Abstract
Background
Orofacial clefts are the most common malformations of the head and neck with a World-wide prevalence of 1/700 births. They are commonly divided into CL(P) and CP based on anatomical, genetic and embryological findings. A Nigerian craniofacial anomalies study “NigeriaCRAN” was set up in 2006 to investigate the role of gene-environment interaction in the etiology of orofacial clefts in Nigeria.
Subjects and Methods
DNA isolated from saliva from the Nigerian probands was used for genotype association studies and direct sequencing on the cleft candidate genes: MSX1, IRF6, FOXE1, FGFR1, FGFR2, BMP4, MAFB, ABCA4, PAX7 and VAX1, and the chromosome 8q region.
Results
A missense mutation A34G in MSX1 was observed in nine cases and four hap map controls. No other apparent etiologic variations were identified. A deviation from HWE was observed in the cases (p= 0.00002). There was a significant difference between the affected side for unilateral CL (p=0.03) and, between bilateral clefts and clefts on either side (p=0.02). A significant gender difference was also observed for CP (p=0.008).
Conclusions
The replication of a mutation previously implicated in other populations suggests a role for the MSX1 A34G variant in the etiology of CL(P).
Keywords: Orofacial clefts, MSX1, Nigeria, A34G
INTRODUCTION
In spite of better quality health care access in developed countries, mortality due to birth defects accounts for an increasing proportion of infant mortality (30–50% of perinatal deaths and 20–30 % of infant mortality) (WHO, 2005). Neonatal death is an issue in Sub-Saharan Africa that cannot be ignored with rates in many places more than ten times greater than those in developed countries. The multiple causes include infectious diseases (HIV / AIDS, tuberculosis and malaria); malnutrition; poverty and a possibility of infanticide in those born with congenital birth defects including orofacial clefts (Strauss, 1985; Akinmoladun et al., 2007).
Orofacial clefts (OFC) require complex multidisciplinary treatment and are associated with elevated infant mortality and significant lifelong morbidity. The development of craniofacial structures is an exquisitely orchestrated process involving the coordinated growth of multiple, independently derived primordia. Perturbations impacting on the growth of these primordia may interfere with the proper morphogenesis of facial structures, resulting in clefting of the lip, the primary or secondary palate, or a combination of these sites (Jugessur et al., 2009).
Clefts of either the lip and/or palate are the most frequent congenital malformation of the head and neck with a combined overall prevalence of approximately 1 in 700 (WHO, 2002). With variation between ethnic groups and geographic regions, high rates are reported for cleft lip with or without cleft palate (CL(P)) in parts of Latin America and Asia (China, Japan) and low in Israel, South Africa, and southern Europe. Rates of isolated cleft palate are high in Canada and parts of northern Europe and low in parts of Latin America and South Africa (Mossey et al., 2009).
In sub-Saharan Africa, the prevalence of orofacial clefts remains uncertain due to the lack of data and the few reported studies suggest that the prevalence is low compared to rates reported in other continents. However, caution must be exercised when comparing rates of other continents to Africa because of the differences in ascertainment, sample sizes and analysis methodology (Butali and Mossey, 2009). Although studies on prevalence of clefts are limited, there are several reports in the literature on the care and management of orofacial clefts in Nigeria (Onah et al., 2008; Olasoji et al., 2009).
Recent advances in the knowledge of OFC etiology have reported strong evidence and a significant association with polymorphisms in certain candidate genes including interferon regulatory factor six (IRF6) (Zucchero et al., 2004, Rahimov et al., 2008), forkehead boxE1 (FOXE1) (Marazita at al., 2004 and Moreno et al., 2009) and muscle segment homeobox (MSX1) (Sakatoka and Maas, 1994; Jezewski et al., 2003, Suzuki et al., 2004, Tongkobpetch et al., 2006, and Modesto et al., 2006).
Recently the first genome wide association study (GWAS) of CL/P identified genetic susceptibility for non-syndromic CL(P) on chromosome 8q24.21 (Birnbaum et al., 2009). Consistent with the findings of Birnbaum et al 2009, another GWAS identified genetic susceptibility on chromosome 8q24 amongst non-syndromic cleft lip with or without (CL(P)) cases (Grant et al., 2009). Both studies used individuals of European ancestry. In a recent expansion of the number of samples used in the Birnbaum paper, a GWAS carried out by Mangold et al. (2010) reported two new loci associated with NSCL/P at 17q22 and 10q25.3(Mangold et al., 2010).
GWAS carried out by Beaty et al (2010) for CL/P replicated the evidence of association at a genome-wide level of significance (i.e. p-value<5*10-8) for SNPS on chromosome 8q24 and IRF6. In addition the study identified several other loci that showed evidence of association attaining genome-wide significance for two loci (ABCA4 on chromosome 1p22.1, MAFB on chromosome 20q12), and was strongly suggestive for PAX7 on chromosome 1p36, VAX1 on chromosome 10q25.3 and NTN1 on chromosome 17p13).
Other candidate genes that have been reported to play a role in clefting include the bone morphogenetic protein (BMP4)(Suzuki et al., 2009), Tumor protein 63 (TP63) (Scapoli et al., 2008), Jagged 2 gene (JAG2) (Vieira et al., 2005 and Scapoli et al., 2008), Fibroblast growth factors and their receptors (FGFs, FGFRs) (Riley et al., 2007), Poliovirus Receptor Related 1 (PVRL 1) (Avila et al., 2005, Turhani et al., 2005). All these candidate gene studies and genetic analyses serve to inform the current study of oral facial clefting in the Nigerian population.
The current study investigated the role of these candidate genes and potential environmental influences in the etiology of orofacial cleft in the Nigerian population using a case-control study design. Direct sequencing and genotyping was used to evaluate the role of a selected group of candidate genes amongst Nigerian probands.
MATERIALS AND METHODS
Patient and Control Recruitment
Ascertainment of eligible cases was through the free surgical repairs project sponsored by a U.S. based charity Smile Train. All participating centers in the NigeriaCRAN project are Smile Train partners and this link ensured recruitment of eligible patients. Prior to the Smile Train support, the financial burden as a result of surgical cost was a limiting factor to recruitment.
