Abstract
The diagnosis of sessile serrated adenomas (SSAs) is challenging, and there is a great deal of interobserver variability amongst pathologists in differentiating SSAs from hyperplastic polyps (HPPs). The aim of this study was (i) to assess the utility of epigenetic changes such as DNA methylation in differentiating SSAs from HPPs and (ii) to identify common methylation based molecular markers potentially useful for early detection of premalignant neoplastic lesions of gastrointestinal tract. A total of 97 primary patient adenoma samples were obtained from The Johns Hopkins Hospital pathology archive with IRB approval and HIPAA compliance. We analyzed the promoter associated CpG island methylation status of 17 genes using nested multiplex methylation specific PCR (MSP). Methylation of CDX2, hMLH1 and TLR2 was detected in SSAs and SSAs with dysplasia but not in HPPs. A subset of genes including EVL, GATAs (4 and 5), HIN-1, SFRPs (1, 2, 4 and 5), SOX17 and SYNE1 were methylated frequently in all premalignant gastrointestinal adenomas including tubular adenomas, villous adenomas, SSAs and SSAs with dysplasia but infrequently in non-premalignant polyps such as HPPs. Methylation of CDX2, hMLH1 and TLR2 may be of diagnostic utility in differentiating, histologically challenging cases of SSAs from HPPs. Genes such as EVL, GATAs, HIN-1, SFRPs, SOX17 and SYNE1, which are frequently methylated in all types of tested premalignant adenomas, may be useful as biomarkers in stool-based strategies for early detection of these adenomas and CRCs in future.
Keywords: gastrointestinal adenoma, methylation, sessile serrated, classical
Sporadic colorectal cancers (CRCs) are thought to arise through the classical adenoma-carcinoma sequence (in approximately two-thirds of the cases), where classical adenomas [tubular adenomas (TAs) and villous adenomas, (VAs)] progress to invasive carcinomas through a series of genetic and epigenetic alterations including APC and KRAS mutations.1,2 Conversely, other polyps such as hyperplastic polyps (HPPs) are generally regarded as non-neoplastic. Recently, several types of serrated lesions have also been recognized as additional premalignant lesions of the gastrointestinal tract, which may progress to malignancy albeit through a different pathway, commonly known as serrated pathway.3,4
“Serrated adenomas” were first identified by Longacre and Fenoglio-Preiser in 1990, as lesions, which share some morphologic features with both premalignant lesions such as classical adenomas (TAs and VAs) and non-premalignant lesions such as HPPs.5 These lesions (“serrated adenomas”) exhibit cytologic dysplasia reminiscent of classical adenomas and a serrated architecture resembling HPPs.5 Because of these overlapping morphological features, they were previously categorized with either VAs6 or HPPs.7
The initial class of “serrated adenomas” as defined by Longacre and Fenoglio-Preiser was heterogenous and included a variety of lesions including sessile serrated adenomas (SSAs), mixed “TA-HPPs” and conventional APC-related tubular or tubulovillous adenoma with a serrated configuration.8 In 2003, Torlakovic et al. and later in 2005, Snover et al. further refined the criteria and proposed the subclassification of colorectal serrated polyps into HPPs, SSAs (serrated lesions with architecturally distorted crypts and lack of conventional cytologic dysplasia) and so called “traditional” serrated adenomas (TSAs, serrated lesions with adenomatous cytological features).9,10 Most of the easily identifiable lesions originally defined by Longacre and Fenoglio-Preiser that have an adenomatous change fall into the category of TSAs.8 Recent studies have shown that “serrated adenomas” (which is now an older term and includes a variety of serrated lesions) may harbor genetic alterations including mutations in BRAF, KRAS or TGFβRII.11 In addition, these lesions also demonstrate hypermethylation of hMLH112–15 and MGMT16 along with microsatellite instability (MSI)12 and may be associated with subsequent malignancy.17 However, unlike classical adenomas, these “serrated adenomas” lack genetic alterations in APC and β-catenin (CTNNB1).18–20 The association with BRAF mutations, hMLH1 methylation, risk of subsequent malignancy and lack of APC/β-catenin mutations have led to the proposition of a novel serrated pathway3 to carcinogenesis, which is thought to be distinct from the classical adenoma-carcinoma1 pathway.
SSAs pose a diagnostic challenge as they lack conventional adenomatous changes as seen in TSAs and, therefore, more closely resemble HPPs. This makes the distinction between SSAs and HPPs difficult and contingent upon the architectural differences; which has led to a great deal of interobserver variability in the diagnosis of these lesions amongst pathologists.21 Recent evidence suggests that SSAs may account for the so called “HPPs,” which grow to a large size, are located on the right side and may evolve into microsatellite unstable CRC.9,22,23 Because of their malignant potential, some authorities now recommend that SSAs should be managed in the same fashion as classical adenomas. The success of such a strategy, however, depends on the ability to correctly diagnose SSAs by the pathologists. In the absence of accurate diagnostic tools, HPPs may be misclassified as SSAs, and these patients will unnecessarily be subjected to enhanced surveillance. Conversely, some people with SSAs mislabeled as HPPs may develop carcinomas that could have been potentially prevented by correct initial pathologic diagnosis. Therefore, there is a need for objective markers to supplement pathology to aid in correct diagnosis of these lesions and to allow for more effective CRC screening and surveillance.
