Abstract
The thyroid and lungs originate as neighboring bud shaped outgrowths from the midline of the embryonic foregut. When and how organ specific programs regulate development into structures of distinct shapes, positions and functions is incompletely understood. To characterize, at least in part, the genetic basis of these events, we have employed laser capture microdissection and microarray analysis to define gene expression in the mouse thyroid and lung primordia at E10.5. By comparing the transcriptome of each bud to that of the whole embryo as well as to each other, we broadly describe the genes that are preferentially expressed in each developing organ as well as those with an enriched expression common to both. The results thus obtained provide a valuable resource for further analysis of genes previously unrecognized to participate in thyroid and lung morphogenesis and to discover organ specific as well as common developmental mechanisms. As an initial step in this direction we describe a regulatory pathway involving the anti-apoptotic gene Bcl2 that controls cell survival in early thyroid development.
Keywords: Organogenesis, Foregut patterning, Microarrays, LCM, Thyroid bud, Lung bud
Highlights
► The transcriptomes of the early thyroid and lung primordia are characterized. ► Organ-specific and common motifs of gene expression are identified and validated. ► The data provide a resource for finding genes regulating foregut patterning. ► A mechanism involving Bcl2 controls cell survival in the thyroid primordium.
Introduction
Bud outgrowth is a morphogenetic feature shared by endodermal derivatives originating from the midline of the gut tube as the thyroid, lungs, liver and pancreas as well as organs that develop from the pharyngeal pouches as the thymus and parathyroids (Wells and Melton, 1999; Zorn and Wells, 2009). Morphologically, these primordia first appear with similar dynamics and structural characteristics and it could consequently be anticipated that some basic developmental programs are conserved among them (Zorn and Wells, 2009). Over the past few years a coherent picture of mechanisms regulating early morphogenesis of the liver and pancreas, which emerge as outgrowths from the posterior foregut, has started to form (Zaret and Grompe, 2008). In contrast, less is known about differentiation programs governing the development of derivatives from other foregut regions. In this context it is therefore interesting to investigate these processes in the thyroid and the lungs as they initially develop in proximity from the median portion of the anterior embryonic foregut. Both these organ primordia express the transcription factor Nkx2-1, which is the first evidence of fate decision of distinct subsets of endodermal cells towards a thyroid or lung fate (Lazzaro et al., 1991). Indeed, Nkx2-1 deficient embryos display thyroid and lung agenesis as relatively isolated malformations in addition to a CNS phenotype (Kimura et al., 1996). Also other murine models targeting the Shh and Fgf pathways underscore that some aspects of the development of these organs involve similar gene sets (Fagman et al., 2004; Ohuchi et al., 2000). However, even though thyroid and lung organogenesis is initiated with similar features and at approximately the same time, the ultimate result is organs with distinct structure, size and functions. The thyroid gland can first be distinguished in mouse embryos around embryonic day (E) 9 as a thickening of the anterior aspect of the ventral endoderm located close to the cardiac outflow tract. Around E10–10.5 these cells form a caudally directed bud that dissociates from the pharyngeal floor (Fagman and Nilsson, 2010). The thyroid primordium thereafter migrates to its final position and starts to form primitive follicles. The lung buds emerge caudally to the thyroid and subsequently extend into the surrounding mesenchyme. In contrast to the thyroid, the lungs remain connected to the gut tube and undergo a highly conserved program of dichotomous branching to generate the respiratory tree (Metzger et al., 2008).
The morphogenesis of these organs is sometimes perturbed. Defective embryonic development of the thyroid gland resulting in structural anomalies (athyreosis, ectopia, hypoplasia) collectively known as thyroid dysgenesis (TD), is a major cause of congenital hypothyroidism (Kratzsch and Pulzer, 2008). This condition affects 1 of 3000 neonates, making it the most common disorder of the endocrine system in newborns (Corbetta et al., 2009). However, the underlying pathogenetic mechanisms are largely obscure. Even though findings from a number of murine gene knockout models have revealed the importance of transcription factors like Nkx2-1, Pax8, Foxe1 and Hhex for thyroid development (De Felice et al., 1998; Kimura et al., 1996; Mansouri et al., 1998; Martinez Barbera et al., 2000) only few germ line mutations have been detected in the corresponding genes of human patients (Al Taji et al., 2007; De Felice and Di Lauro, 2004; Narumi et al., 2010). This has led to hypotheses of a pathogenetic role of epigenetic alterations or early somatic mutations (Vassart and Dumont, 2005). The latter possibility is partly difficult to investigate. If early somatic mutations cause athyreosis by elimination of progenitor cells or prevention of differentiation, no tissue is amenable to genetic analysis. The only hitherto published case where somatic mutations have been analyzed in dysgenic (sublingual) thyroid tissue revealed no somatic mutations in Nkx2-1, Pax8 or Foxe1 (Stoppa-Vaucher et al., 2010). Interestingly, a number of nonrecurrent copy number variations have recently been identified in TD patients. Some of these encompassed genomic regions containing several genes. It has therefore been proposed that genomic mechanisms leading to TD are heterogeneous and that a broader outlook on genes that participate in the regulation of thyroid morphogenesis is needed (Thorwarth et al., 2010). In the case of the lungs, several congenital anomalies have been described (Berrocal et al., 2004; Freedom et al., 2006) but knowledge on candidate genes involved in their pathogenesis is still scarce (Correia-Pinto et al., 2010).
In order to obtain insights into both the common and the diverse, organ specific mechanisms responsible for lung and thyroid morphogenesis, we have sought to compare their gene expression patterns in early development. To that end we have combined isolation of mRNA from primordial cells by laser capture microdissection (LCM) with microarray analysis and compared the transcriptional landscape of the thyroid and lung buds at the earliest stages of morphogenesis. The results obtained provide a valuable resource to further characterization of individual genes, sets of genes and signaling pathways that may determine organogenesis of the thyroid and lungs as well as be involved in the pathogenesis of dysgenetic conditions. As a proof of concept we show two novel features emerging from this analysis. The first is that Foxa2, previously considered as a pan-endodermal transcription factor (Ang et al., 1993; Monaghan et al., 1993), is specifically excluded from the thyroid bud, while being present in the lung. The second is the high expression of the anti-apoptotic gene Bcl2 in the thyroid bud, where it is controlled by the thyroid enriched transcription factor Pax8. In the absence of Pax8 an apoptotic pathway is activated demonstrating a previously unrecognized role of apoptotic vs. anti-apoptotic factors in thyroid organogenesis.
Materials and methods
Mice
Animals were kept in an animal house under controlled conditions of temperature, humidity, and light and were supplied with standard food and water ad libitum. All animal experiments were performed in accordance with regulations and guidelines of Italy and the European Union and were approved by the local ethical committee. Wild-type C57/Bl6 and CD1 mice were purchased from Charles River Laboratories (Calco, LC, Italy). Pax8 null mice (Pax8−/−) have been described previously (Mansouri et al., 1998). Pax8Cre/Floxed is a conditional knockout strain obtained by crossing Pax8Cre/+ mice, expressing the Cre recombinase gene under the control of the endogenous Pax8 locus (Bouchard et al., 2004), with Pax8Floxed/+ mice, which carry a mutated Pax8 allele where exons 3 and 4 are flanked by LoxP sequences. Detailed information on the generation and characterization of the conditional Pax8 knockout strain will be detailed elsewhere (Marotta, De Felice and Di Lauro, unpublished results).
Embryo dissection and embedding for LCM
E10.5 embryos were obtained by crossing wildtype C57BL/6 mice. Embryonic age (E) was calculated by considering the morning when a vaginal plug was detected as E0.5. Embryos were dissected on ice under aseptic conditions in cold DEPC treated phosphate-buffered saline (pH 7.2) (PBS-DEPC). After cryoprotection in 30% sucrose in PBS-DEPC (overnight at 4 °C), embryos were embedded in OCT compound (Sakura, Zoeterwoude, the Netherlands) and stored at − 80 °C.
Laser capture microdissection (LCM)
Tissue sections (8 μm) were cut on a cryostat (Leica Microm HM 500 M, Wetzlar, Germany) on polylysine slides (Menzel-Gläser, Braunschweig, Germany), stored on dry ice (< 1 h) and stained with eosin (70% ethanol 30 s, dH2O DEPC 20 s, 70% ethanol 20 s, 95% ethanol 20 s, eosinY 2 s, 95% ethanol 10 s, 95% ethanol 10 s, 100% ethanol 30 s, 100% ethanol 60 s, xylene 5 min, drying 5 min). LCM was performed immediately thereafter using the PixCell II system (Arcturus Engineering, Santa Clara, CA) under a 20× objective (laser spot size 7.5 μm, output power 100 mW, pulse duration 1.5 ms). Thyroid and lung buds from three pools of 5–7 embryos each were captured on thermoplastic CapSure HS caps (Arcturus).