Subjects were ascertained through the Nigerian craniofacial anomalies project (NigeriaCRAN); a multi-center collaboration between the University of Dundee, Scotland and 11 hospitals in Nigeria1 All participating hospitals obtained ethical permission. Eligible infants were those born between 1st of September 2006 and 31st of August 2008 to Nigerian parents in Nigeria. Births from Caucasians, Asians and other African countries were excluded. Nigerian children born with cleft lip and palate from the 1st of September 2006 to 31st of August 2008 in Nigeria and their parents were included in the study as cases. During the cleft clinic visit, the children were reviewed by the cleft surgeon and the diagnosis was confirmed and any recognized or diagnosed syndromes excluded. Controls were Nigerian children born alive without any congenital birth defects after the 1st of September 2006 in Nigeria and their parents. These children were examined by a pediatrician to ensure that they did not have any congenital birth defects. A protocol of 2 controls to one case was implemented, and controls were matched to cases by birth month, year, and gender. Controls were recruited in five centers that are located across the five geopolitical regions and representative of the areas where the cases were recruited. Every family (case and control) recruited into the study was assigned a code which is the unique identifier number (UNID) for that family and the study co-ordinator informed the designated cleft surgeon of the intention to approach the patient for NigeriaCRAN project. Signed informed consent was obtained from all families that participated in the study. A nurse or a surgical registrar was designated to interview the mothers to obtain information using the study questionnaire. The information obtained included maternal medical history, lifestyle and occupational and environmental exposures-particularly in the peri-conceptional (before pregnancy and first trimester). The administration of questionnaire to case and control mothers was carried out within the first year of child birth. The aim was to standardize the collection of information. The completed questionnaires were returned to the study coordinator in the hospital and all completed questionnaires were checked, compiled and forwarded on to the study coordinator in Dundee.
DNA extraction
A saliva sample was taken from the child by the study coordinator or surgeon at the clinic or time of surgery using saliva sponges (Oragene). The parent’s saliva samples were taken by arrangement with the cleft team using the 2ml self collecting Oragene kits. All samples were sent directly to the Human Virology laboratory at the Nigerian Institute of Medical Research (NIMR) in Lagos, Nigeria for DNA extraction and storage at −20°C. The extraction of DNA was carried out according to the protocol provided by the product manufacturer (http://www.dnagenotek.com/focus_gr_purification.htm). Following material transfer agreement, the DNA samples were transferred from NIMR to the University of Iowa for the candidate gene sequencing, genotyping and analysis.
A total of 118 cases including 2 cases (76 triads and 42 dyads i.e. mother and child) with a family history of clefts were collected. The family cases were obtained from the mothers as part of the family history of cleft information contained in the questionnaire data information and no physical examination was carried out on the relative with clefts to ascertain the types of clefts. These case individuals had the following phenotypes: 69 CL(P), 38CL, 11 CP. Further classification of the groups into types of clefts, laterality and gender is presented in Table 1. For a case - control comparison, the study attempted to recruit two controls per case by geopolitical region, birth month and gender. A total of 166 control samples (164 dyads i.e. mother and child and 2 triads) were recruited from individuals at hospitals in each geo-political zone (which represents 76.4% of the planned number of controls for the study).
Table 1.
Characteristics of the non-syndromic orofacial cleft case families
| CL/P | CL(P) | CL | CP | Controls | ||
|---|---|---|---|---|---|---|
| sporadic | 116 | 68 | 38 | 10 | ||
| familial | 2 | 1 | 0 | 1 | ||
| Laterality | ||||||
| Right side cleft | 17 | 7 | ||||
| Left side cleft | 18 | 21 | ||||
| Bilateral | 34 | 10 | ||||
| p-values | 0.02a | 0.03b | ||||
| Gender | ||||||
| male | 57 | 38 | 16 | 3 | 88 | |
| female | 61 | 31 | 22 | 8 | 78 | |
| p-values | 0.139c | 0.008d | ||||
| No of probands | 118 | 69 | 38 | 11 | 166 | |
CL/P = orofacial clefts, CL(P) = cleft lip with/without cleft palate and CP = cleft palate.
The superscript are the p-values following comparison using Fisher’s exact test where “a” for between bilateral clefts and either side of clefts, b is between left and right sided cleft lip only, c is between males and females with clefts and d is between males and females with cleft palate.
Table 1 shows the number of non-syndromic clefts, cleft type and controls in males and females. Overall, there were more CL(P) (69) than CL (38) and CP (11). There were more males (38) with CL(P) than females (31), more CL (22) in females than males (16) and more females (8) with CP (3) than males. Overall, there were more males than females from the study. A total of 166 matched controls, 88 males and 78 females were recruited (70.3% recruitment rate). There was a significant difference between the sides for CL (p=0.03) and, between bilateral clefts and clefts on either side (p=0.02) using the Fisher’s exact probability test. A significant gender difference was also observed for CP (p=0.008), but there was no statistically significant gender difference for CL (p=0.139) using the Fisher’s exact probability test.
PCR and Direct sequencing
Ten microliters of the 20ng/ul DNA was sent to the Functional biosciences inc. laboratory in Wisconsin, U.S.A for sequencing the two exons of MSX1 (CR1–4). The PCR conditions were initial 96°C for 5min, 94°C for 30sec, 60°C for 45sec, 72°C for 1min 30 sec for 39 cycles followed by 72°C 5 min and 4°C. Two rounds of PCR amplification were carried out prior to sequencing. The samples were then sequenced in both directions to confirm variants using Big Dye 3.1 on an ABI 3730xl. Sequencing was attempted for the genes: IRF6, FOXE1, and BMP4, but failed secondary to poor quality DNA that was likely damaged in preparation, storage or shipment.