DNA methylation induced silencing of cancer-related genes is an early and frequent event in CRC.2 A subset of colon cancers have a high number of hypermethylated genes, and this hypermethylator CpG island methylation phenotype (CIMP) may also be associated with a unique disease pathology and prognosis (CIMP).24–26 CIMP tumors are characterized by presentation in patients at older age, female sex, predilection for proximal colon, mucinous or poor differentiation and MSI and BRAF mutations.24,27,28 SSAs have also been suggested as the precursors of these CIMP tumors in the colon.3,13,17,22,27,29 However, most of the initial studies that reported a high level of CpG island methylation in “serrated adenomas” either did not use the refined criteria as proposed by Torlakovic et al. in 2003 or were based on lesions that mostly fit into the category of TSAs.14,16 Additionally, most studies have used a limited panel of 3–5 loci of methylation markers.15,29 However, Wynter et al.30 and Yang et al.11 did recognize the increased frequency of methylation in SSAs and used the recent histopathologic classification criteria proposed by Torlakovic et al.10 Wynter et al.30 confirmed the occurrence of either BRAF or KRAS mutations as an early event in serrated pathway and also suggested that epigenetic phenomenon may play an important role in promoting progression from HPPs to advanced serrated lesions. Yang et al.11 also recognized CIMP-high status, hMLH1 methylation and absence of KRAS mutation in SSAs and advanced serrated lesions with dysplasia. SSAs with dysplasia, also known as admixed polyp, mixed hyperplastic/conventional adenomas or mixed SSAs-TAs, may represent a further advanced lesion in the serrated pathway as they occur frequently in the vicinity of right sided carcinomas and serrated adenocarcinomas.4 However, methylation changes in these mixed lesions have not been studied extensively yet.
Previous studies have suggested that epigenetic alterations may contribute to the malignant potential of SSAs.11,30 We hypothesized that epigenetic differences between HPPs and SSAs may contribute to the malignant potential of SSAs and such epigenetic signatures could also be potentially useful for diagnostic purposes to differentiate SSAs from HPPs. Moreover, the data on CpG island methylation status of genes in advanced lesions, which harbor a noninvasive malignancy such as SSAs with dysplasia, are limited, and it is still not clear whether these lesions epigenetically resemble SSAs or classical adenomas. These lesions are also important to investigate as they may be the immediate precursor of serrated carcinoma and may provide further insight into the pathogenetic mechanisms leading to serrated carcinogenesis.4,31 In this study using a panel of 17 genes, we demonstrate that the frequency of gene methylation increases progressively from HPPs to SSAs to SSAs with dysplasia. This study also highlights some important similarities between SSAs, the key lesions of the serrated neoplasia pathway and classical colorectal adenomas (TAs and VAs) which represent the classical adenoma-carcinoma pathway and delineates a subset of genes which are frequently methylated in these lesions and may be useful for early detection of CRC. Finally, we show that WNT signaling pathway, which is known to be frequently dysregulated in classical adenomas, is also being dysregulated in SSAs, a phenomenon which could partly be explained by epigenetic mechanisms.
Material and Methods
Adenoma samples
A total of 97 primary patient adenoma samples (from 94 patients) were analyzed including 18 TAs, 22 VAs (including 2 tubulovillous adenomas), 29 SSAs, 19 SSAs with dysplasia and 9 HPPs. Formalin-fixed and paraffin-embedded slides were obtained from the pathology archives of The Johns Hopkins Hospital. The study was conducted in accordance with all rules and regulations of The Johns Hopkins University Institutional Review Board and HIPAA compliance. All samples were first reviewed by an expert gastrointestinal pathologist and were selected based on the following diagnostic criteria and availability of tissue. Criteria proposed by Torlakovic et al. were used to diagnose SSAs, SSAs with dysplasia and HPPs. Standard criteria were used to diagnose TAs and VAs. SSAs were identified based on features of prominent basilar crypt dilation, abundant intracellular and extracellular mucin, dystrophic goblet cells and abnormal proliferation. HPPs (all microvesicular type) were identified based on the features of thickened surface basal membrane, thickening and extension of the muscularis mucosae, presence of Kulchitsky (endocrine) cells and decreased overall architectural distortion. Polyps with mixed features of SSAs and HPPs were included in the SSA category. Dysplasia in SSAs was recognized as classical epithelial changes of increased nuclear to cytoplasmic ratio, nuclear crowding, loss of apical mucin and prominence of mitotic figures.
CRC cell lines
Four human CRC cell lines were used including HCT116, HT29, RKO and DKO. The cell lines were purchased from American Type Culture Collection (Manassas, VA) and cultivated and maintained in the appropriate media.
Methylation analysis
Because the aim of this study was to identify genes that could help differentiate HPPs and SSAs and to find differences and similarities between classical adenomas and adenomas of ser-rated pathway, a panel of 17 genes was selected. These genes are involved in a number of different pathways relevant to colorectal carcinogenesis including WNT signaling, gut embryogenesis, cell cycle regulation, mismatch repair, DNA damage repair, cell structure and signal transduction (Table 1). Moreover, all these genes have previously been shown to be methylated in CRC by either a conventional candidate gene approach [APC1A, CDKN2A/P16, CDX2, hMLH1, MGMT, SFRP1-5, GATA4-5 and HIN-1]32–37 or recently identified by genome-wide high throughput strategies as being frequently hypermethylated in CRC (EVL, RAB32, SOX17, SYNE1 and TLR2).38,39 As these genes are frequently methylated in CRC, we further tested their methylation status in TAs, VAs, SSAs, SSAs with dysplasia and HPPs.
Table 1.