RNA isolation, amplification and labeling
Total RNA from cells obtained by LCM was isolated using the PicoPure RNA isolation kit (Arcturus). To obtain RNA from whole embryos, three pools of three wild type embryos each were lysed in TRIZOL (Invitrogen, Carlsbad, CA) and total RNA was purified with the QIAGEN RNeasy kit (Qiagen, Hilden, Germany). RNA quality and integrity was determined by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) on RNA Pico Chips (Agilent Technologies). RNA integrity numbers (RIN) were > 7 indicating high RNA quality. All biological replicates were normalized to the same input amount of total RNA (13 ng). Three biological replicates of labeled cRNA from whole embryos, thyroid and lung buds were obtained by two rounds of in vitro transcription-based linear amplification using the Two-Cycle Target Amplification and Labeling kit (Affymetrix, Santa Clara, CA) according to manufacturer's instructions with the modification of 1 μg of RNA-binding protein T4 (Gp32) being added during first strand cDNA synthesis to increase accessibility of the mRNA template to the reverse transcriptase and reduce formation of higher order RNA structures (Rapley, 1994). cRNA quality, size-distribution and quantity were analyzed by an Agilent 2100 Bioanalyzer.
Sample hybridization and microarray data analysis
Biotinylated cRNA was hybridized Mouse 430 2.0 Genome Arrays (Affymetrix), containing over 39,000 genes and open reading frames from Mus musculus (genome databases GenBank, dbEST and RefSeq). Chips were scanned on the Affymetrix Complete GeneChip® Instrument System, generating digitized image data (DAT) files. DAT files were analyzed with AGCC (Affymetrix) producing CEL files. Robust multichip average (RMA) normalization (Irizarry et al., 2003) and data analysis were performed using GeneSpring 11.0.2 (Agilent Technologies). To assess the reproducibility of RNA amplification, three different input amounts of total RNA from a pool of three E10.5 WT embryos were amplified, labeled and hybridized to arrays as described above. Scatter plot comparisons of gene expression levels of amplified cRNA from different RNA input confirmed a high degree of reproducibility (data not show).
The bioB probeset is a control hybridization spike added into the hybridization cocktail, independent of RNA sample preparation at a concentration approximating one RNA molecule (1.5 pM). An expression value below the bioB value is thus considered to indicate absence of gene expression (Lonergan et al., 2007) and probesets overcoming the bioB expression value were called present. Differentially expressed probesets (DEps) between organ buds and whole embryos were filtered for fold change ≥ 3. Statistical analysis was performed using an unpaired T-test, with p ≤ 0.05. DEps were sorted as “up” or “down” regulated. Each group of upregulated probesets was filtered for presence calls to obtain lists of probesets enriched in thyroid or lung buds as compared to the whole embryo. These lists were further cross-analyzed to generate the intersection (common genes) and the union (pool) of the enriched bud transcriptomes. Functional annotation was performed by Gene Ontology (GO) focused on Biological Process terms. Enriched GO terms (i.e. terms with a significantly higher than expected number of associated genes) were filtered (p ≤ 0.05) by the hypergeometric test and corrected using False Discovery Rate (FDR) (Benjamini and Yekutieli, 2001). For the pool of genes (union), the enriched GO categories were organized according to DAVID (Dennis et al., 2003) and the first two levels were plotted. The array data reported in this paper have bee deposited in ArrayExpress with ID E-MEXP-3133.
In situ hybridization
E10.5 embryos from matings of wildtype CD1 mice were fixated in 4% paraformaldehyde (overnight, 4 °C), cryoprotected in 30% sucrose in PBS (overnight, 4 °C), embedded in OCT (Sakura), quick-frozen over dry ice/ethanol slurry and stored at − 80 °C. Frozen sections (10 μm) were obtained as described above on Superfrost slides (Mentzel Gläser). Templates for riboprobes were generated by PCR from commercially available plasmids (Open Biosystems Huntsville, Alabama or Gene Service Cambridge, UK) or from cDNA obtained by reverse transcription of mRNA from E10.5 mouse embryos using forward and reverse primers extended at their 5′ ends with either T7 or SP6 promoter sequences (T7, GGATTTAATACGACTCACTATAGGGAGA; Sp6, CGATTTAGGTGACACTATAGA) (see Supplemental information), following a protocol from the GenePaint consortium (http://www.genepaint.org) (7 min initial template denaturation at 95 °C, 35 cycles with 1 min denaturation (95 °C), 1 min primer annealing (52–68 °C), 1 min elongation (72 °C), final elongation for 7 min at 72 °C). The resulting PCR products were resolved on agarose gels and desired bands excised and purified using the QIAGEN gel extraction kit (QIAGEN). After PCR reamplification products were purified using QIAquick PCR purification kit (QIAGEN), analyzed on an agarose gel and verified by sequencing. Nkx2.1 antisense riboprobe was a 625 bp fragment derived from the 3′ non-translated region of the Nkx2.1 cDNA (bp 1694–2319) (Guazzi et al., 1990). Digoxygenin-labeled riboprobes (sense and antisense) were obtained using a DIG-labeling RNA kit (Roche Diagnostics Basel, Switzerland) following the manufacturer's instructions. In situ hybridization of frozen sections was performed following the protocol described by Little et al.(2007). No signal was detected with the sense riboprobes (not shown). Images were obtained using an Axioplan2 microscope equipped with an Axiocam digital camera (Zeiss, Oberkochen, Germany). Images were processed using the Axion Vision software and Image J software.
Immunofluorescence
10 μm thick frozen sections were collected on polylysine glass slides (Menzel-Gläser). After permeabilization in 0.1% Triton X-100 in PBS for 20 min, sections were incubated in blocking buffer (PBS with 2% normal donkey serum (Jackson ImmunoResearch, Jacksonville, MI)) for 1 h at room temperature followed by overnight incubation at 4 °C with primary antibodies (rabbit pAbs against Nkx2-1 (Lazzaro et al., 1991), Foxa2 (Seven Hills Bioreagents, Cincinnati, OH), Bcl2 (BD Pharmingen, San Diego, CA), Caspase-3 (Cell Signaling Technologies, Beverly, MA), rat mAb against E-cadherin (Calbiochem Merck KGaA, Darmstadt, Germany)) diluted in PBS with 2% normal donkey serum. Secondary antibodies (Rhodamin RedX conjugated donkey anti-rabbit, FITC conjugated donkey anti rat (Jackson Immunolabs, West Grove, PA) were added in blocking buffer for 60 min at room temperature. All incubation steps were separated by washing in 0.1% Triton X-100 in PBS for 3 × 5 min. Images were obtained in a Zeiss AttoArc II epifluorescence microscope or a Bio Radiance 2000 Laser Scanning Microscope.
TUNEL assay
Thyroids dissected from 1-month old mice were fixed overnight at 4 °C in 4% paraformaldehyde in PBS, dehydrated, cleared in xylene and embedded in paraffin blocks. For TUNEL staining the In Situ Cell Death fluorescent kit (Roche Diagnostic, Mannheim, Germany) was used, following the manufacturer's instructions. Briefly, 4 μm thyroid sections were dewaxed, hydrated and incubated for 8 min in a 0.1% Triton X-100, 0.1% sodium citrate solution. Subsequently, 50 μL of TUNEL reaction mixture was added to each section and the slides were incubated for 60 min at 37 °C. Sections were counterstained with DAPI before mounting. Microscopy and imaging were performed in a Zeiss AxionPlan II epifluorescence (FluoArc) Microscope. Images were processed using Axion Vision software and the Image J software.
Results and discussion
Establishing the transcriptomes of the thyroid and lung primordia
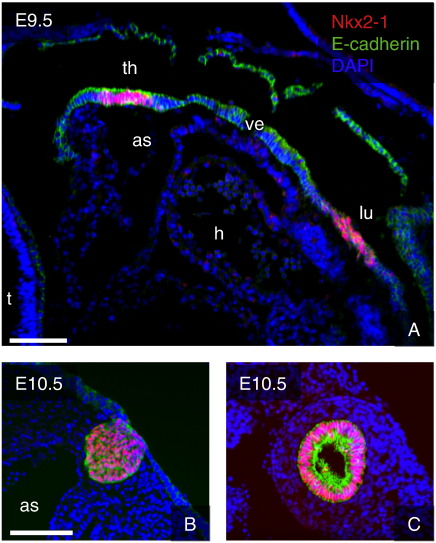
The global transcriptome of organs that form as outgrowths from the anterior foregut at a stage as early as E10.5 has to the best of our knowledge not been described before as the organ primordia are not amenable to manual dissection. To overcome this limitation we have successfully combined LCM with microarray analysis of mRNA expression. The thyroid and lungs are both derived from the ventral aspect of the anterior endoderm. At E9.5 the specified organ anlagen can be distinguished as neighboring regions of the foregut epithelium that express the transcription factor Nkx2-1 (Fig. 1A). However, by morphological criteria alone it is difficult to precisely delimit the progenitor areas at this early stage. One day later, at E10.5, the developing thyroid has formed a caudally directed bud that is closely apposed to the cardiac outflow region. At the same stage outgrowth of the right and left lung buds is evident and these are surrounded by concentrically arranged layers of mesenchymal cells whereas further dichotomous branching has not yet occurred (Metzger et al., 2008) (Figs. 1B and C). Thus, since it is possible to unambiguously recognize thyroid and lung buds at this stage by routine eosin staining of frozen sections, selective isolation of epithelial populations by LCM was carried out, without a need of prior immunostaining of specific organ markers. The precision and selectivity of the microdissection was subsequently confirmed by in situ hybridization experiments (see below).
Fig. 1.
