007/s00109-010-0643-0. [DOI] [PubMed] [Google Scholar]
- [43].Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature. 1985;313:404–6. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- [44].Murphy DM, Buckley PG, Bryan K, Das S, Alcock L, Foley NH, et al. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS One. 2009;4:e8154. doi: 10.1371/journal.pone.0008154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Edsjo A, Holmquist L, Pahlman S. Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin Cancer Biol. 2007;17:248–56. doi: 10.1016/j.semcancer.2006.04.005. [DOI] [PubMed] [Google Scholar]
- [46].Henriksen JR, Haug BH, Buechner J, Tomte E, Lokke C, Flaegstad T, et al. Conditional expression of retrovirally delivered anti-MYCN shRNA as an in vitro model system to study neuronal differentiation in MYCN-amplified neuroblastoma. BMC Dev Biol. 2011;11:1. doi: 10.1186/1471-213X-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Negroni A, Scarpa S, Romeo A, Ferrari S, Modesti A, Raschella G. Decrease of proliferation rate and induction of differentiation by a MYCN antisense DNA oligomer in a human neuroblastoma cell line. Cell Growth Differ. 1991;2:511–8. [PubMed] [Google Scholar]
- [48].Tonelli R, Purgato S, Camerin C, Fronza R, Bologna F, Alboresi S, et al. Anti-gene peptide nucleic acid specifically inhibits MYCN expression in human neuroblastoma cells leading to cell growth inhibition and apoptosis. Mol Cancer Ther. 2005;4:779–86. doi: 10.1158/1535-7163.MCT-04-0213. [DOI] [PubMed] [Google Scholar]
- [49].Edsjo A, Nilsson H, Vandesompele J, Karlsson J, Pattyn F, Culp LA, et al. Neuroblastoma cells with overexpressed MYCN retain their capacity to undergo neuronal differentiation. Lab Invest. 2004;84:406–17. doi: 10.1038/labinvest.3700061. [DOI] [PubMed] [Google Scholar]
- [50].Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, et al. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci U S A. 2007;104:7957–62. doi: 10.1073/pnas.0700071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Thiele CJ, Israel MA. Regulation of N-myc expression is a critical event controlling the ability of human neuroblasts to differentiate. Exp Cell Biol. 1988;56:321–33. doi: 10.1159/000163498. [DOI] [PubMed] [Google Scholar]
- [52].Laneve P, Gioia U, Andriotto A, Moretti F, Bozzoni I, Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010;38:6895–905. doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Schulte JH, Schowe B, Mestdagh P, Kaderali L, Kalaghatgi P, Schlierf S, et al. Accurate prediction of neuroblastoma outcome based on mirna expression profiles. Int J Cancer. 2010 doi: 10.1002/ijc.25436. [DOI] [PubMed] [Google Scholar]
- [55].Loven J, Zinin N, Wahlstrom T, Muller I, Brodin P, Fredlund E, et al. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci U S A. 2010;107:1553–8. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Evangelisti C, Florian MC, Massimi I, Dominici C, Giannini G, Galardi S, et al. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J. 2009;23:4276–87. doi: 10.1096/fj.09-134965. [DOI] [PubMed] [Google Scholar]
- [57].Guidi M, Muinos-Gimeno M, Kagerbauer B, Marti E, Estivill X, Espinosa-Parrilla Y. Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol Biol. 2010;11:95. doi: 10.1186/1471-2199-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gao CF, Xie Q, Su YL, Koeman J, Khoo SK, Gustafson M, et al. Proliferation and invasion: plasticity in tumor cells. Proc Natl Acad Sci U S A. 2005;102:10528–33. doi: 10.1073/pnas.0504367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–45. e1–7. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- [60].Das S, Foley N, Bryan K, Watters KM, Bray I, Murphy DM, et al. MicroRNA mediates DNA demethylation events triggered by retinoic acid during neuroblastoma cell differentiation. Cancer Res. 2010;70:7874–81. doi: 10.1158/0008-5472.CAN-10-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Love WK, Berletch JB, Andrews LG, Tollefsbol TO. Epigenetic regulation of telomerase in retinoid-induced differentiation of human leukemia cells. Int J Oncol. 2008;32:625–31. [PMC free article] [PubMed] [Google Scholar]
- [62].Rowling MJ, McMullen MH, Schalinske KL. Vitamin A and its derivatives induce hepatic glycine N-methyltransferase and hypomethylation of DNA in rats. J Nutr. 2002;132:365–9. doi: 10.1093/jn/132.3.365. [DOI] [PubMed] [Google Scholar]
- [63].Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–82. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- [64].Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–20. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66:2249–61. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Buckley PG, Das S, Bryan K, Watters KM, Alcock L, Koster J, et al. Genome-wide DNA methylation analysis of neuroblastic tumors reveals clinically relevant epigenetic events and large-scale epigenomic alterations localized to telomeric regions. Int J Cancer. 2010;128:2296–305. doi: 10.1002/ijc.25584. [DOI] [PubMed] [Google Scholar]
- [67].Buckley PG, Das S, Bryan K, Watters KM, Alcock L, Koster J, et al. Genome-wide DNA methylation analysis of neuroblastic tumors reveals clinically relevant epigenetic events and large-scale epigenomic alterations localized to telomeric regions. Int J Cancer. 2010 doi: 10.1002/ijc.25584. [DOI] [PubMed] [Google Scholar]
- [68].Bredt DS, Snyder SH. Transient nitric oxide synthase neurons in embryonic cerebral cortical plate, sensory ganglia, and olfactory epithelium. Neuron. 1994;13:301–13. doi: 10.1016/0896-6273(94)90348-4. [DOI] [PubMed] [Google Scholar]
- [69].Ciani E, Severi S, Contestabile A, Bartesaghi R. Nitric oxide negatively regulates proliferation and promotes neuronal differentiation through N-Myc downregulation. J Cell Sci. 2004;117:4727–37. doi: 10.1242/jcs.01348. [DOI] [PubMed] [Google Scholar]
- [70].Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- [71].Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–9. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- [72].Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]