Summary of gene names and pathways
| Pathways | Genes investigated |
|---|---|
| WNT signaling Pathway | APC1A (Adenomatosis Polyposis Coli), SFRP1 (Secretory Frizzled Receptor Protein), SFRP2, SFRP4, SFRP5, SOX17 (SRY-related HMG-box transcription factor) |
| Gut embyogenesis | GATA4 (GATA binding protein), GATA5, CDX2 (Caudal type homeobox) |
| Cell cycle regulation | CDKN2A/P16 (cyclin dependent kinase inhibitor), HIN-1 (high in normal) and RAB32 (Ras related protein rab32) |
| Mismatch repair | hMLH1 (Human mutL homologue) |
| DNA damage repair | MGMT (O6-methylguanine-DNA methyltransferase) |
| Signal transduction | EVL (Ena/Vasp like protein) and TLR2 (Toll like receptor2) |
| Cytoskeletal protein | SYNE1 (Spectrin repeat containing, nuclear envelope 1) |
Primer sequences were either previously published or designed by authors.40 DNA was extracted following a standard extraction protocol. Bisulfite modification was done using the EZ DNA methylation Kit™ (Zymo Research) as per manufacturer's instructions. Methylation specific PCR was used for methylation analysis.41 A nested multiplex strategy was utilized, as previously described.42 This strategy was chosen to overcome the limitation of small amount of DNA available from adenomas and comparatively larger amount of DNA required for methylation analysis of 17 genes. Genes were divided into 3–4 panels consisting of 3–6 genes and verified initially using cell lines. These panels were:
Panel 1 → APC1A, CDX2, SFRP1, SFRP2, SFRP4 and SFRP5
Panel 2 → CDKN2A/P16, hMLH1, MGMT
Panel 3 → HIN-1, GATA4 and GATA5
Panel 4 → EVL, RAB32, TLR2, SOX17 and SYNE1
Annealing temperatures were standardized for each panel to obtain optimum product intensity. In the first stage, external primers located in the flanking region were used to amplify the DNA sequences of interest for 35 cycles. Each multiplex PCR was carried out in a volume of 25 μl with 1μl of JumpStart Red Taq DNA polymerase (Sigma, St. Louis, MO), 10 pmol of each external primer and 4 μl of bisulfite modified DNA. PCR conditions included an initial denaturation at 95°C for 5 min followed by 35 cycles of (95°C × 30 sec, annealing temp × 30 sec, 72°C × 30 sec) final elongation at 72°C for 5 min. In the and second stage, 1:500–1:1,000 dilutions of first stage products were used and reamplified for 30 cycles using an internal primer set. Except for the annealing temperatures and cycle number, PCR cycling conditions were the same for first and second stages. Annealing temperatures and primer sequences are summarized in Supporting Information Table 1. In vitro methylated DNA (IVD) was used as a positive control for PCR. IVD was created by treating lymphocytes from a healthy volunteer with SssI methylase (New England Biolabs, Ipswich, MA) as directed by the manufacturer. DKO, which is a Double Knockout derivative of the CRC cell line HCT116 with knockout of the major DNA methyltransferases (DNMT1–/– and DNMT3b–/–), was used as an additional negative control. It lacks methylation at 95% of the known CpG sites.43 Amplification products (8 μl of 25 μl total volume) were separated using electrophoresis on 2.5% agarose gels, stained with GelStar™, a Nucleic Acid Gel Stain (Cambrex Bio Science), illuminated with UV light and photographed.
BRAF and KRAS mutational analysis
Sequencing for BRAF and KRAS were performed as described by Yachida et al.44 Regions of serrated epithelium and adenomatous epithelium from unstained sections were dissected from unstained 10-μ-thick sections. Genomic DNA was extracted from each sample by phenol-chloroform, and 20 ng was used for PCR amplification of KRAS2 exon 2 and BRAF exon 15 using intronic primers flanking these exons. PCR products were sequenced in both directions by use of a M13F primer (5′-GTAAAACGACGGCCAGT-3′) and a M13R primer (5′-CAGGAAACAGCTATGACC-3′) that were incorporated into the forward and reverse primer of each primer pair, respectively (Agencourt Bioscience Corporation, Beverly, MA). Sequence data were analyzed using Sequencher™ 4.8 software (Gene Codes, Ann Arbor, MI). Verification of all mutations was accomplished by bidirectional sequencing of a second PCR product derived independently from the original template.
Immunohistochemical staining
Immunohistochemical labeling was performed using standard methods. Unstained 5-μm sections were cut from paraffin blocks, and the slides were deparaffinized by routine techniques followed by incubation in 1× sodium citrate buffer (diluted from 10× heat-induced epitope retrieval buffer Ventana-Bio Tek Solutions, Tucson, AZ) before steaming for 20 min in 80°C. Slides were cooled for 5 min and incubated with β-catenin (monoclonal antibody, catalog No. 610154, 1:1000 dilution; Transduction Laboratories, Lexington, KY) using a Bio Tek TechMate 1000 automated stainer (Ventana-Bio Tek Solutions). Immunolabeling was detected per kit instructions (Ventana IVIEW Detection Kits, catalog No. 750091). β-catenin labeling was evaluated with respect to membranous and/or nuclear localization, and the location of nuclear labeling (if present). For example, membranous labeling was always considered normal, as was nuclear labeling confined to the crypt bases where the stem cell compartment normally resides (described in detail by Wu et al).45 By contrast, we considered β-catenin labeling as abnormal if nuclear labeling was accompanied by a loss of membranous labeling and was seen outside the crypt bases where the β-catenin positive progenitor population normally resides.
Statistical analysis
Categorical variables were analyzed using Chi square tests, whereas continuous variables were analyzed by the Student's t test, assuming unequal variances or ANOVA (analysis of variance) if more than two categories were compared. p values of less than 0.05 were considered significant. Chi square trend test was used while comparing the methylation frequencies between HPPs, SSAs and SSAs with dysplasia assuming that there is a progressive increase in risk of malignancy from HPPs to SSAs to SSAs with dysplasia. Methylation index was calculated for each sample using the formula—(Number of genes methylated)/(Number of genes tested) × 100. The means for each adenoma type were then compared. Correlation coefficient (Spearman's rank correlation coefficient) was used to find correlation between nuclear β-catenin staining and WNT signaling pathway gene methylation. All statistical analysis was performed using the STATA 9.2 software package (College Station, TX). Unsupervised hierarchical clustering was performed for gene methylation frequencies in specified subtypes. The heat map was produced with R statistical software using euclidean distances and Ward's algorithm.