A. Anterior foregut region at E9.5. The thyroid and lung primordia can be distinguished as discrete regions of the ventral endoderm expressing Nkx2-1, located in close vicinity of the developing heart. B. At E10.5 the thyroid primordium forms a caudally directed bud that is closely apposed to the aortic sac. C. The lung buds at E10.5 form hollow, blind-ended epithelial tubes surrounded by concentric layers of mesenchyme. Sagittal sections. As, aortic sac; h, heart; lu, lung primordium; th, thyroid primordium; ve, ventral endoderm. Scale bars = 100 μm in A, 75 μm in B and C.
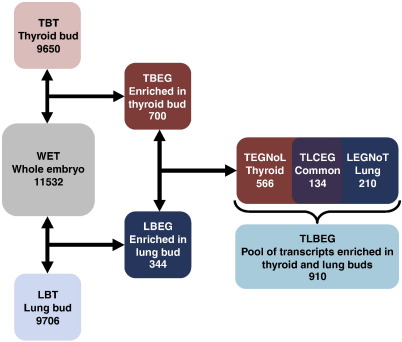
To define the gene expression profile of the developing thyroid and lung, RNA obtained from 5 to 7 pooled buds was amplified, labeled and hybridized to Affymetrix MOE430 mouse whole genome arrays. In parallel, a comparable amount of RNA, extracted from whole embryos at the same developmental stage, was amplified and hybridized to arrays as described above. These procedures yielded 9650 and 9706 probe sets hybridizing to thyroid and lung buds RNA, respectively, and 11,532 probe sets hybridizing to whole embryo RNA (see Fig. 2). Statistical validation and filtering of the signals were carried out as described in the Materials and methods section. In the following we will refer to these transcriptome sets as TBT (Thyroid Bud Transcriptome), LBT (Lung Bud Transcriptome) and WET (Whole Embryo Transcriptome) (see Supplementary file 1).
Fig. 2.
Schematic outline of the general strategy for finding genes with enriched expression in the developing thyroid and lung at E10.5. First, levels of transcripts that are expressed (i.e. overcoming bioB filtering, see Materials and methods section) in thyroid (Thyroid Bud Transcriptome, TBT) and lung buds (Lung Bud Transcriptome, LBT) dissected by LCM are compared to the corresponding one in the whole E10.5 embryo (Whole Embryo Transcriptome, WET). In this way enriched genes, i.e. those with a level of expression at least 3-fold over that in the whole embryo are identified, forming two discrete sets of transcripts; genes enriched in the thyroid bud (Thyroid Bud Enriched Genes, TBEG) and genes enriched in the lung buds (Lung Bud Enriched Genes, LBEG). When these sets are compared some genes are common to both organs (Thyroid and Lung Common Enriched Genes, TLCEG), whereas others are exclusive to either the thyroid (Thyroid Enriched Genes Not Lung, TEGNoL) or the lung (Lung Enriched Genes Not Thyroid, LEGNoT). The union of these sets constitutes a pool of genes expressed in either or both organs (Thyroid and Lung Bud Enriched Genes, TLBEG).
Whereas LCM has been widely employed to isolate neoplastic cells, a much smaller number of studies have applied this technique on embryonic primordia (Brown et al., 2009; Brunskill et al., 2008; Masuda et al., 2009; Purcell et al., 2009; Scheidl et al., 2002). Furthermore, most reports have focused on a limited and predefined number of mRNA species whereas fewer have combined LCM with microarray analysis. By an interesting alternative approach to LCM, Gu et al have characterized the transcriptome of the developing mouse pancreas at E10.5 by manually dissecting the pancreatic region from Pdx1-GFP reporter embryos. Cells have then been dissociated and sorted by FACS to remove contaminating mesenchyme (Gu et al., 2004). By a similar strategy Sherwood et al have manually dissected organ primordia (esophagus, lung, stomach, liver, pancreas, intestine) in wild type embryos at E11.5 and subsequently isolated endodermal cells from the mesenchyme by flow cytometric sorting of EpCAM positive cells (Sherwood et al., 2009). Regardless of the method employed a major technical hurdle has been the limited amount of RNA that is obtained which requires amplification of RNA to allow for successful microarray analysis.
Identification of genes enriched in the thyroid and lung buds
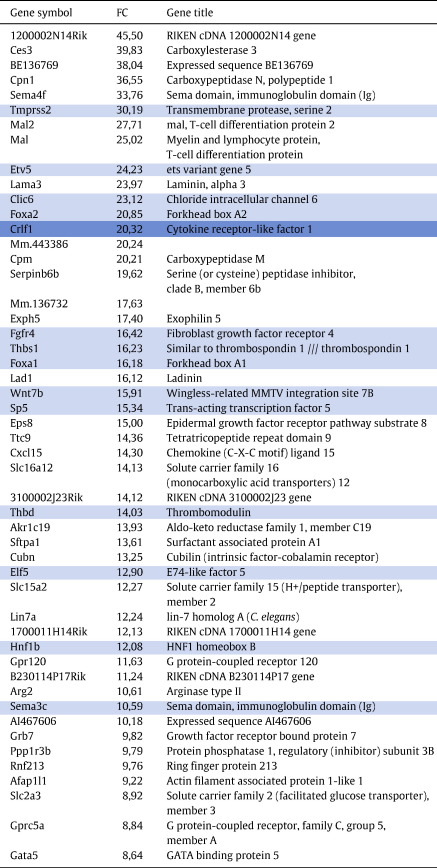
To find candidates genes of specific relevance in regulation of thyroid and lung development, we focused further attention on transcripts with a high relative expression in each bud. To this end we compared the expression level of each transcript in either TBT or LBT with that in the WET and sorted out transcripts with an expression level, in either bud, at least 3-fold higher than that in the whole embryo. Hereafter we will refer to them in short as “enriched genes”. A similar strategy was recently employed to study the transcriptome of the temporomandibular joint and find novel developmental regulators (Purcell et al., 2009). By this procedure we detected 700 probe sets (representing 553 genes) enriched (FC > 3) in the thyroid bud (defined as Thyroid Bud Enriched Genes, TBEG) and 344 probe sets (287 genes) enriched in the lung bud (defined as Lung Bud Enriched Genes, LBEG). Since some of these transcripts are common to both organs as the union of the two sets (Thyroid and Lung Bud Enriched Genes, TLBEG) consisted of 910 probesets (840 genes) (see Supplementary file 2).
A potential pitfall of this approach lies in the comparison of entirely epithelial organ primordia to the entire embryo that is a mixture of all three germ layers. This is likely to cause a bias towards finding an enrichment of general epithelial determinants in addition to organ specific transcripts. However, by comparing the datasets of enriched genes in the thyroid and lung buds, common epithelial denominators can be sorted out as well as those that might contribute more specifically to the development of each organ. Indeed, we found that 134 enriched probe sets (113 genes) were common to both primordia (Thyroid and Lung Common Enriched genes, TLCEG) whereas 566 probe sets (455 genes) were specific to the thyroid (Thyroid Enriched Genes Not Lung, TEGNoL) and 210 probe sets (179 genes) to the lung (Lung Enriched Genes Not Thyroid, LEGNoT) (see Supplementary file 3). Fig. 2 depicts a flowchart of the general strategy for generating and comparing these sets of transcripts. Since several genes were identified by more than one probeset, for the remainder of this paper we will refer to genes and not probesets, considering the probeset with the highest fold difference (FC) as compared to the WET for each gene.
In both TBEG and LBEG a strong enrichment was noticed of key epithelial markers like Cdh1 (E-cadherin; thyroid FC 14, lung FC 9) and EpCAM (Epithelial Cell Adhesion Molecule; thyroid FC 8, lung FC 7) (see Table 3), which is furthermore specific to the endoderm (Sherwood et al., 2007). On the contrary, no signal was present for the endothelial antigen Cd34 (Young et al., 1995) or for markers of embryonic mesenchyme like Pdgfrα (Platelet derived growth factor receptor-α) (Orr-Urtreger and Lonai, 1992), Mest (Mesodermal specific transcript) (Sado et al., 1993) and Cdh11 (OB-cadherin) (Simonneau et al., 1995) even though they were all abundant in the WET. This suggests that no significant contamination of adjacent non-epithelial cells occurred during the LCM procedure.
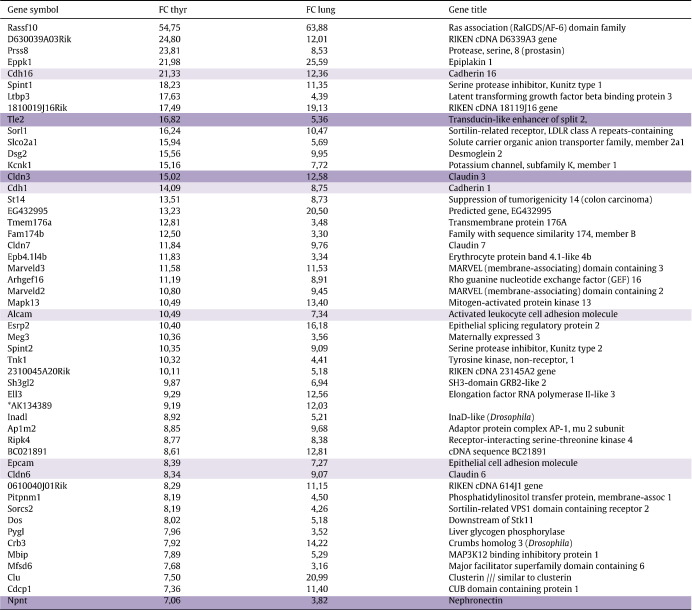
Table 3.