Genotyping
In order to investigate family based associations, genotyping of the case and control families were done using TaqMan technology. A total of 9 candidate genes (Chromosome 8q, ABCA4, BMP4, FGFR1, FGFR2, FOXE1, IRF6, MAFB, MSX1) were investigated using 18 markers (see Results, Table 4). The master mix contained 1.5µl of TaqMan master mix which is the polymerase, 0.0375µl of TaqMan probe (the marker for each gene investigated) and 1.4625 of double distilled H20 (ddH20) making a 3µl of the master mix. At the completion of the PCR process, the plates were read using the 7900HT fast real time PCR system controlled by the SDS software version 2.4. All the files with samples and markers were imported into Progeny version 7.6.04 from Progeny software LLC U.S.A (www.progenygenetics.com/articles.html) to check for Mendelian errors and discrepancies in each file.
Table 4.
Family based association test (FBAT) results for case and control probands.
| Marker | Alleles | freq cases (n=88) |
freq controls (n=88) |
OR (95% CI) |
X2df = 1 p value |
Case HWE | Control HWE |
|---|---|---|---|---|---|---|---|
| IRF6_ rs642961 | 1 | 0.93 | 0.90 | 1.04 (0.78–1.39) | 0.04 (0.832) | 0.583 | 0.212 |
| 2 | 0.07 | 0.10 | |||||
| MSX1_ rs4075 | 1 | 0.81 | 0.70 | 1.25 (0.94–1.67) | 2.2.1(0.138) | 0.131 | 0.319 |
| 2 | 0.19 | 0.30 | |||||
| MSX1_ rs12532 | 1 | 0.62 | 0.52 | 1.22 (0.92–1.63) | 1.81 (0.179) | 0.647 | 0.865 |
| 2 | 0.38 | 0.48 | |||||
| FGFR1_rs13317 | 1 | 0.22 | 0.20 | 1.04 (0.78–1.39) | 1.04 (0.832) | 0.409 | 1 |
| 2 | 0.78 | 0.80 | |||||
| FGFR1_C_2080144_10 | 1 | 0.41 | 0.35 | 1.13 (0.85–1.51) | 1.13 (0.437) | 0.619 | 0.360 |
| 2 | 0.59 | 0.65 | |||||
| ABC4_rs560426 | 1 | 0.51 | 0.51 | 1(0.75–1.33) | 0.01(0.944) | 0.422 | 0 |
| 2 | 0.49 | 0.49 | |||||
| ABC4_rs481931 | 1 | 0.88 | 0.89 | 0.98(0.74–1.31) | 0.01(0.944) | 0.832 | 0 |
| 2 | 0.12 | 0.11 | |||||
| ABC4_rs4147811 | 1 | 0.88 | 0.90 | 0.96(0.72–1.28) | 0.05(0.832) | 0.159 | 0 |
| 2 | 0.12 | 0.10 | |||||
| 8qrs_987525 | 1 | 0.66 | 0.63 | 1.06(0.80–1.42) | 0.13(0.724) | 0.848 | 0.770 |
| 2 | 0.34 | 0.37 | |||||
| MAFB_rs13041247 | 1 | 0.22 | 0.27 | 0.91(0.68–1.21) | 0.41(0.525) | 0.897 | 0.105 |
| 2 | 0.78 | 0.73 | |||||
| FGFR1_C_2080139_10 | 1 | 0.84 | 0.74 | 1.16 (0.86–1.55) | 1.16 (0.351) | 0.771 | 0.980 |
| 2 | 0.16 | 0.26 | |||||
| FGFR1_rs881301 | 1 | 0.17 | 0.26 | 0.83 (0.62–1.10) | 1.63 (0.202) | 0.980 | 0.698 |
| 2 | 0.83 | 0.74 | |||||
| FOXE1_rs647843 | 1 | 0.31 | 0.35 | 0.92 (0.69–1.23) | 0.25 (0.621) | 0.970 | 1.0 |
| 2 | 0.69 | 0.65 | |||||
| FOXE1_rs894673 (9q22.33) | 1 | 0.31 | 0.33 | 0.96 (0.72–1.28) | 0.05 (0.832) | 0.878 | 1.0 |
| 2 | 0.69 | 0.67 | |||||
| FOXE1_rs3758249_G>A | 1 | 0.31 | 0.34 | 0.94 (0.71–1.26) | 0.13 (0.72) | 1 | 0.980 |
| 2 | 0.69 | 0.66 | |||||
| FGFR2_rs10466215 | 1 | 0.21 | 0.18 | 1.06 (0.78–1.42) | 0.13 (0.720) | 0.775 | 1.0 |
| 2 | 0.79 | 0.82 | |||||
| BMP4_rs276188 | 1 | 0.78 | 0.85 | 0.87 (0.65–1.16) | 0.85 (0.358) | 0.512 | 0.970 |
| 2 | 0.22 | 0.15 | |||||
| BMP4_rs17563 | 1 | 0.94 | 0.93 | 1.02(0.77–1.36) | 0.01 (0.944) | 1 | 1.0 |
| 2 | 0.06 | 0.07 |
OR=odd ratio HWE = Hardy Weinberg equilibrium
Table 4 shows the allele frequencies for 88 cases and 88 controls (these are matched case and control probands with complete genotype and maternal environment factor data). The odds ratio was calculated between cases and controls for each marker investigated using the chi-square test. There was no significant result observed for any of the markers investigated.
Statistical Analysis
The Family based association test (FBAT) was used to show evidence for linkage disequilibrium using transmission disequilibrium testing. In this test, heterozygous parents (who do not have the phenotype but may carry the genotypes) are tested to determine the percent of transmission of each of the two alleles.