Results
Clinicopathologic data
Summary of the clinicopathologic information on the different adenoma types is presented in Table 2. There were no significant differences in the sex distribution across the different adenoma subtypes; however, significant differences were seen with respect to age, side and size distribution. SSAs, SSAs with dysplasia and HPPs were from relatively younger patients when compared with TAs and VAs (anova, p value < 0.001). TAs, SSAs and SSAs with dysplasia were predominantly right sided, whereas VAs and HPPs were predominantly left sided (chi2, p value < 0.001). VAs were relatively larger in size when compared with other adenomas. There was no significant difference in the incidence of synchronous polyps at the time of colonoscopy across different adenoma types (chi2, p value = 0.287). These findings may reflect a selection bias due to retrospective nature of the study, as the lesions were selected based on their morphology rather than clinical characteristics.
Table 2.
Clinicopathologic information for the various adenoma samples used in the study
| TA(N = 18) | VA(N = 22) | SSA(N = 29) | SSA with dysplasia(N = 19) | HPP(N = 9) | p value * | ||
|---|---|---|---|---|---|---|---|
| Age | Mean ± S.D. | 68.4 ± 11.9 | 71.6 ± 10.7 | 59.0 ± 12.33 | 62.15 ± 11.31 | 60.3 ± 10.9 | 0.001 |
| Sex | Male | 11 | 12 | 16 | 9 | 4 | 0.9 |
| Female | 7 | 10 | 13 | 10 | 5 | ||
| Side | Right | 12 | 6 | 17 | 17 | 2 | <0.001 |
| Left | 6 | 16 | 12 | 2 | 7 | ||
| Size | <5 mm | 14 | 7 | 20 | 12 | 8 | 0.008 |
| ≥5 mm | 4 | 15 | 9 | 7 | 1 | ||
| <10 mm | 16 | 12 | 26 | 15 | 9 | 0.007 | |
| ≥10 mm | 2 | 10 | 3 | 4 | 0 | ||
| Synchronous polyp | Yes | 9 | 10 | 14 | 7 | 1 | 0.287 |
| No | 9 | 12 | 14 | 12 | 8 |
p value was calculated for age using anova and for sex, side, size and synchronous polyps using Chi2.
TA, tubular adenoma; VA, villous adenoma; SSA, sessile serrated adenoma; HPP, hyperplastic polyp.
Methylation frequencies of all the genes in different adenoma types are summarized in Table 3. A total of 95% of the methylation specific PCR reactions amplified successfully.
Table 3.
Methylation frequency of tested genes in different adenoma types
| Gene | TA (N = 18) | VA (N = 22) | SSA (N = 29) | SSA with dysplasia (N = 19) | HPP (N = 9) | p value for trend tests for comparison of HPP vs. SSA vs. SSA with dysplasia |
|---|---|---|---|---|---|---|
| APC1A | 61.1 | 86.4 | 30.77 | 47.37 | 12.5 | 0.073 |
| CDX2 | 52.9 | 31.8 | 30.43 | 73.68 | 0 | <0.001 |
| SFRP1 | 94.4 | 100 | 96 | 100 | 50 | 0.001 |
| SFRP2 | 76.5 | 90.91 | 88.46 | 94.44 | 50 | 0.022 |
| SFRP4 | 41.2 | 80 | 84.62 | 83.33 | 62.5 | 0.325 |
| SFRP5 | 44.4 | 80 | 84 | 88.24 | 33.33 | 0.021 |
| CDKN2A/P16 | 17.7 | 36.36 | 66.67 | 73.68 | 14.29 | 0.019 |
| hMLH1 | 11.8 | 18.18 | 32 | 68.42 | 0 | <0.001 |
| MGMT | 47.1 | 68.18 | 29.63 | 52.63 | 14.29 | 0.044 |
| HIN-1 | 76.5 | 75 | 77.78 | 89.47 | 44.44 | 0.014 |
| GATA4 | 94.4 | 90.91 | 96.55 | 94.74 | 77.78 | 0.197 |
| GATA5 | 76.5 | 81.82 | 61.54 | 89.47 | 44.44 | 0.01 |
| EVL | 68.8 | 90.91 | 77.78 | 93.33 | 37.5 | 0.005 |
| RAB32 | 77.8 | 63.64 | 44.44 | 64.71 | 33.33 | 0.319 |
| TLR2 | 16.7 | 50 | 50 | 77.78 | 0 | <0.001 |
| SOX17 | 93.8 | 100 | 100 | 100 | 28.57 | <0.001 |
| SYNE1 | 72.2 | 86.36 | 72 | 93.75 | 33.33 | 0.005 |
| Mean methylation index | 58.19 ± 25.34 | 71.75 ± 17.38 | 67.15 ± 19.64 | 79.14 ± 25.13 | 30.69 ± 16.05 | <0.001 |
TA, tubular adenoma; VA, villous adenoma; SSA, sessile serrated adenoma; HPP, Hyperplastic polyps.
Methylation analysis of SSAs, SSAs with dysplasia and HPPs
There is a progressive increase in the mean methylation index from HPPs to SSAs to SSAs with dysplasia (Mean methylation index ± standard deviation HPPs 30.7 ± 16.0%, SSAs = 67.1 ± 19.6%, SSAs with dysplasia = 79.1 ± 25.1%, anova p value < 0.001) as is between TAs and VAs (Mean methylation index TAs = 58.2 ± 25.3% vs. VAs = 71.7 6 ± 17.4%, t test p value 0.052). Interestingly, methylation of CDX2, hMLH1 and TLR2 was seen in SSAs and SSAs with dysplasia but not in HPP. There is also a progressive and significant increase in the methylation frequencies for 13 of the 17 genes tested from HPPs to SSAs to SSAs with dysplasia (Table 3 Trend test: CDX2 p value = < 0.001, SFRP1 p value = 0.001, SFRP2 p value = 0.022, SFRP5 p value = 0.021, CDKN2A/P16 p value = 0.019, hMLH1 p value < 0.001, MGMT p value = 0.044, HIN-1 p value = 0.014, GATA5 p value = 0.010, EVL p value = 0.005, TLR2 p value < 0.001, SOX17 p value < 0.001, SYNE1 p value = 0.005). For the remaining four genes, namely APC1A, SFRP4, GATA4 and RAB32, although there is also an increase in methylation frequency from HPPs to SSAs to SSAs with dysplasia, the trend test is not statistically significant.