Genes that are enriched in both the thyroid and lung buds at E10.5 (TLCEG). The 50 genes with the highest relative enrichment (fold change) as compared to expression in the whole embryo at the same stage are listed. Dark shading denotes genes where expression is validated by in situ hybridization (see Fig. 3). Light shading denotes genes for which previously published data show expression in either organ at E10.5–E11.5. For references, see Supplemental information.
Genes that are enriched in both the thyroid and lung buds at E10.5 (TLCEG). The 50 genes with the highest relative enrichment (fold change) as compared to expression in the whole embryo at the same stage are listed. Dark shading denotes genes where expression is validated by in situ hybridization (see Fig. 3). Light shading denotes genes for which previously published data show expression in either organ at E10.5–E11.5. For references, see Supplemental information.
Genes enriched in the E 10.5 thyroid bud
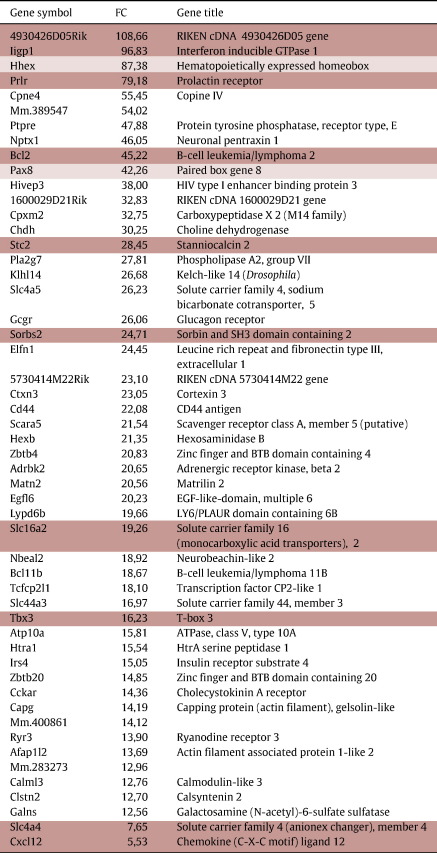
Of 455 genes present in TEGNoL, 72 displayed a FC > 10 and the top entries are listed in Table 1. Among these are well-established key regulators of thyroid development like Hhex (FC 87) and Pax8 (FC 42) (Parlato et al., 2004). Foxe1, considered to be a signature transcription factor of the embryonic thyroid, is absent in TEGNoL, as no Foxe1 probesets were present on the arrays. At E10.5 the thyroid bud is surrounded by mesenchyme (Fig. 1B), presumed to be mainly of neural crest origin (Jiang et al., 2000). No neural crest markers like Gja1 (connexin43) (Lo et al., 1997), tcfap2a (AP-2) (Mitchell et al., 1991) or Twist (Fuchtbauer, 1995) were present in the TBT, whereas strong signal levels were detected in the WET. This suggests that no contamination of mesenchymal cells of mesodermal (see above) or neural crest origin occurred during LCM isolation of the thyroid bud.
Table 1.
Genes specifically enriched in the thyroid bud at E10.5 (TEGNoL). The 50 genes with the highest relative enrichment (fold change) as compared to expression in the whole embryo at the same stage are listed. Data are also shown for two genes with lower fold change that have been validated by in situ hybridization. Dark shading denotes genes where expression is validated by in situ hybridization (see Fig. 3). Light shading denotes genes for which previously published data show expression in the thyroid at E10.5–E11.5. For references, see Supplemental information.
Genes specifically enriched in the thyroid bud at E10.5 (TEGNoL). The 50 genes with the highest relative enrichment (fold change) as compared to expression in the whole embryo at the same stage are listed. Data are also shown for two genes with lower fold change that have been validated by in situ hybridization. Dark shading denotes genes where expression is validated by in situ hybridization (see Fig. 3). Light shading denotes genes for which previously published data show expression in the thyroid at E10.5–E11.5. For references, see Supplemental information.
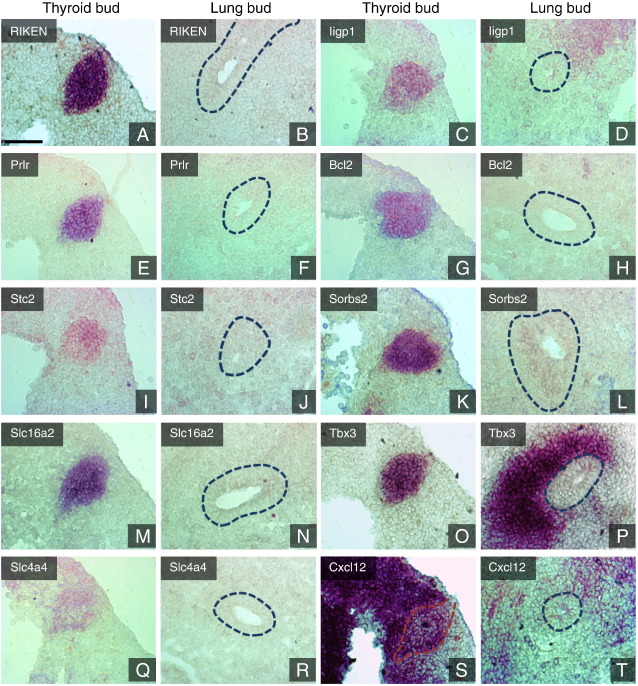
For further validation, 10 genes representing a broad range of FC values and biological functions (receptors, enzymes, ion channels, transcription factors and morphogens) were selected and their expression was investigated by ISH (Fig. 3). All of these showed distinct signals in the thyroid bud. As expected, no expression of these genes was seen in the lung bud epithelium and only restricted regions of the embryo outside the thyroid anlage showed a positive signal, confirming the validity of the list of enriched genes. For example, Slc16a2/Mct8, a thyroid hormone transporter, was present also in a restricted region of the telecephalon (not shown), whereas Cxcl12 was abundant in the mesenchyme adjacent to the developing heart (Fig. 3S). The expression of Tbx3 is particularly intriguing, since it was found in the mandibular component of the first pharyngeal arch (not shown) and also highly expressed in the mesenchyme surrounding the lung bud and trachea (Fig. 3P) whereas the lung bud epithelium itself is negative, consistent with previous findings (Chapman et al., 1996). This finding also underscores that LCM was performed with high selectivity, given that Tbx3 was not present among the lung enriched genes (see below). Furthermore, the striking juxtaposition of Tbx3 expressing mesenchyme to Tbx3 negative epithelium strongly suggests a role for this transcription factor in mesenchyme–endoderm interaction.
Fig. 3.
In situ hybridization validation of thyroid bud enriched genes at E10.5. A, B. RIKEN cDNA 4930426D05 gene. C, D. Interferon inducible GTPase 1. E, F. Prolactin receptor. G, H. B-cell leukemia/lymphoma. I, J. Stanniocalcin 2, K, L. Sorbin and SH3 domain containing 2. M, N. Solute carrier family 16 (monocarboxylic acid transporters), member 2; Monocarboxylate transporter 8. O, P. T-box 3. Apart from strong accumulation in the thyroid bud prominent expression also is seen in the mesenchyme surrounding the lung bud. Q, R. Solute carrier family 4 (anion exchanger), member 4. S, T. Chemokine (C-X-C motif) ligand 12. Strong enrichment is seen in the thyroid bud as well as in the surrounding mesenchyme. Sagittal sections. Lung bud epithelium is outlined by dotted lines. Scale bar = 75 μm.