RESULTS
Direct Sequencing
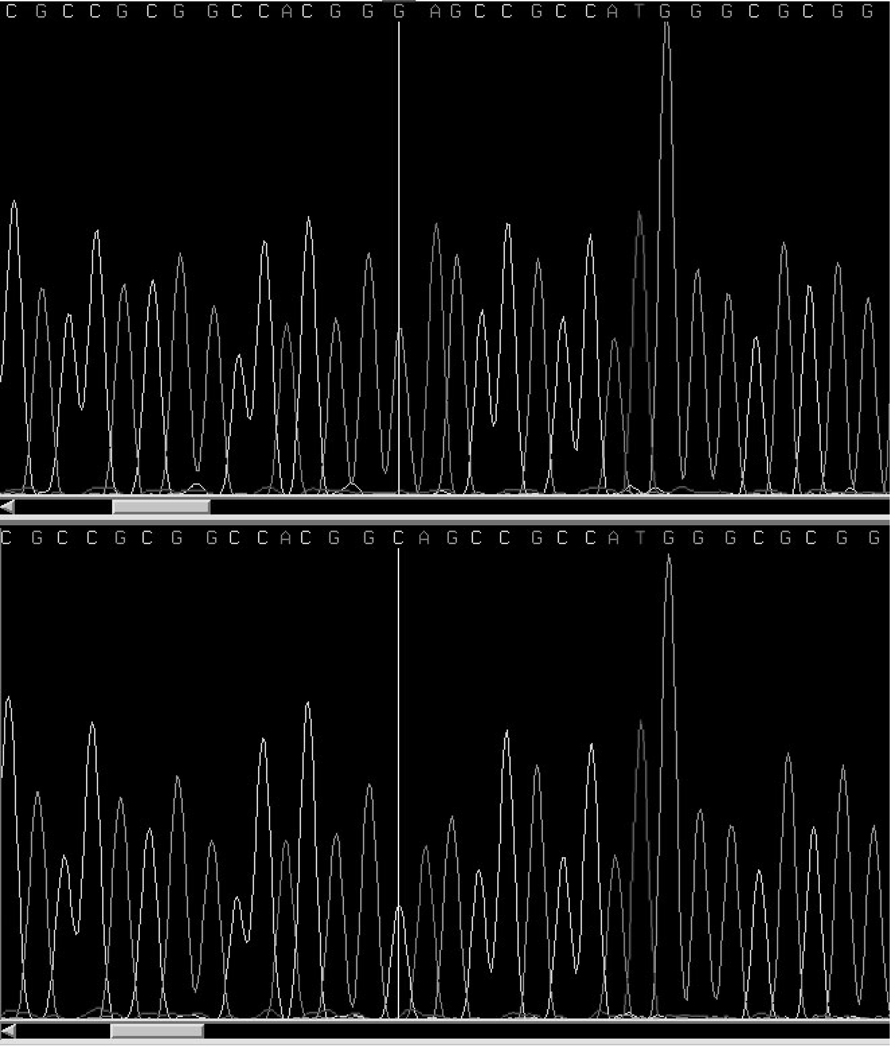
The two exons of MSX1 were sequenced in 59 Nigerian case probands (samples with successfully amplified DNA products following repeated PCR of the 118 cases recruited) using four primers that overlap to ensure that the entire gene is sequenced. These were the same primers (A, B, G, and H) represented graphically (figure 1) in the Jezewski et al. (2003). In exon 1, 1 non-synonymous missense variant (A34G) and one synonymous variant (G116G) were observed (Figure 1). Nine patients have the A34G missense variant; five are homozygous for the rare allele and four of them heterozygous for the wild type. The G116G was observed in 3 probands, two being heterozygote for the wild type and one homozygote for the rare allele.
Figure 1.
MSX1 DNA sequence trace showing the change in nucleotide from CC to GG at nucleotide position 68. This change is a nonsynonomous coding mutation with amino acid change from alanine to glycine at position 34 (A34G).
The control samples failed to amplify after several attempts following PCR and in order to validate the A34G variants in the Nigerian population; the exon 1 was sequenced in 25 unrelated HapMap samples of Nigerian origin (Yoruba) obtained in Ibadan, South West region of Nigeria. Four individuals have the A34G variant, two of them are heterozygous for the wild type and the other two are homozygous for the rare type.
Table 2 shows the sequencing results for MSX1 genes and conserved regions in 59 cleft cases and 25 Hap map Yoruba samples. In exon 1, there was a change in nucleotide at position 68C>G relative to the start codon. This change is a non-synonymous coding mutation which changes the amino acid from alanine to glycine at position 34 (A34G). In the same CR1, there was also a change in nucleotide at position 297 C>T relative to the start codon. The change is a synonymous mutation with no change in amino acid for the glycine at position 116 (G116G). Deletion of a T nucleotide was observed in nucleotide position 150 in the exon 1.
Table 2.
Variant list for MSX1 in the Nigerian population
| Nucleotide variant |
Amino acid variant |
Variant frequencya/Population | |
|---|---|---|---|
| Cases | Hap map samples | ||
| 68 C>G | A34G | 14/118 | 8/50 |
| 297 C>T | G116G | 6/118 | - |
| 150 T-deletion | - | 22/118 | - |
Number of variants is number of chromosomes for each allele type, not individuals.
Table 3 shows the expected frequencies of the alleles for the cases and Hap map controls calculated from the observed genotypes. In the cases, there is significant deviation from Hardy Weinberg equilibrium (p=0.00002).
Table 3.
Hardy Weinberg equilibrium.
| p | Q | p-e2 | 2pq | q –e2 | χ -e2 | df | P | |
|---|---|---|---|---|---|---|---|---|
| C | G | CC | CG | GG | ||||
| Cases | 104 | 14 | 50 | 4 | 5 | 21.681 | 2 | 0.00002 |
| Expected | 104 | 14 | 46 | 12 | 1 | |||
| Controls | 44 | 6 | 21 | 2 | 2 | 3.011 | 2 | 0.221 |
| Expected | 43 | 7 | 19 | 5 | 1 |
Table 4 shows the allele frequencies for 88 case families and 88 control families. These are samples with complete genotype and environmental data out of the total 118 cases and 166 controls recruited). The odds ratio was calculated between cases and controls for each marker investigated using the Fisher’s exact test. There was no significant result observed for any of the markers investigated.
DISCUSSION
Epidemiology
The prevalence of cleft lip and palate in Africa has often being reported as low relative to other populations (Iregbulem, 1982). However, there is only one birth defects surveillance system in Africa (the South African birth defects surveillance system SABDSS) currently monitoring birth defects. In most studies that have reported prevalence, data were obtained from hospital records which are often prone to bias. A systematic review by Butali and Mossey (2009) concluded that cleft lip and palate in Africa is under ascertained. The authors suggested that ascertainment can be improved by establishing birth defects registries through collaboration amongst various experts involved with cleft care in Africa. Little or no research on the genetic or environmental factors associated with craniofacial anomalies has been carried out in the African population (WHO, 2004).