Gene clustering analysis
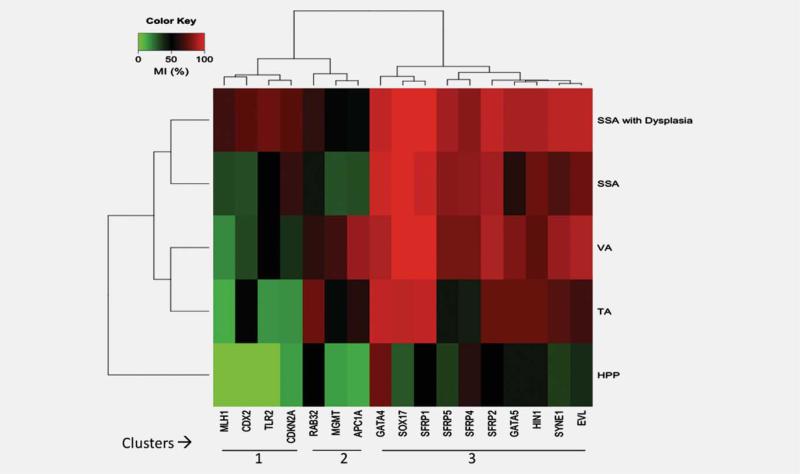
Figure 1 illustrates the unsupervised clustering analysis of tested genes and polyp types based on the methylation frequencies of these genes. Unsupervised clustering analysis revealed broadly three gene clusters. Cluster 1 includes genes with highest frequency of methylation in SSAs and SSAs with dysplasia including hMLH1, CDX2, TLR2 and CDKN2A/P16. Cluster 2 includes genes with highest frequency of methylation in TAs and VAs including, RAB32, MGMT. and APC1A. Cluster 3 includes genes with uniformly high frequency of methylation in all premalignant adenomas including TAs, VAs, SSAs and SSAs with dysplasia. These genes include EVL, GATAs (GATA4 and 5), HIN-1, SFRPs (SFRP1, 2, 4 and 5), SOX17 and SYNE1. Additionally, clustering reveals three subclusters of adenomas including serrated adenoma cluster (SSAs and SSAs with dysplasia) and classical adenoma cluster (TAs and VAs). Interestingly, HPPs cluster separately from these two clusters.
Figure 1.
Unsupervised clustering analysis of methylation frequencies of tested genes against different adenoma types (SSAs-Sessile serrated adenomas, TA-Tubular adenomas, VA-Villous adenomas, HPP-Hyperplastic polyps. Numbers 1, 2 and 3 refer to the genes belonging to cluster 1 (hMLH1, CDX2, TLR2 and CDKN2A), cluster 2 (RAB32, MGMT and APC1A,) and cluster 3 (GATA4, SOX17, SFRP1, SFRP5, SFRP4, SFRP2, GATA5, HIN-1, SYNE1 and EVL). Cluster 1 genes are methylated at highest frequency in sessile serrated adenomas (SSAs) and sessile serrated adenomas with dysplasia (SSAs with Dysplasia). Genes such as hMLH1, CDX2 and TLR2 which are methylated in SSAs and SSAs with dysplasia but not in HPPs may have diagnostic utility in pathologically challenging cases of SSAs and HPPs. Cluster 2 genes are methylated at highest frequency in tubular adenomas (TAs) and villous adenomas (VAs). Cluster 3 genes are methylated at high levels in all categories of adenomas including SSAs, SSAs with dysplasia, TAs and VAs. All genes are infrequently methylated in HPPs.
BRAF and KRAS2 mutations in SSAs
The SSA samples were assayed for commonly occurring mutations in BRAF and KRAS2. Nine of sixteen SSAs were found to be positive for BRAF mutations (codon V600E). Three SSAs were wild type and four samples failed to amplify. BRAF mutations were thus seen in 75% of informative SSA cases (9/12). For KRAS2, mutations were assayed in exon 2 codons 12 and 13, and exon 3 codon 61. For KRAS2 exon 2, codons 12 and 13 mutations, 15 SSAs were found to be wild type while one failed to amplify. For exon 2 codon 61 mutations, eight samples were found to be wild type and rest failed to amplify.
Immunolabeling patterns for β-catenin and methylation of WNT signaling genes in SSAs, SSAs with dysplasia and HPPs
β-catenin staining was performed in 36 samples including ten SSAs, 17 SSAs with dysplasia and nine HPPs. All nine HPPs showed positive membranous staining for β-catenin. Interestingly, six of the ten SSAs (60%) showed positive nuclear staining for β-catenin, and the remaining four showed weak membranous labeling. Surprisingly, nuclear staining for β-catenin was found to be positive in all 17 of the SSAs with dysplasia (100%). We further correlated the nuclear labeling of β-catenin with methylation status of WNT signaling pathway genes including APC1A, SFRPs (1,2,4 and 5) and SOX17 in SSAs, SSAs with dysplasia and HPPs. Methylation of SFRPs (1, 2 and 5 ) and SOX17 was found to significantly correlate with nuclear labeling of β-catenin. SOX17 showed the strongest correlation (correlation coefficient 0.73 p value < 0.0001). Table 4 summarizes the correlation between nuclear labeling of β-catenin and methylation status of WNT signaling pathway genes.
Table 4.