Bcl2 is strongly accumulated in the developing thyroid and is regulated by Pax8
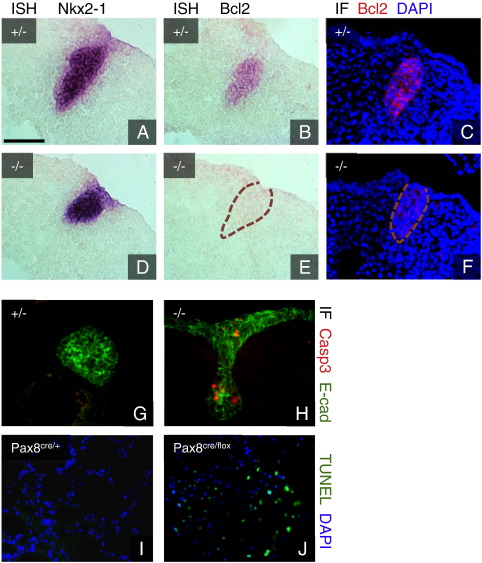
Among the genes with the strongest accumulation in the thyroid bud is the anti-apoptotic regulator Bcl2 (FC 45) (Brunelle and Letai, 2009). In situ hybridization and immunofluorescence staining confirmed the strong expression of this gene and its protein product in the thyroid primordium at E10.5 (Figs. 4B and C) in contrast to other endodermal regions (data not shown). In Pax8 deficient embryos the thyroid is specified and bud formation takes place. However, the thyroid thereafter regresses by a yet uncharacterized mechanism that has been suggested to involve an increased rate of apoptosis (Parlato et al., 2004). This prompted us to investigate if expression of Bcl2 is altered in Pax8 null embryos. In accordance with previous studies, the thyroid bud was found to be smaller and misshapen at E10.5 in the absence of Pax8. Whereas the expression of Nkx2-1 is unaffected (Fig. 4D), Bcl2 transcripts are virtually undetectable in the thyroid bud of mutant embryos (Fig. 4E), in contrast to control littermates. Also Bcl2 protein expression is clearly reduced (Fig. 4F), indicating that Bcl2 expression in the thyroid bud depends on Pax8. In agreement with this is the finding that Pax8 is able to bind and transactivate the Bcl2 promoter in vitro (Hewitt et al., 1997). As a reduction of Bcl2 levels may negatively affect cell survival, the presence of apoptotic cells was investigated by staining for activated effector caspase-3. Whereas no positive cells were noticed in wild type thyroid primordia, scattered apoptotic cells were consistently seen in the thyroid buds of Pax8 deficient embryos at E11.5, indicative of an increased apoptotic rate (Figs. 4G and H). Interestingly, widespread apoptosis was evident at 1 day after birth in mice where Pax8 is conditionally ablated in the thyroid, confirming the role of Pax8 in controlling survival in both thyroid progenitors and follicular cells (Figs. 4I and J). Pax genes, chiefly those of the Pax2/5/8 class, have previously been found to control apoptosis and a cooperative role of Pax2 and Pax8 has been described in kidney development (Bouchard et al., 2002). Our data strongly suggest that Pax8 controls cell survival in the thyroid during development by positive regulation of Bcl2 expression, thus blocking apoptosis. Interestingly, two Pax2/5/8 proteins promote cell survival in C. elegans by transcriptional regulation of ced-9, the Bcl2 ortholog (Park et al., 2006). Furthermore we show that this role of Bcl2 is likely organ specific within the foregut, since the thyroid bud shows very high levels of Bcl2, in contrast to the neighboring lung bud. Most importantly, the presence of high levels of Bcl2 controlled by Pax8 at the very beginning of thyroid organogenesis suggests a scenario of a death pathway ready to ensue in the developing thyroid that is kept under control by a tissue-specific circuit leading to high expression of an anti-apoptotic facor. It is conceivable that such a complex regulation might be in place to ensure prompt removal of precursors if subsequent steps, such as differentiation, fail to proceed properly. Thyroid development has not been studied in detail in Bcl2 deficient mice even though a structurally intact thyroid gland has been reported (Veis et al., 1993). This suggests that absence of Bcl2 alone is not sufficient to shift the balance of cell survival towards apoptosis in thyroid progenitor cells. Hypothetically, Pax8 might repress the expression of proapoptotic regulators by yet unknown mechanisms in addition to positive regulation of Bcl2 transcription.
Fig. 4.
Pax8 controls expression of Bcl2 and apoptosis in the thyroid. A–C. In Pax8 heterozygous embryos, expression of Nkx2-1 and Bcl2 transcripts (A, B) as well as Bcl2 protein (C) is abundant. D–F. Whereas Nkx2-1 transcripts (D) are unaffected in Pax8 deficient embryos, levels of Bcl-2 transcripts (E) and protein (F) are strongly downregulated. G, H. In contrast to control embryos (G), scattered caspase-3 positive cells are seen in the thyroid bud of E11.5 Pax8 deficient embryos (H). I, J. In newborn mice TUNEL positive apoptotic cells are absent in control pups (I) but abundant in the thyroid gland when Pax8 has been conditionally deleted (J). Sagittal sections. Scale bar = 75 μm.
Genes enriched in the E10.5 lung bud
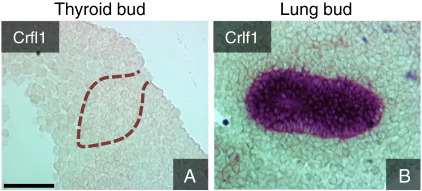
Of 179 genes present in the LEGNoT, 43 displayed a FC > 10 (Table 2). Among these are several genes that are known to be abundant in distal tip epithelium and govern budding morphogenesis of the lung. These include Etv5 (FC 24), Foxa2 (FC 21), Foxa1 (FC 16), Wnt7b (FC 16), Elf5 (FC 13), Shh (FC 6), Bmp4 (FC 4) and Spry2 (FC 4), reviewed in Morrisey and Hogan(2010). On the contrary, genes that are known to be restricted to the surrounding mesenchyme like Fgf10, Foxf1 and Tbx3 (Morrisey and Hogan, 2010) are not present, further indicating that only lung bud epithelium was isolated by LCM. However, also many genes that are not before known to be strongly enriched in the lung buds were detected. As further proof of principle we validated the expression of one such gene, Crfl1 (FC 20), by ISH and found a strong signal in the lung bud whereas no signal was present in the thyroid (Fig. 5).
Table 2.
Genes specifically enriched in the lung bud at E10.5 (LEGNoT). The 50 genes with the highest relative enrichment (fold change) as compared to expression in the whole embryo at the same stage are listed. Dark shading denotes gene where expression is validated by in situ hybridization (see Fig. 3). Light shading denotes genes for which previously published data show expression in the lung at E10.5–E11.5. For references, see Supplemental information.
Genes specifically enriched in the lung bud at E10.5 (LEGNoT). The 50 genes with the highest relative enrichment (fold change) as compared to expression in the whole embryo at the same stage are listed. Dark shading denotes gene where expression is validated by in situ hybridization (see Fig. 3). Light shading denotes genes for which previously published data show expression in the lung at E10.5–E11.5. For references, see Supplemental information.
Fig. 5.
In situ hybridization validation of cytokine receptor-like factor 1 expression in the thyroid (A) and lung (B) buds of E10.5 embryos. Sagittal sections. Thyroid bud epithelium is outlined by dotted lines. Scale bar = 75 μm.
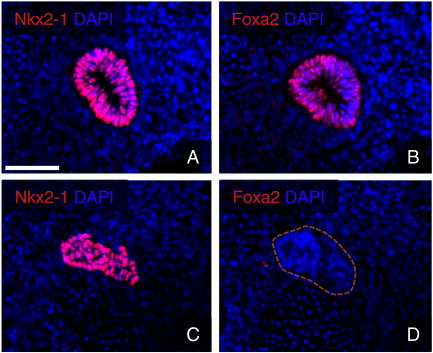
We noticed a strong enrichment of Foxa2 expression (FC 21) in the lung bud, as was anticipated given that Foxa2 is generally considered to be a ubiquitous marker of the endoderm lineage (Ang et al., 1993; Monaghan et al., 1993). Surprisingly, no significant transcript levels were detected in the thyroid. This finding was confirmed by immunofluorescence staining that showed nuclear Foxa2 to be abundant in the lung bud (Fig. 6B) as well as other known expression domains like the floor plate and notochord (not shown). In contrast, no signal could be detected in the developing thyroid (Fig. 6D). It was recently shown that Nkx2.1 and Foxa1/Foxa2 physically interact in a DNA independent manner, which may either inhibit or augment the transcription of target genes in lung epithelial cells (Minoo et al., 2007). The current finding of a lack of Foxa1/Foxa2 expression already at early stages of thyroid development may provide an explanation, at least in part, of how Nkx2-1 differentially regulates its target genes in the developing thyroid and lung.
Fig. 6.
Foxa2 is not expressed in the developing thyroid gland. Nkx2-1 is strongly expressed in the thyroid (A) and lung (C) primordia at E11.5. In contrast, Foxa2 expression is seen only in the lung (B) but not in the thyroid (D). Scale bar = 75 μm.
Foxa1 and Foxa2 are also involved in an intricate signaling network governing bud formation and branching morphogenesis in early lung development. Primary bud outgrowth is driven by Fgf10 from the mesenchyme signaling via FgfrIIb (De Moerlooze et al., 2000; Sekine et al., 1999). In embryos deficient of these genes early lung morphogenesis fails to proceed indicating the fundamental importance of this signaling system in primary bud formation. Interestingly, also the thyroid gland is lacking in embryos deficient of Fgf10 signaling, suggesting that this mechanism of primary bud outgrowth is conserved between the thyroid and lung (Ohuchi et al., 2000). Sox2 that is negatively regulated by Fgf10 (Que et al., 2007) is downregulated at the sites of nascent thyroid and lung bud formation in chicken embryos (Ishii et al., 1998), supporting the notion of mechanistic conservation. Further bud development in the lung is determined by an intricate signaling network where distally expressed genes like Spry2, BMP4, Shh, Foxa1, Foxa2, and wnt7b provide negative and positive signals to adjust the budding morphogenetic program (Cardoso and Lu, 2006; Morrisey and Hogan, 2010). Indeed, we find all these genes to be strongly enriched in the lung but not in the thyroid bud (Tables 1, 2 and Supplemental information). It thus seems that even if some basic mechanisms of primary bud development are common to both organs, such as the involvement of Fgf signaling, the key genetic networks in further development rapidly diverge.