In the present study there is evidence of incomplete ascertainment of clefts and therefore this study sample is prone to biases. The proportion of CP is low consistent with similar reports from previous studies in Africa (Iregbulem,1982 and Ogle,1993) and different compared to other studies that reported a high prevalence of CP in Northern Europe (Tolarova and Cervenka, 1998). There is also a high proportion of bilateral clefts, particularly in the CL(P) group, when they are compared to previous literature reports citing a low prevalence (Hagberg et al., 1998). Poorer survival and biases in referral based on severity may affect the observed rates in populations where true birth prevalences cannot be determined. The greater number of CL(P) in males, more CP in females and the CL(P) to CL ratio of 2:1 are however consistent with the literature (Mossey and Little, 2002).
Genetic
Various ethnic groups in Nigeria believe that witchcraft, evil spirit or the devil, the mother, and occasionally the child are responsible for orofacial clefts outcomes (Oginni et al., 2010). These cultural and ethnic beliefs may be a reason for infanticide in those born with orofacial clefts (Strauss, 1985; Akinmoladun et al., 2007).
The present study is the first to investigate candidate genes in orofacial clefts in the African population. Although the small sample size limits statistical power, there is tendency towards genetic association at a few candidate gene loci.
MSX1 sequencing
Multiple missense coding mutations within MSX1 have been identified in patients exhibiting clefting and tooth agenesis (van den Bogaard et al., 2000) as well as in familial and sporadic nonsyndromic oral facial clefting cases (Lidral et al., 1998; Jezewski et al., 2003; Vieira et al. 2004; Suzuki et al., 2004 and; Tongkobpetch et al.,2006).There are also multiple reports of highly penetrant MSX1 coding mutations exhibiting Mendelian inheritance of tooth agenesis (oligodontia) phenotypes (Vastardis et al., 1996; Lidral et al., 2002; De Muynck et al., 2004; Chishti et al., 2006; and Xuan et al., 2008). Finnerty et al. (2009) performed bioinformatics analysis of the Msx gene family and identified seven ancient, strongly conserved coding domains within the Msx1 orthologs. The protein domain composition of these Msx1 orthologs was found to have diverged substantially from Msx2 and Msx3 paralogs within the vertebrate lineage. Several of the known MSX1 coding mutations cited in the studies above occur at deeply conserved sites within these Msx Homology (MH) domains. The A34G missense variant is found within the most divergent portion of the Msx1 sequence, upstream of the first of the MH deeply conserved domains (see Finnerty et al., 2009, figure 6).
In the human genome UCSC browser, A34G is rs36059701 found within a run of seven alanines (and one threonine at T33) on the N-terminal side of the MSX1 protein. There is also another poly-alanine tract within MSX1 on the C-terminal side of the homeodomain. While neither of these are purely homopolymeric amino acid tracts, they are generally conserved within Msx1 orthologs from mammalian lineages (See supplemental Figure 1, Finnerty et al. (2009) for a curated alignment). Neither of these polyalanine tracts is found in either Msx2 or Msx3 protein sequences, thus again distinguishing Msx1 from Msx2 and Msx3 in this regard.
Previous literature reports (Messaid and Rouleau 2009; Lavoie et al., 2003) suggest that polyalanine tracts of a moderate size (< 20 alanines) operate as protein-protein interaction domains important during the formation of transcriptional complexes. Lavoie et al (2003) also reported that these types of alanine repeats were conserved among mammals but were rarer and shorter in fish and reptiles. Thus it is conceivable that Msx1 N-terminal poly-alanine tract represents a more modern Msx1 protein motif, which evolved and stabilized during the mammalian lineage. Consistent with this possibility is previous functional analyses of the Msx1 protein that demonstrated the N-terminal region contributes to repression activity and augments its interaction with the general transcription complex (Catron et al., 1995; Zhang et al.,1996). Thus the A34G missense mutation may actually be disrupting a Msx1 ortholog-specific protein-protein interaction domain that is important during transcriptional repression activity.
The direct evidence for the role of the A34G variant in clefting in the present report comes from the distortion of HWE on a small sample size. This result suggests that homozygotes for this variant may be overrepresented in cleft cases even though there was no evidence for allelic distortion and thus function of MSX1 dimers impaired. There is considerable existing genetic evidence beyond the present report to support this possibility. The A34G mutation that changes the amino acid from Alanine to Glycine at position 34 and has been previously reported by various studies in different population by Lidral et al. (1998) in Caucasians from Iowa; Jezewski et al.(2003) in Iowan, Asian (Filipino), South American; Vieira et al. (2004) in South Americans in Chile; Suzuki et al.(2004) in Vietnamese; Tongkobpetch et al. (2006) in Thai population. However, there is as yet no population frequency data available in the databases (DbSNP and 1000genomes). Although this mutation has been previously been presumed to be benign (Jezewski et al., 2003; Suzuki et al.,2004), homozygous individuals have only been reported in the Thai study (Tongkobpetch et al., 2006).
In addition, Modesto et al. (2006) reported a significant association between this polymorphism and the disease state, even after removing syndromic cases. Xuan et al. (2008), identified a novel coding mutation (A221E), likely in phase with the minor allele of A34G, both cosegregating with oligodontia in a three generation pedigree. Lidral et al. (1998) reported that the fourth allele of the MSX1 intronic CA repeat was associated with clefting by case-control allele frequency comparisons, as well as by Transmission Disequilibrium Test (TDT) and Affected Family Based Association Controls (AFBAC) nuclear family based analyses. In this same study, they tested for association with several additional sequence polymorphisms, including A34G. Haplotype comparisons of the CL(P) or CP probands with the controls showed evidence of nearly significant LD with two different haplotypes. One of these haplotypes included the A34G minor allele. This haplotype was not found on any of the control chromosomes leading them to speculate that this coding substitution may affect protein function. Modesto et al. (2006) also investigated the A34G allele amongst a set of patients with and without tooth agenesis (TA). The A34G substitution was observed more frequently in patients with both OFC and TA than found in controls (p=0.0008).