Correlation between nuclear labeling of β-catenin and methylation of WNT signaling pathway genes in tested samples of HPPs (N = 9), SSAs (N = 10) and SSAs with dysplasia (N = 17)
| Gene | Spearman coefficient | p value |
|---|---|---|
| APC1A | 0.29 | 0.1285 |
| SFRP1 | 0.67 | 0.0002* |
| SFRP2 | 0.41 | 0.0357* |
| SFRP4 | 0.13 | 0.5017 |
| SFRP5 | 0.4497 | 0.0275* |
| SOX17 | 0.7372 | <0.0001* |
SFRP1, SFRP2 and SFRP5 show a weak positive (>0.4) correlation, whereas SOX17 shows a strong positive (>0.7) correlation with nuclear β catenin staining.
Discussion
Our study demonstrates for the first time methylation of EVL, RAB32, SOX17, TLR2 and CDX2 in TAs, VAs, SSAs and SSAs with dysplasia. There is a progressive increase in mean methylation index from HPPs to SSAs to SSAs with dysplasia, and we show that there is increase in methylation between TA and VA concordant with their increasing malignant potential. Interestingly, methylation of CDX2, hMLH1 and TLR2 is seen in SSAs and SSAs with dysplasia but not in HPPs. These results are also consistent with other studies, which have shown downregulation of CDX2 and hMLH1 in SSAs.10,45,46
SSAs are critical to diagnose as these are the lesions which tend to occur at the margins of small carcinomas and are more closely associated with the development of microsatellite unstable cancers.8 Recently, it has been recommended that these lesions should be managed in the same way as classical adenomas.9 However, as previously noted, there is a great deal of interobserver variability in the pathological diagnosis of these lesions.21 In such circumstances, some people with HPPs labeled as SSAs may undergo unnecessary enhanced surveillance, whereas others who have SSAs mis-diagnosed as HPPs may develop a subsequent malignancy, which could have been prevented by enhanced surveillance and proper management. These genes including CDX2, hMLH1 and TLR2 that are methylated exclusively in SSAs with or without dysplasia and not in HPPs could be of potential diagnostic utility in those cases in which it is difficult to differentiate SSAs from HPPs. These findings, if validated in larger prospective studies may provide a means to correctly stratify patients for proper clinical management in the future. Although, histology will remain the gold standard to diagnose these lesions as it is universally available, methylation studies may, however, be used in a subset of cases that are challenging to diagnose histologically. At this time, the use of methylation strategies to differentiate SSAs from HPPs may appear to be expensive and may be more useful for research purposes, but in future as the technology advances, it may become more practical and cost effective. Although the increasing amount of methylation in HPPs, SSAs and SSAs with dysplasia may be interpreted as “progression,” it may also occur independently. Whether methylation increases in the same lesion over time will require further investigation.
Clustering analysis revealed three clusters of genes including genes with highest levels of methylation in SSAs and SSAs with dysplasia (Cluster 1), genes with highest levels of methylation in classical adenomas (TAs and VAs) (Cluster 2) and genes with high levels of methylation in all premalignant adenomas TAs, VAs, SSAs and SSAs with dysplasia (Cluster 3). Cluster 3 genes may potentially have important clinical and functional implications because these genes show high levels of methylation in all tested premalignant adenomas including TAs, VAs, SSAs and SSAs with dysplasia but are unmethylated or have low frequency of methylation in HPPs. These genes may be good candidates for investigation as biomarkers for early detection of CRC in stool-based DNA methylation assays. Jass et al. and Lazarus et al. have also proposed that SSAs may grow into subsequent adenocarcinoma more rapidly at least in some patients.3,17 This has led to the idea that rapid transformation of SSAs may be responsible for the so called interval tumor phenomenon, where a large right-sided adenocarcinoma is identified in a region that appeared lesion free at the colonoscopy performed in the preceding 1 or 2 years. In such a scenario, a stool-based assay, which could potentially be performed more frequently than colonoscopy, may be useful, and the cluster 3 genes will be good candidates for investigation in such a study. One such study has already shown the utility of methylation of SFRP1, which is also enlisted in our cluster 3 genes, in early detection of cancer.47 However, further investigation will be required to illustrate clinical utility of other markers for such assays.
Functionally, cluster 3 genes highlight the fact that the two pathways of colorectal carcinogenesis, namely the classical adenoma carcinoma pathway and the serrated neoplasia pathway, may share some features rather than being mutually exclusive. Perhaps, common methylation of cluster 3 genes may be one such feature, and this could also be exploited in various screening strategies. Cluster analysis further suggested that many of the cluster 3 genes belonged to the WNT signaling pathway genes including SFRP1, SFRP2, SFRP4, SFRP5 and SOX17. This prompted us to further investigate the functional consequences of such epigenetic changes in SSAs. We correlated the methylation status of these genes with nuclear labeling of β-catenin, which was used as readout for WNT signaling activation. Surprisingly, a fairly strong correlation was seen between nuclear labeling of β-catenin and methylation of WNT signaling pathway genes in SSAs and SSAs with dysplasia. Our results are consistent with those of Wu et al., who recently reported increased nuclear labeling of β-catenin in SSAs. Conversely, some of the previous reports that failed to identify β-catenin mutations in serrated adenomas used the criteria proposed by Fenoglio-Preiser and the lesions tested by them more closely resembled TSAs. We, therefore suggest that as a result our findings are not directly comparable with theirs.20 Given the fact that the correlation between nuclear labeling of β-catenin and methylation of WNT signaling pathway genes is not perfectly concordant, WNT signaling dysregulation in SSAs may also be due to other mechanisms as suggested by Sawyer et al.18 This may also mean contribution by methylation of some of the other genes which were not analyzed in this study. However, ours is the first study to suggest that DNA methylation may lead to dysregulation of WNT signaling in SSAs and SSAs with dysplasia.