Previous studies confirm that also several of the transcription factors we find to be enriched in the lung bud at E10.5 (Table 2 and Supplemental information) are indeed expressed in the distal bud epithelium and regulate lung morphogenesis. Whereas Etv5 (Liu et al., 2003), Elf5 (Metzger et al., 2007), Foxp1/Foxp2 (Shu et al., 2007), Irx1/Irx2 (van Tuyl et al., 2006), and Gata6 (Keijzer et al., 2001) regulate different aspects of lung development, deficiency of other transcription factors (Hnf1b, Foxa2) results in embryonic lethality at early stages preceding lung bud outgrowth. The global transcriptome of the lung buds at such early development as E10.5 has to the best of our knowledge not previously been described. In a study focusing of transcription factors in foregut derivatives at E11.5, an exclusive expression of Foxp2 and Irx1 was found in the developing lungs. Also the transcription factors Etv5, Gata6, Hoxa1, Hoxb5, Irx2/Irx3/Irx5, Jun, Sox9, Sp5 and Trps1 were accumulated in the developing lung (Sherwood et al., 2009). Interestingly, we detect a significant enrichment (FC > 3) in 9 of these 13 transcripts (not Hoxb5, Irx3/Irx5, Trps1) in lung buds already at E10.5 (see Table 2 and Supplemental information). At this stage some degree of proximodistal differentiation of cells along the developing respiratory tree is evident with Sox2, netrin1 and netrin4 expression in proximal cells whereas Sox9 and Id2 are confined to distal buds (Morrisey and Hogan, 2010). Indeed, we find a strong enrichment of the distal markers Sox9 and Id2 in LCM dissected E10.5 lung buds but none of the proximal markers. This indicates that our data truly reflect the transcriptome of developing lung endoderm that is committed to a distal fate. The membrane proteins Sema4f, Slc15a2 and Unc5b are selective markers of the lung bud lineage as compared to other endodermal derivatives at E11.5 (Sherwood et al., 2009). Also at E10.5 we find Sema4f and Slc15a2 among the most highly enriched transcripts of the lung bud (Table 2). However, Unc5b shows a similar level of enrichment in the thyroid and lung buds and is therefore not exclusive to the lung lineage (see Supplemental information). Taken together, the highly conserved expression pattern of transcription factors and surface markers in our study at E10.5 when compared to published data at E11.5 strongly supports a high validity of our characterization of the lung lineage transcriptome at the earliest stage described so far.
Genes common to the developing thyroid and lung
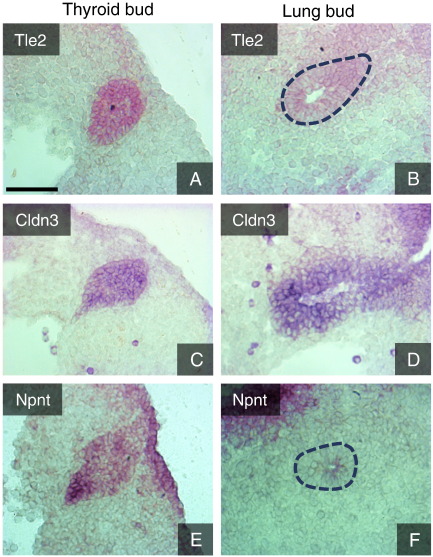
Many components of tight junctions (Cldn3, Cldn6, Cldn7), adherens junctions (Cdh1, Cdh16) and desmosomes (Eppk1, Dsg2) are found among the 113 genes present in TLCEG (Table 3). The expression of several genes in this category in either or both organ buds has previously been demonstrated. Additionally, we validated by ISH the expression of Tle2, Cldn3 and Npnt in both epithelial buds (Fig. 7). Cldn3 (thyroid FC 15, lung FC 13) was abundant in both primordia. The signal for the other two transcripts was evident in the thyroid (Tle FC 17, Npnt FC 7) but faint in the lung bud, which also had a lower enrichment of these genes (Tle FC 5, Npnt FC 4).
Fig. 7.
In situ hybridization validation of genes enriched in the thyroid and lung primordia at E10.5. A, B. Transducin-like enhancer of split 2. C, D. Claudin 3. E, F. Nephronectin. Sagittal sections. Scale bar = 75 μm.
Apart from Hhex we find only three additional transcription factors (Foxo3, Grhl2, jun) to be enriched in both the thyroid and lung buds (see Supplemental information). Even though the Foxo3 expression encompasses wider regions than only the thyroid and lung, mice deficient of this transcription factor are viable and grossly normal whereas they develop ovarian failure and secondary infertility (Castrillon et al., 2003). This suggests that other members of the large Fox family of transcription factors are able to functionally compensate in other organs. Also Grhl2 that is indeed strongly expressed in the developing lung seems to be more widespread in endodermal regions as well as in ectodermal cells (Auden et al., 2006). This is reflected by the drastic phenotype with neural tube and facial closure defects in Grhl2 deficient embryos that are not viable beyond E11.5 (Rifat et al., 2010). In keeping with the notion of a more general function in epithelial cells is the recent finding that Grhl2 is a transcriptional activator of epithelial cell–cell junction components E-cadherin and Cldn4 (Werth et al., 2010), both of which are enriched in the E10.5 thyroid and lung buds. C-jun is also expressed in a wide range of organs during development and null mutants die at midgestation with increased apoptosis in the liver and cardiac anomalies (Hilberg et al., 1993). Taken together, apart from Hhex and Nkx2-1, the transcription factors that we find in TLBEG seem to be primarily involved in generic developmental processes as the acquisition of an epithelial phenotype rather than specifically related to the emergence of midline foregut derivatives. For Nkx2-1 no hybridization signal could be detected either with thyroid or lung RNA, even though corresponding probesets are present on the arrays. This is in contrast to the well established presence of Nkx2-1 mRNA in both thyroid and lung and cannot presently be explained. Speculatively, this could be due to an alternative 3′ end of the Nkx2-1 RNA early in embryogenesis.
In silico validation of the identified gene set
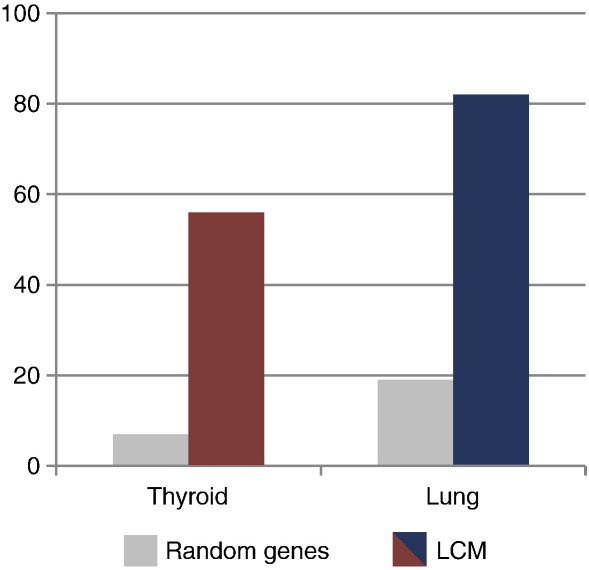
Publicly available images of ISH gene expression patterns from serial sections of E14.5 mouse embryos are available in the GenePaint database (www.genepaint.org; Max Planck Institute, Hannover) (Visel et al., 2004). In an attempt to broadly validate expression of genes highly enriched in the E10.5 mouse thyroid or lung buds also at a later stage of embryonic development (E14.5), images in the GenePaint database of genes with a FC > 10 were evaluated. For approximately 70% of the genes images were available (thyroid 72%, lung 75%). It could be anticipated that strong expression for a number of genes in any set is due to chance alone rather than to true enrichment of organ specific genes in the lists. The expression patterns of 100 randomly selected genes were therefore also analyzed in the developing thyroid and lung. For 66% of these, images could be retrieved from the database and analyzed. The percentage of genes showing strong enrichment (defined as a signal in the thyroid or lung primordia that is either the strongest in the embryo or stronger than that seen in the liver, heart, brain or mandibular mesenchyme representing, respectively, derivatives of endoderm, mesoderm, ectoderm or neural crest) in the organ buds is shown in Fig. 8. For both the TBEG and LBEG sets the percentage of genes with a strong accumulation is clearly higher than in the cohort of randomly picked genes. This further demonstrates that our approach has identified a highly enriched and organ specific transcriptome of the embryonic primordia and that the expression of some of these genes persists also in later development.
Fig. 8.
In silico validation of gene expression in the thyroid and lung primordia at E14.5 by semiquantification of transcript levels in publicly deposited images from automated in situ hybridization of mouse embryo sections (GenePaint database (Max Planck Institute, Hannover) (Visel et al., 2004)). Genes from the TBEG and LBEG sets with a relative enrichment > 10 in the primordia were analyzed. The graph shows the percentage of genes that show a strong expression in the organ buds, defined as a signal in the thyroid (red) or lung (blue) that is either the strongest in the embryo or stronger than that in the liver, heart, brain or mandibular mesenchyme. As a control, the same analysis was performed on 100 randomly selected genes (gray).