In the present study, nine of the case probands and four of the Hap map controls have the A34G mutation. All of the case individuals with the mutation have complete cleft lip and palate and these include three with bilateral CL(P), one right CL(P) and five left complete CL(P). The clefting status of the HapMap controls is unknown. There is an excess of homozygous for the rare type amongst the cases and a deviation from Hardy Weinberg equilibrium (p=0.00002). This may stem from the small sample size in the study, ascertainment bias or possible biological effect of the A34G missense mutation in the MSX1 gene. All these considerations suggest a role for the Msx1-type polyalanine tracts within transcriptional repression complexes that may be partially disrupted by the A34G missense mutation. More clinical and functional studies will be needed before a definite genotype-phenotype correlation will be confirmed.
Hardy Weinberg equilibrium
There was a deviation from Hardy Weinberg equilibrium in the distribution of MSX1 A34G variant genotypes in the cases genotypes from the case control study. While possible due to chance, sequencing or genotyping errors the latter two were carefully checked and repeated. It may be that A34G is acting as a weak recessive allele and is implicated in oral facial clefts.
FBAT / TDT
The comparison between markers for the 11 candidate genes investigated did not reveal any statistically significant result between cases (all clefts) and controls with p >0.05. This implies that there are no differences in the markers between cleft cases and the controls. However, the sample size and low minor allele frequency for many of the SNPs in genes previously associated with CL/P (IRF6, MAFB) limits this statistical interpretation.
Limitation of present study
The present study was carried out in a single population and African ethnic group with a small number of eligible cases recruited. There is incomplete ascertainment of cases and inadequate sub-groups for analysis and all clefts were analysed in one group. Poor quality DNA further limited some analyses. While no definitive conclusion can be drawn from the study it does serve as a useful introduction to studies of clefts in Africa. Sudden closures of participating hospitals during the period of the project; delays in getting collection kits; loss of patients to follow-up; and chances that cleft palate (CP) may not be detected were some of the reasons for a low recruitment of cases. The NigeriaCRAN project should be replicated in other sub-Saharan countries in order to establish a DNA bio-bank and carry out African-wide whole genome scan to pick up new candidate genes.
CONCLUSION
This study is the first in sub-Saharan Africa to investigate candidate genes in the aetiology of orofacial clefts. The epidemiological data revealed a statistically significant difference between the frequency of occurrence of left versus right sided clefts (p=0.003), between bilateral and unilateral clefts (p=0.02) and an excess of females with CP (p=0.008). The replication of a missense mutation that has been previously reported in other populations suggests a role for the MSXI A34G variant in the aetiology of CL(P) that requires further investigation.
Acknowledgment
We are grateful to all the families that participated in this project, members of the NigeriaCRAN collaborating teams, nurses and registrars who helped with recruitments. We also appreciate the administrative assistance and support of Rosemary Inglis and Paul Hughes in Dundee, Jamie L'Heureux, Nancy Davin and Susie McConnell in Iowa for their support with sample transfer to Iowa. Our gratitude goes to the West African Research Association for the travel grant to obtain control samples, CLAPA and SCALP for providing funds for the DNA collection kits, Smile Train Charity for funding surgical repairs in Nigeria and University of Dundee for supporting the NigeriaCRAN collaboration. The work was supported by NIH grant P50 DE-1615 and DE08559.
Footnotes
(Lagos University Teaching hospital, University of Benin Teaching hospital, Ladoke Akintola University Teaching hospital, Aminu Kano Teaching hospital, National Orthopedic hospital Enugu, University of Port Harcourt Teaching Hospital, University of Maiduguri Teaching hospital, Obafemi Awolowo University Teaching hospital, Ebonyi State University Teaching hospital, University of Ilorin Teaching hospital and Usmanu Dan Fodio University teaching hospital).
REFERENCES
- Akinmoladun VI, Owotade FJ, Afolabi AO. Bilateral transverse facial cleft as an isolated deformity: Case report. Ann Afr Med. 2007 Mar;6(1):39–40. doi: 10.4103/1596-3519.55730. [DOI] [PubMed] [Google Scholar]
- Avila JR, Jezewski PA, Vieira AR, Orioli IM, Castilla EE, Christensen K, Daack-Hirsch S, Romitti PA, Murray JC. PVRL1 variants contribute to non-syndromic cleft lip and palate in multiple populations. Am J Med Genet A. 2006 Dec 1;140(23):2562–2570. doi: 10.1002/ajmg.a.31367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral A, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox A, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Doheny K, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, Scott AF. A genome wide association study of cleft lip with / without cleft palate using case-parent trios of European and Asian ancestry identifies MAFB and ABCA4 as novel candidate genes. Nat Genet. 2010 Jun;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco R, Chatraburty R, Barton SA, Carreno H, Parades M, Jara I. Evidence of a sex dependant association between the MSX1 focus and non syndromic cleft lip with or without cleft palate in the chilean population. Hum Biol. 2001;73:81–89. doi: 10.1353/hub.2001.0002. [DOI] [PubMed] [Google Scholar]