Another interesting finding of this study is the description of epigenetic changes in SSAs with dysplasia. These are have also been termed “mixed SSAs-TAs” and have morphological features of both the lesions. Our data suggest that these lesions show highest levels of methylation for most of the tested genes and may be the true hypermethylator polyps of the colon. These lesions could be the next step in the progression of SSAs to carcinoma which is similar to the suggestions made by Makinen et al.4 However, it remains to be elucidated whether such a progression occurs in each individual SSA.
BRAF and KRAS2 sequencing was done only in a subset of SSAs. BRAF mutations were found in 75% of the informative samples, while all the tested lesions were wild type for KRAS2. This is consistent with the fact that BRAF and KRAS mutations are mutually exclusive.48 As most of our tested lesions were positive for BRAF, KRAS mutations might be underrepresented in these lesions. The serrated pathway is also believed to follow two routes; one identified by BRAF and other by KRAS mutations.49 Our results may be more representative of the first serrated pathway which predominantly shows BRAF mutations. The pathway involving KRAS mutations is thought to more commonly involve TSAs.4 These observations of others may explain lack of KRAS mutations in SSAs tested in our study.
There were several limitations to this study. These include lack of immunohistochemical data for all the genes to show their downregulation in these lesions. Moreover, TSAs, which may represent another premalignant lesion in the serrated pathway, were not analyzed in this study. Additionally, the number of HPPs analyzed in this study was small (n 9). TSAs with conventional dysplasia can be difficult to differentiate from VAs and concerns may be raised that MGMT methylated cases could potentially be TSAs with conventional dysplasia. Because all our lesions were reviewed by an expert gastrointestinal pathologist, the possibility of any such occurrence although plausible is felt to be remote.
In conclusion, our data suggest that there is a progressive increase in methylation frequencies of genes from HPPs to SSAs to SSAs with dysplasia. Methylation of CDX2, hMLH1 and TLR2, which is seen frequently in SSAs and SSAs with dysplasia but not in HPPs, may have diagnostic utility to differentiate SSAs from HPPs. The classical adenoma carcinoma pathway and serrated neoplasia pathway may share some common features especially dysregulation of WNT signaling. WNT signaling dysregulation in SSAs and SSAs with dysplasia may be explained by epigenetic mechanisms such as DNA methylation of WNT signaling pathway genes. Common epigenetic abnormalities in all the premalignant polyps of the colorectum may be exploited to devise stool-based DNA methylation assays in future.
Acknowledgements
The authors thank Marco A. Riojas and Ghaith Habboub for technical assistance.
Grant sponsor: Wendy Will Case Cancer Fund Inc; Grant number: 303847; Grant sponsors: Ross Clinician Scientist Award, National Cancer Institute; Grant number: K23CA127141; Grant sponsors: American Surgical Association Fellowship Foundation
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR. Serrated route to colorectal cancer: back street or super highway? J Pathol. 2001;193:283–5. doi: 10.1002/1096-9896(200103)193:3<283::AID-PATH799>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Makinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131–50. doi: 10.1111/j.1365-2559.2006.02548.x. [DOI] [PubMed] [Google Scholar]
- 5.Longacre TA, Fenoglio-Preiser CM. Mixed hyperplastic adenomatous polyps/serrated adenomas. A distinct form of colorectal neoplasia. Am J Surg Pathol. 1990;14:524–37. doi: 10.1097/00000478-199006000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Jass JR, Filipe MI, Abbas S, Falcon CA, Wilson Y, Lovell D. A morphologic and histochemical study of metaplastic polyps of the colorectum. Cancer. 1984;53:510–15. doi: 10.1002/1097-0142(19840201)53:3<510::aid-cncr2820530324>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 7.Goldman H, Ming S, Hickock DF. Nature and significance of hyperplastic polyps of the human colon. Arch Pathol. 1970;89:349–54. [PubMed] [Google Scholar]
- 8.Torlakovic EE, Gomez JD, Driman DK, Parfitt JR, Wang C, Benerjee T, Snover DC. Sessile serrated adenoma (SSA) vs. traditional serrated adenoma (TSA). Am J Surg Pathol. 2008;32:21–9. doi: 10.1097/PAS.0b013e318157f002. [DOI] [PubMed] [Google Scholar]
- 9.Snover DC, Jass JR, Fenoglio-Preiser C, Batts KP. Serrated polyps of the large intestine: a morphologic and molecular review of an evolving concept. Am J Clin Pathol. 2005;124:380–91. doi: 10.1309/V2EP-TPLJ-RB3F-GHJL. [DOI] [PubMed] [Google Scholar]
- 10.Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol. 2003;27:65–81. doi: 10.1097/00000478-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Yang S, Farraye FA, Mack C, Posnik O, O'Brien MJ. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol. 2004;28:1452–9. doi: 10.1097/01.pas.0000141404.56839.6a. [DOI] [PubMed] [Google Scholar]
- 12.Hawkins NJ, Ward RL. Sporadic colorectal cancers with microsatellite instability and their possible origin in hyperplastic polyps and serrated adenomas. J Natl Cancer Inst. 2001;93:1307–13. doi: 10.1093/jnci/93.17.1307. [DOI] [PubMed] [Google Scholar]
- 13.Jass JR. Serrated adenoma of the colorectum and the DNA-methylator phenotype. Nat Clin Pract Oncol. 2005;2:398–405. doi: 10.1038/ncponc0248. [DOI] [PubMed] [Google Scholar]
- 14.Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol. 2003;162:815–22. doi: 10.1016/S0002-9440(10)63878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petko Z, Ghiassi M, Shuber A, Gorham J, Smalley W, Washington MK, Schultenover S, Gautam S, Markowitz SD, Grady WM. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res. 2005;11:1203–9. [PubMed] [Google Scholar]