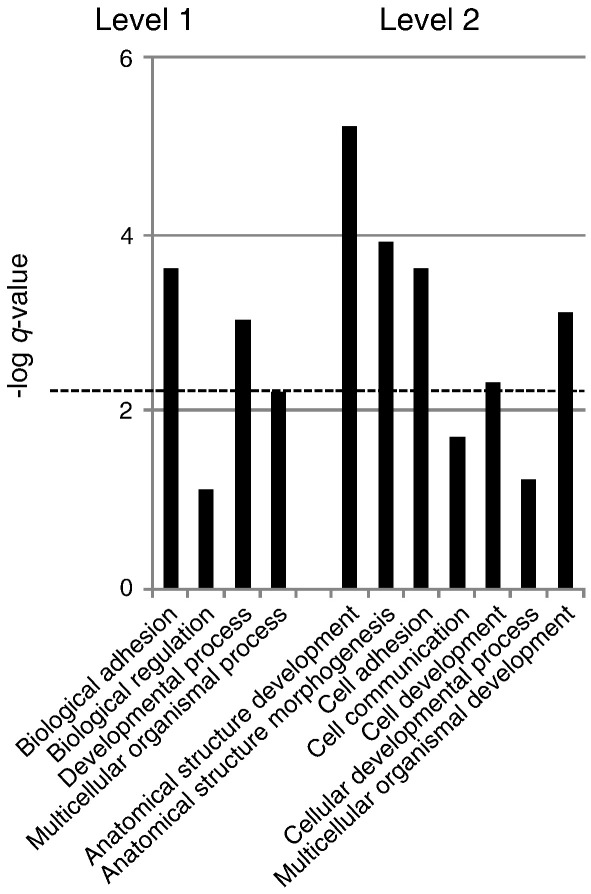
To broadly characterize the lists of genes enriched in the developing thyroid and lung on a functional level, the genes present in the TLBEG list were grouped in Gene Ontology (GO) categories of biological processes. Fig. 9 shows that the distribution of TLBEG genes displays a significant enrichment in categories correlated with developmental processes. Thus, our procedure identified, as we anticipated, genes that are likely to contribute to the developmental fate of both the thyroid and lung bud.
Fig. 9.
Functional characterization of genes enriched in the developing thyroid and lungs (TLBEG). The graph shows high level (1 and 2) GO categories organized according to DAVID that are significantly enriched in the E10.5 thyroid and lung buds. The categories above the dotted line reach statistic significance (q-value 0.05).
Conclusions
We have shown in this paper the gene expression profiles of two endodermal buds, thyroid and lung, at an early developmental stage. A comparison of such transcriptomes with the one obtained with total embryo RNA, identifies genes that by several criteria are likely to be involved in further development of the two buds studied here. This will provide a fertile ground for further characterization of genes and mechanisms that are associated with the pathogenesis of congenital defects of the thyroid and lungs. Furthermore, this study highlights the relevance of a regulatory circuit converging on the anti-apoptotic gene Bcl2, suggesting a novel role for the apoptotic process in thyroid development.
The following are the supplementary materials related to this article.
Supplemental information.
Supplementary file 1.
Supplementary file 2.
Supplementary file 3.
Acknowledgments
Henrik Fagman was supported by an EMBO long-term fellowship, Västra Götalandsregionen under the LUA/ALF agreement and the Assar Gabrielssons Foundation.
This work was supported in part by Telethon, Grant GGP05161, “Molecular Genetics of Thyroid Dysgenesis,” and by the European Community, Integrated Project CRESCENDO Grant LSHM-CT-2005-01865.
References
- Al Taji E., Biebermann H., Limanova Z., Hnikova O., Zikmund J., Dame C., Gruters A., Lebl J., Krude H. Screening for mutations in transcription factors in a Czech cohort of 170 patients with congenital and early-onset hypothyroidism: identification of a novel PAX8 mutation in dominantly inherited early-onset non-autoimmune hypothyroidism. Eur. J. Endocrinol. 2007;156:521–529. doi: 10.1530/EJE-06-0709. [DOI] [PubMed] [Google Scholar]
- Ang S.L., Wierda A., Wong D., Stevens K.A., Cascio S., Rossant J., Zaret K.S. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development (Cambridge, England) 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Auden A., Caddy J., Wilanowski T., Ting S.B., Cunningham J.M., Jane S.M. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns. 2006;6:964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- Berrocal T., Madrid C., Novo S., Gutierrez J., Arjonilla A., Gomez-Leon N. Congenital anomalies of the tracheobronchial tree, lung, and mediastinum: embryology, radiology, and pathology. Radiographics. 2004;24:e17. doi: 10.1148/rg.e17. [DOI] [PubMed] [Google Scholar]
- Bouchard M., Souabni A., Mandler M., Neubuser A., Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16(22):2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M., Souabni A., Busslinger M. Tissue-specific expression of cre recombinase from the Pax8 locus. Genesis. 2004;38:105–109. doi: 10.1002/gene.20008. [DOI] [PubMed] [Google Scholar]
- Brown J.D., Dutta S., Bharti K., Bonner R.F., Munson P.J., Dawid I.B., Akhtar A.L., Onojafe I.F., Alur R.P., Gross J.M., Hejtmancik J.F., Jiao X., Chan W.Y., Brooks B.P. Expression profiling during ocular development identifies 2 Nlz genes with a critical role in optic fissure closure. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1462–1467. doi: 10.1073/pnas.0812017106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle J.K., Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill E.W., Aronow B.J., Georgas K., Rumballe B., Valerius M.T., Aronow J., Kaimal V., Jegga A.G., Yu J., Grimmond S., McMahon A.P., Patterson L.T., Little M.H., Potter S.S. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev. Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso W.V., Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development (Cambridge, England) 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- Castrillon D.H., Miao L., Kollipara R., Horner J.W., DePinho R.A. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- Chapman D.L., Garvey N., Hancock S., Alexiou M., Agulnik S.I., Gibson-Brown J.J., Cebra-Thomas J., Bollag R.J., Silver L.M., Papaioannou V.E. Expression of the T-box family genes, Tbx1–Tbx5, during early mouse development. Dev. Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Corbetta C., Weber G., Cortinovis F., Calebiro D., Passoni A., Vigone M.C., Beck-Peccoz P., Chiumello G., Persani L. A 7-year experience with low blood TSH cutoff levels for neonatal screening reveals an unsuspected frequency of congenital hypothyroidism (CH) Clin. Endocrinol. (Oxf.) 2009;71:739–745. doi: 10.1111/j.1365-2265.2009.03568.x. [DOI] [PubMed] [Google Scholar]
- Correia-Pinto J., Gonzaga S., Huang Y., Rottier R. Congenital lung lesions—underlying molecular mechanisms. Semin. Pediatr. Surg. 2010;19:171–179. doi: 10.1053/j.sempedsurg.2010.03.003. [DOI] [PubMed] [Google Scholar]
- De Felice M., Di Lauro R. Thyroid development and its disorders: genetics and molecular mechanisms. Endocr. Rev. 2004;25:722–746. doi: 10.1210/er.2003-0028. [DOI] [PubMed] [Google Scholar]
- De Felice M., Ovitt C., Biffali E., Rodriguez-Mallon A., Arra C., Anastassiadis K., Macchia P.E., Mattei M.G., Mariano A., Scholer H., Macchia V., Di Lauro R. A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat. Genet. 1998;19:395–398. doi: 10.1038/1289. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L., Spencer-Dene B., Revest J.M., Hajihosseini M., Rosewell I., Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal–epithelial signalling during mouse organogenesis. Development (Cambridge, England) 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Fagman H., Grande M., Gritli-Linde A., Nilsson M. Genetic deletion of sonic hedgehog causes hemiagenesis and ectopic development of the thyroid in mouse. Am. J. Pathol. 2004;164:1865–1872. doi: 10.1016/S0002-9440(10)63745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagman H., Nilsson M. Morphogenesis of the thyroid gland. Mol. Cell. Endocrinol. 2010;323:35–54. doi: 10.1016/j.mce.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Freedom R.M., Yoo S.J., Goo H.W., Mikailian H., Anderson R.H. The bronchopulmonary foregut malformation complex. Cardiol. Young. 2006;16:229–251. doi: 10.1017/S104795110600031X. [DOI] [PubMed] [Google Scholar]
- Fuchtbauer E.M. Expression of M-twist during postimplantation development of the mouse. Dev. Dyn. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- Gu G., Wells J.M., Dombkowski D., Preffer F., Aronow B., Melton D.A. Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development. 2004;131:165–179. doi: 10.1242/dev.00921. [DOI] [PubMed] [Google Scholar]
- Guazzi S., Price M., De Felice M., Damante G., Mattei M.G., Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 1990;9:3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt S.M., Hamada S., Monarres A., Kottical L.V., Saunders G.F., McDonnell T.J. Transcriptional activation of the bcl-2 apoptosis suppressor gene by the paired box transcription factor PAX8. Anticancer Res. 1997;17:3211–3215. [PubMed] [Google Scholar]
- Hilberg F., Aguzzi A., Howells N., Wagner E.F. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ishii Y., Rex M., Scotting P.J., Yasugi S. Region-specific expression of chicken Sox2 in the developing gut and lung epithelium: regulation by epithelial–mesenchymal interactions. Dev. Dyn. 1998;213:464–475. doi: 10.1002/(SICI)1097-0177(199812)213:4<464::AID-AJA11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Jiang X., Rowitch D.H., Soriano P., McMahon A.P., Sucov H.M. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Keijzer R., van Tuyl M., Meijers C., Post M., Tibboel D., Grosveld F., Koutsourakis M. The transcription factor GATA6 is essential for branching morphogenesis and epithelial cell differentiation during fetal pulmonary development. Development (Cambridge, England) 2001;128:503–511. doi: 10.1242/dev.128.4.503. [DOI] [PubMed] [Google Scholar]