- Brancaccio A, Minichiello A, Grachtchouk M, Antonini D, Sheng H, Pariato R, Dathan N, Dlugosz AA, Missero C. Requirement of forkhead gene FOXE 1, a target of sonic hedgehog signalling, in hair follicle morphogenesis. Human Mol Genet. 2004;13:2595–2606. doi: 10.1093/hmg/ddh292. [DOI] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Potzsch S, Steegers-Theunissen RP, Potzsch B, Moebus S, Horsthemke B, Kramer F, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffman P, Knapp M, Nothen MM, Mangold E. Key susceptibilty locus for nonsyndromic cleft lip with or withgout cleft palate on chromosome 8q24. Nature Genet. 2009;41(4):473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Butali A, Mossey PA. Epidemiology of orofacial clefts in africa: methodological challenges in ascertainment. Pan African Medical Journal. 2009;2:2–5. doi: 10.4314/pamj.v2i1.51705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catron KM, Zhang H, Marshall SC, Inostroza JA, Wilson JM, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995 Feb;15(2):861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti MS, Muhammad D, Haider M, Ahmad W. A novel missense mutation in MSX1 underlies autosomal recessive oligodontia with associated dental anomalies in Pakistani families. J Hum Genet. 2006;51(10):872–878. doi: 10.1007/s10038-006-0037-x. [DOI] [PubMed] [Google Scholar]
- De Muynck S, Schollen E, Matthijs G, Verdonck A, Devriendt K, Carels C. A novel MSX1 mutation in hypodontia. Am J Med Genet. 2004 Aug 1;128A(4):401–403. doi: 10.1002/ajmg.a.30181. [DOI] [PubMed] [Google Scholar]
- Finnerty JR, Mazza ME, Jezewski PA. Domain duplication, divergence, and loss events in vertebrate Msx paralogs reveal phylogenomically informed disease markers. BMC Evol Biol. 2009 Jan 20;9:18. doi: 10.1186/1471-2148-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SFA, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield PJ, Glessner JT, Thomas KA, Garris M, Frackelton EC, Otieno FG, Chiavacci RM, Nah H, Kirschner RE, Hakonarson H. A genome -wide association study identifies a locus fr non-syndromic cleft lip with or without cleft palate on 8q24. Journal of Pediatr. 2009 Dec;155(6):909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Hagberg C, Larson O, Milerad J. Incidence of cleft lip and palate and risks of additional malformations. Cleft Palate Craniofac J. 1998 Jan;35(1):40–45. doi: 10.1597/1545-1569_1998_035_0040_ioclap_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Iregbulem IM. The incidence of cleft lip and cleft palate in Nigeria. Cleft Palate J. 1982;19:201–205. [PubMed] [Google Scholar]
- Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE. Complete sequencing shows a role for MSX1 in nonsyndromic cleft lip and palate. J Med Genet. 2003;40:399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 2009 Oct;15(7):437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Debeane F, Trinh QD, Turcotte JF, Corbeil-Girard LP, Dicaire MJ, Saint- Denis A, Pagé M, Rouleau GA, Brais B. Polymorphism, shared functions and convergent evolution of genes with sequences coding for polyalanine domains. Hum Mol Genet. 2003 Nov 15;12(22):2967–2979. doi: 10.1093/hmg/ddg329. [DOI] [PubMed] [Google Scholar]
- Lidral AC, Romiti PA, Basart AM, Doetschman T, Leysens NJ, daack-hirsch S, Semina EV, Johnson LR, Machida J, Burds A, Parnell TJ, Rubenstein JL, Murray JC. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–568. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Reising BC. The role of MSX1 in human tooth agenesis. J Dent Res. 2002 Apr;81(4):274–278. doi: 10.1177/154405910208100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, Almeida de Assis N, Al Chawa T, Mattheisen M, Steffens M, Barth S, Kluck N, Paul A, Becker J, Lauster C, Schmidt C, Braumann B, Scheer M, Reich RH, Hemprich A, Pötzsch S, Blaumeiser B, Moebus S, Krawczak M, Schreiber S, Meitinger T, Wichmann H, Steegers-Theunissen RP, Kramer F, Cichon S, Propping P, Wienker TF, Rubini FA, Mossey PA, Hoffmann P, Nöthen MN. Susceptibility loci for non-syndromic cleft lip with or without cleft palate on chromosome 17q22 and 10q25.3. Nature Genetics. 2010 Jan;42(1):24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein TH, Maher BS, Daack-hirsch S, Shultz S, Mansilla MA, Field LL, Ye I, Prescott N, Malcolm S, Winter R, Ray A, Moreno L, Valencia C, Neiswanger K, Wyszynski DF, Bailey-Wilson JE, Albacha-Hejazi H, Beaty TH, Mcintosh I, Hetmanski JB, Tuncbilek G, Edwards M, Harkin I, Scott R, Roddick IG. Meta-analysis of 13 gene scans reveals multiple cleft lip / palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaed C, Rouleau GA. Molecular mechanisms underlying polyalanine diseases. Neurobiol Dis. 2009 Jun;34(3):397–405. doi: 10.1016/j.nbd.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Modesto A, Moreno LM, Krahn K, King S, Lidral AC. MSX1 and orofacial clefting with and without tooth agenesis. J Dent Res. 2006;85:542–546. doi: 10.1177/154405910608500612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, Johnson MK, Brauer D, Krahn K, Daack-hirsch s, L'Heureux J, Valencia-Ramirez C, Rivera D, López AM, Moreno MA, Hing A, Lammer EJ, Jones M, Christensen K, Lie RT, Jugessur A, Wilcox AJ, Chines P, Pugh E, Doheny K, Arcos-Burgos M, Marazita ML, Murray JC, Lidral AC. FOXE1 association with isolated cleft lip with or without cleft palate; and isolated cleft palate. Hum Mol Genet. 2009 Dec. 1518(24):4879–4896. doi: 10.1093/hmg/ddp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J. Chapter 12: “Epidemiology of oral clefts: an international perspective”. In: Wyszynski DF, editor. Cleft lip and palate. From origin to treatment. Oxford University Press; 2002. Aug, pp. 127–158. ISBN: 0-19-513906-2. [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon M, Shaw WC. Cleft lip and palate. Lancet. 2009 Nov 21;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. 2009. [DOI] [PubMed] [Google Scholar]
- Oginni F, Asuku M, Oladele A, Obuekwe O, Nnabuko R. Knowledge And Cultural Beliefs About The Etiology And Management Of Orofacial Clefts In Nigeria's Major Ethnic Groups. The Cleft Palate-Craniofacial Journal. 2010 Jul;47(4):327–334. doi: 10.1597/07-085.1. [DOI] [PubMed] [Google Scholar]