- 16.Dong SM, Lee EJ, Jeon ES, Park CK, Kim KM. Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol. 2005;18:170–8. doi: 10.1038/modpathol.3800261. [DOI] [PubMed] [Google Scholar]
- 17.Lazarus R, Junttila OE, Karttunen TJ, Makinen MJ. The risk of metachronous neoplasia in patients with serrated adenoma. Am J Clin Pathol. 2005;123:349–59. doi: 10.1309/VBAG-V3BR-96N2-EQTR. [DOI] [PubMed] [Google Scholar]
- 18.Sawyer EJ, Cerar A, Hanby AM, Gorman P, Arends M, Talbot IC, Tomlinson IP. Molecular characteristics of serrated adenomas of the colorectum. Gut. 2002;51:200–6. doi: 10.1136/gut.51.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uchida H, Ando H, Maruyama K, Kobayashi H, Toda H, Ogawa H, Ozawa T, Matsuda Y, Sugimura H, Kanno T, Baba S. Genetic alterations of mixed hyperplastic adenomatous polyps in the colon and rectum. Jpn J Cancer Res. 1998;89:299–306. doi: 10.1111/j.1349-7006.1998.tb00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto T, Konishi K, Yamochi T, Makino R, Kaneko K, Shimamura T, Ota H, Mitamura K. No major tumorigenic role for beta-catenin in serrated as opposed to conventional colorectal adenomas. Br J Cancer. 2003;89:152–7. doi: 10.1038/sj.bjc.6601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farris AB, Misdraji J, Srivastava A, Muzikansky A, Deshpande V, Lauwers GY, Mino-Kenudson M. Sessile serrated adenoma: challenging discrimination from other serrated colonic polyps. Am J Surg Pathol. 2008;32:30–5. doi: 10.1097/PAS.0b013e318093e40a. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein NS, Bhanot P, Odish E, Hunter S. Hyperplastic-like colon polyps that preceded microsatellite-unstable adenocarcinomas. Am J Clin Pathol. 2003;119:778–96. doi: 10.1309/DRFQ-0WFU-F1G1-3CTK. [DOI] [PubMed] [Google Scholar]
- 23.Torlakovic E, Snover DC. Serrated adenomatous polyposis in humans. Gastroenterology. 1996;110:748–55. doi: 10.1053/gast.1996.v110.pm8608884. [DOI] [PubMed] [Google Scholar]
- 24.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–5. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 27.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–87. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 28.Whitehall VL, Wynter CV, Walsh MD, Simms LA, Purdie D, Pandeya N, Young J, Meltzer SJ, Leggett BA, Jass JR. Morphological and molecular heterogeneity within nonmicrosatellite instability-high colorectal cancer. Cancer Res. 2002;62:6011–4. [PubMed] [Google Scholar]
- 29.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–35. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–80. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldstein NS. Small colonic microsatellite unstable adenocarcinomas and high-grade epithelial dysplasias in sessile serrated adenoma polypectomy specimens: a study of eight cases. Am J Clin Pathol. 2006;125:132–45. [PubMed] [Google Scholar]
- 32.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, Baylin SB, Herman JG. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–71. [PubMed] [Google Scholar]
- 33.Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–33. [PubMed] [Google Scholar]
- 34.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–94. [PubMed] [Google Scholar]
- 35.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–71. [PubMed] [Google Scholar]
- 37.Shigematsu H, Suzuki M, Takahashi T, Miyajima K, Toyooka S, Shivapurkar N, Tomlinson GE, Mastrangelo D, Pass HI, Brambilla E, Sathyanarayana UG, Czerniak B, et al. Aberrant methylation of HIN-1 (high in normal-1) is a frequent event in many human malignancies. Int J Cancer. 2005;113:600–4. doi: 10.1002/ijc.20622. [DOI] [PubMed] [Google Scholar]
- 38.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, van Engeland M, Ting AH, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Glockner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, Ahuja N, Baylin SB. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–72. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandes JC, Carraway H, Herman JG. Optimal primer design using the novel primer design program: MSPprimer provides accurate methylation analysis of the ATM promoter. Oncogene. 2007;26:6229–37. doi: 10.1038/sj.onc.1210433. [DOI] [PubMed] [Google Scholar]
- 41.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Engeland M, Weijenberg MP, Roemen GM, Brink M, de Bruine AP, Goldbohm RA, van den Brandt PA, Baylin SB, de Goeij AF, Herman JG. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133–7. [PubMed] [Google Scholar]
- 43.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin SB, Vogelstein B. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 44.Yachida S, Mudali S, Martin SA, Montgomery EA, Iacobuzio-Donahue CA. Beta-catenin nuclear labeling is a common feature of sessile serrated adenomas and correlates with early neoplastic progression after BRAF activation. Am J Surg Pathol. 2009;33:1823–32. doi: 10.1097/PAS.0b013e3181b6da19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu JM, Montgomery EA, Iacobuzio-Donahue CA. Frequent beta-catenin nuclear labeling in sessile serrated polyps of the colorectum with neoplastic potential. Am J Clin Pathol. 2008;129:416–23. doi: 10.1309/603UQKM7C2KELGJU. [DOI] [PubMed] [Google Scholar]
- 46.Mochizuka A, Uehara T, Nakamura T, Kobayashi Y, Ota H. Hyperplastic polyps and sessile serrated ‘adenomas’ of the colon and rectum display gastric pyloric differentiation. Histochem Cell Biol. 2007;128:445–55. doi: 10.1007/s00418-007-0326-2. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W, Bauer M, Croner RS, Pelz JO, Lodygin D, Hermeking H, Sturzl M, Hohenberger W, Matzel KE. DNA stool test for colorectal cancer: hypermethylation of the secreted frizzled-related protein-1 gene. Dis Colon Rectum. 2007;50:1618–26. doi: 10.1007/s10350-007-0286-6. discussion 26–7. [DOI] [PubMed] [Google Scholar]
- 48.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 49.Jass JR, Whitehall VL, Young J, Leggett BA. Emerging concepts in colorectal neoplasia. Gastroenterology. 2002;123:862–76. doi: 10.1053/gast.2002.35392. [DOI] [PubMed] [Google Scholar]