- Kimura S., Hara Y., Pineau T., Fernandez-Salguero P., Fox C.H., Ward J.M., Gonzalez F.J. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Kratzsch J., Pulzer F. Thyroid gland development and defects. Best Pract. Res. Clin. Endocrinol. Metab. 2008;22:57–75. doi: 10.1016/j.beem.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Lazzaro D., Price M., de Felice M., Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development (Cambridge, England) 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- Little M.H., Brennan J., Georgas K., Davies J.A., Davidson D.R., Baldock R.A., Beverdam A., Bertram J.F., Capel B., Chiu H.S., Clements D., Cullen-McEwen L., Fleming J., Gilbert T., Herzlinger D., Houghton D., Kaufman M.H., Kleymenova E., Koopman P.A., Lewis A.G., McMahon A.P., Mendelsohn C.L., Mitchell E.K., Rumballe B.A., Sweeney D.E., Valerius M.T., Yamada G., Yang Y., Yu J. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr. Patterns. 2007;7:680–699. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jiang H., Crawford H.C., Hogan B.L. Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev. Biol. 2003;261:10–24. doi: 10.1016/s0012-1606(03)00359-2. [DOI] [PubMed] [Google Scholar]
- Lo C.W., Cohen M.F., Huang G.Y., Lazatin B.O., Patel N., Sullivan R., Pauken C., Park S.M. Cx43 gap junction gene expression and gap junctional communication in mouse neural crest cells. Dev. Genet. 1997;20:119–132. doi: 10.1002/(SICI)1520-6408(1997)20:2<119::AID-DVG5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Lonergan W., Whistler T., Vernon S.D. Comparison of target labeling methods for use with Affymetrix GeneChips. BMC Biotechnol. 2007;7:24. doi: 10.1186/1472-6750-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A., Chowdhury K., Gruss P. Follicular cells of the thyroid gland require Pax8 gene function. Nat. Genet. 1998;19:87–90. doi: 10.1038/ng0598-87. [DOI] [PubMed] [Google Scholar]
- Martinez Barbera J.P., Clements M., Thomas P., Rodriguez T., Meloy D., Kioussis D., Beddington R.S. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development (Cambridge, England) 2000;127:2433–2445. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- Masuda T., Kai N., Sakuma C., Kobayashi K., Koga H., Yaginuma H. Laser capture microdissection and cDNA array analysis for identification of mouse KIAA/FLJ genes differentially expressed in the embryonic dorsal spinal cord. Brain Res. 2009;1249:61–67. doi: 10.1016/j.brainres.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Metzger D.E., Xu Y., Shannon J.M. Elf5 is an epithelium-specific, fibroblast growth factor-sensitive transcription factor in the embryonic lung. Dev. Dyn. 2007;236:1175–1192. doi: 10.1002/dvdy.21133. [DOI] [PubMed] [Google Scholar]
- Metzger R.J., Klein O.D., Martin G.R., Krasnow M.A. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P., Hu L., Xing Y., Zhu N.L., Chen H., Li M., Borok Z., Li C. Physical and functional interactions between homeodomain NKX2.1 and winged helix/forkhead FOXA1 in lung epithelial cells. Mol. Cell. Biol. 2007;27:2155–2165. doi: 10.1128/MCB.01133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P.J., Timmons P.M., Hebert J.M., Rigby P.W., Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- Monaghan A.P., Kaestner K.H., Grau E., Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development (Cambridge, England) 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., Hogan B.L. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumi S., Muroya K., Asakura Y., Adachi M., Hasegawa T. Transcription factor mutations and congenital hypothyroidism: systematic genetic screening of a population-based cohort of Japanese patients. J. Clin. Endocrinol. Metab. 2010;95:1981–1985. doi: 10.1210/jc.2009-2373. [DOI] [PubMed] [Google Scholar]
- Ohuchi H., Hori Y., Yamasaki M., Harada H., Sekine K., Kato S., Itoh N. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A., Lonai P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development. 1992;115:1045–1058. doi: 10.1242/dev.115.4.1045. [DOI] [PubMed] [Google Scholar]
- Park D., Jia H., Rajakumar V., Chamberlin H.M. Pax2/5/8 proteins promote cell survival in C. elegans. Development (Cambridge, England) 2006;133:4193–4202. doi: 10.1242/dev.02614. [DOI] [PubMed] [Google Scholar]
- Parlato R., Rosica A., Rodriguez-Mallon A., Affuso A., Postiglione M.P., Arra C., Mansouri A., Kimura S., Di Lauro R., De Felice M. An integrated regulatory network controlling survival and migration in thyroid organogenesis. Dev. Biol. 2004;276:464–475. doi: 10.1016/j.ydbio.2004.08.048. [DOI] [PubMed] [Google Scholar]
- Purcell P., Joo B.W., Hu J.K., Tran P.V., Calicchio M.L., O'Connell D.J., Maas R.L., Tabin C.J. Temporomandibular joint formation requires two distinct hedgehog-dependent steps. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18297–18302. doi: 10.1073/pnas.0908836106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J., Okubo T., Goldenring J.R., Nam K.T., Kurotani R., Morrisey E.E., Taranova O., Pevny L.H., Hogan B.L. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley R. Enhancing PCR amplification and sequencing using DNA-binding proteins. Mol. Biotechnol. 1994;2:295–298. doi: 10.1007/BF02745882. [DOI] [PubMed] [Google Scholar]
- Rifat Y., Parekh V., Wilanowski T., Hislop N.R., Auden A., Ting S.B., Cunningham J.M., Jane S.M. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 2010;345:237–245. doi: 10.1016/j.ydbio.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Sado T., Nakajima N., Tada M., Takagi N. A novel mesoderm-specific cDNA isolated from a mouse embryonal carcinoma cell line. Develop Growth Differ. 1993;35:551–560. doi: 10.1111/j.1440-169X.1993.00551.x. [DOI] [PubMed] [Google Scholar]
- Scheidl S.J., Nilsson S., Kalen M., Hellstrom M., Takemoto M., Hakansson J., Lindahl P. mRNA expression profiling of laser microbeam microdissected cells from slender embryonic structures. Am. J. Pathol. 2002;160:801–813. doi: 10.1016/S0002-9440(10)64903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K., Ohuchi H., Fujiwara M., Yamasaki M., Yoshizawa T., Sato T., Yagishita N., Matsui D., Koga Y., Itoh N., Kato S. Fgf10 is essential for limb and lung formation. Nat. Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Sherwood R.I., Chen T.Y., Melton D.A. Transcriptional dynamics of endodermal organ formation. Dev. Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood R.I., Jitianu C., Cleaver O., Shaywitz D.A., Lamenzo J.O., Chen A.E., Golub T.R., Melton D.A. Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev. Biol. 2007;304:541–555. doi: 10.1016/j.ydbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Shu W., Lu M.M., Zhang Y., Tucker P.W., Zhou D., Morrisey E.E. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development (Cambridge, England) 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- Simonneau L., Kitagawa M., Suzuki S., Thiery J.P. Cadherin 11 expression marks the mesenchymal phenotype: towards new functions for cadherins? Cell Adhes. Commun. 1995;3:115–130. doi: 10.3109/15419069509081281. [DOI] [PubMed] [Google Scholar]
- Stoppa-Vaucher S., Lapointe A., Turpin S., Rydlewski C., Vassart G., Deladoey J. Ectopic thyroid gland causing dysphonia: imaging and molecular studies. J. Clin. Endocrinol. Metab. 2010;95:4509–4510. doi: 10.1210/jc.2010-0882. [DOI] [PubMed] [Google Scholar]
- Thorwarth A., Mueller I., Biebermann H., Ropers H.H., Grueters A., Krude H., Ullmann R. Screening chromosomal aberrations by array comparative genomic hybridization in 80 patients with congenital hypothyroidism and thyroid dysgenesis. J. Clin. Endocrinol. Metab. 2010 doi: 10.1210/jc.2009-2195. [DOI] [PubMed] [Google Scholar]
- van Tuyl M., Liu J., Groenman F., Ridsdale R., Han R.N., Venkatesh V., Tibboel D., Post M. Iroquois genes influence proximo-distal morphogenesis during rat lung development. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;290:L777–L789. doi: 10.1152/ajplung.00293.2005. [DOI] [PubMed] [Google Scholar]
- Vassart G., Dumont J.E. Thyroid dysgenesis: multigenic or epigenetic … or both? Endocrinology. 2005;146:5035–5037. doi: 10.1210/en.2005-1238. [DOI] [PubMed] [Google Scholar]
- Veis D.J., Sorenson C.M., Shutter J.R., Korsmeyer S.J. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 1993;75:229–240. doi: 10.1016/0092-8674(93)80065-m. [DOI] [PubMed] [Google Scholar]
- Visel A., Thaller C., Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.M., Melton D.A. Vertebrate endoderm development. Annu. Rev. Cell Dev. Biol. 1999;15:393–410. doi: 10.1146/annurev.cellbio.15.1.393. [DOI] [PubMed] [Google Scholar]
- Werth M., Walentin K., Aue A., Schonheit J., Wuebken A., Pode-Shakked N., Vilianovitch L., Erdmann B., Dekel B., Bader M., Barasch J., Rosenbauer F., Luft F.C., Schmidt-Ott K.M. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development (Cambridge, England) 2010;137:3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- Young P.E., Baumhueter S., Lasky L.A. The sialomucin CD34 is expressed on hematopoietic cells and blood vessels during murine development. Blood. 1995;85:96–105. [PubMed] [Google Scholar]
- Zaret K.S., Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn A.M., Wells J.M. Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information.
Supplementary file 1.
Supplementary file 2.
Supplementary file 3.