- Ogle OE. Incidence of cleft lip and palate in a newborn Zairian sample. Cleft Palate Craniofac J. 1993;30:250–251. [PubMed] [Google Scholar]
- Olasoji HO, Hassan A, Ligali TO. Challenges of cleft care in Africa. Afr J Med Med Sci. 2009 Dec;38(4):303–310. [PubMed] [Google Scholar]
- Onah II, Opara KO, Olaitan PB, Ogbonnaya IS. Cleft lip and palate repair: the experience from two West African sub-regional centres. J Plast Reconstr Aesthet Surg. 2008 Aug;61(8):879–882. doi: 10.1016/j.bjps.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jegessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC NISC comparative sequencing programme. Disruption of an AP-2α binding site in am IRF6 enhancer is strongly associated with cleft lip. Nat Genet. 2008;40(11):1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dodé C, Mohammadi M, Marazita ML, Murray JC. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Maas R. MSX1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Scapoli L, Martinelli M, Arlotti M, Palmieri A, Masiero E, Pezzetti F, Carinci F. Genes causing clefting syndromes as candidates for non-syndromic cleft lip with or without cleft palate: a family-based association study. Eur J Oral Sci. 2008;116(6):507–511. doi: 10.1111/j.1600-0722.2008.00574.x. [DOI] [PubMed] [Google Scholar]
- Slayton RL, Williams L, Murray JC, Wheeler JJ, Lidral AC, Nishimura CJ. Genetic association studies of cleft lip and / or palate with hypodontia outside the cleft region. Cleft Palate Craniofac J. 2003 May;40(3):274–279. doi: 10.1597/1545-1569(2003)040<0274:GASOCL>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss RP. Culture, rehabilitation, and facial birth defects: international case studies. Cleft Palate J. 1985 Jan;22(1):56–62. [PubMed] [Google Scholar]
- Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, Natsume N, Shimozato K, Ohbayashi N, Suzuki Y, Niimi T, Minami K, Yamamoto M, Altannamar TJ, Erkhembaatar T, Furukawa H, Daack-Hirsch S, L'heureux J, Brandon CA, Weinberg SM, Neiswanger K, Deleyiannis FW, de Salamanca JE, Vieira AR, Lidral AC, Martin JF, Murray JC. Mutations in BMP4 Are Associated with Sub-epithelial, Microform, and Overt Cleft Lip. Am J Hum Genet. 2009 March 13;84(3):406–411. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Jezewski P, Machida J, Watanabe Y, Min Shi MS, Cooper ME, Viet LT, Nguyen TDT, Hai H, Natsume N, Shimozuto K, Marazita ML, Murray JC. In a Vietnamese population, MSX1 variants contribute to cleft lip and palate. Genetics in Medicine. 2004;6:117–125. doi: 10.1097/01.gim.0000127275.52925.05. [DOI] [PubMed] [Google Scholar]
- Tolarova MM, Cervenka J. Classification and birth prevalence of orofacial clefts. Nat Genet. Dec 1995;11(4):415–421. [PubMed] [Google Scholar]
- Tongkobpetch S, Siriwan P, Shotelersuk V. MSX1 mutation contribute to nonsyndromic cleft lip in Thai population. J Human Genet. 2006;51:671–676. doi: 10.1007/s10038-006-0006-4. [DOI] [PubMed] [Google Scholar]
- Turhani D, Item CB, Watzinger E, Sinko K, Watzinger F, Lauer G, Ewers R. Mutation analysis of CLPTM 1 and PVRL 1 genes in patients with non-syndromic clefts of lip, alveolus and palate. J Craniomaxillofac Surg. 2005 Oct;33(5):301–306. doi: 10.1016/j.jcms.2005.04.004. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, de Costa D, Krapels IP, Liu F, van Duijn C, Sinke RJ, Lindhout D, Steegers-Theunissen RP. The MSX1 allele 4 homozygous child exposed to smoking at periconception is most sensitive in developing nonsyndromic orofacial clefts. Hum Genet. 2008 Dec;124(5):525–534. doi: 10.1007/s00439-008-0569-6. [DOI] [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996 Aug;13(4):417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Castillo Taucher S, Aravena T, Astete C, Sanz P, Tastets ME, Monasterio L, Murray JC. Mutational analysis of the muscle segment homeobox gene 1 (MSX1) in Chilean patients with cleft lip/palate. Rev Med Chil. 2004 Jul;132(7):816–822. doi: 10.4067/s0034-98872004000700005. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Felix TM, Rahimov F, Harrington J, Schultz RR, Watanabe Y, Johnson M, Fang J, O_Brien SE, Orioli IM, Castilla EE, Fitzpatrick DR, Jiang R, Marazita ML, Murray JC. Medical sequencing of candidate genes for nonsyndromic cleftlip and palate. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Vol. 12. Baura, Brazil: 2002. Global registry and data base on craniofacial anomailes. Report of a WHO Registry Meeting on Craniofacial Anomalies; pp. 1–3. 2001. [Google Scholar]
- Mossey Peter., editor. WHO Meeting on International Collaborative Research on Craniofacial Anomalies (4th :2004: Geneva, Switzerland). Addressing the global challenges of craniofacial anomalies:report of a WHO meeting on International Research on Craniofacial Anomalies; 2–4 December, 2004; Geneva. [Google Scholar]
- WHO. 2nd International Conference on Birth Defects and Disabilities in the Developing World. Hospital-based registries for monitoring birth defects: The National Collaborative Perinatal Neonatal Network; 11–15 September 2005; Beijing, China. [Google Scholar]
- Xuan K, Jin F, Liu YL, Yuan LT, Wen LY, Yang FS, Wang XJ, Wang GH, Jin Y. Identification of a novel missense mutation of MSX1 gene in Chinese family with autosomal-dominant oligodontia. Arch Oral Biol. 2008 Aug;53(8):773–779. doi: 10.1016/j.archoralbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Zhang H, Catron KM, Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc Natl Acad Sci USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-hirsch S, Nepomuceno B, Ribiero L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli LM, Castilla E, Moreno LM, Arcos-Burgos M, Lidral AC, Leigh Feild L, Liu Y, Ray A, Goldstein TH, Schutte BC, Min Shi MS, Johnson MK, Kondo S, Schultz RE, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risks of isolated cleft lip or palate. New Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]