Abstract
PURPOSE
Merkel cell polyomavirus (MCPyV) is prevalent in the general population, integrates into most Merkel cell carcinomas (MCCs) and encodes oncoproteins required for MCC tumor growth. We sought to characterize T-cell responses directed against viral proteins that drive this cancer as a step toward immunotherapy.
METHODS
Intracellular cytokine cytometry, IFN-γ-ELISPOT, and a novel HLA-A*2402-restricted MCPyV tetramer were used to identify and characterize T-cell responses against MCPyV oncoproteins in tumors and blood of MCC patients and control subjects.
RESULTS
We isolated virus-reactive CD8 or CD4 T cells from MCPyV-positive MCC tumors (2 of 6), but not from virus-negative tumors (0 of 4). MCPyV-specific T-cell responses were also detected in the blood of MCC patients (14 of 27) and control subjects (5 of 13). These T cells recognized a broad range of peptides derived from capsid proteins (2 epitopes) and oncoproteins (24 epitopes). HLA-A*2402-restricted MCPyV oncoprotein processing and presentation by mammalian cells led to CD8-mediated cytotoxicity. Virus-specific CD8 T cells were markedly enriched among tumor-infiltrating lymphocytes as compared to blood, implying intact T-cell trafficking into the tumor. While tetramer-positive CD8 T cells were detected in the blood of 2 of 5 HLA-matched MCC patients, these cells failed to produce IFN-γ when challenged ex vivo with peptide.
CONCLUSIONS
Our findings suggest that MCC tumors often develop despite the presence of T cells specific for MCPyV T-Ag oncoproteins. The identified epitopes may be candidates for peptide-specific vaccines and tumor- or virus-specific adoptive immunotherapies to overcome immune evasion mechanisms in MCC patients.
Introduction
Merkel cell carcinoma (MCC) is an aggressive neuroendocrine skin cancer. Its reported incidence has quadrupled in the past 20 years to ~1600 cases/year in the US (1). In 2008, the Merkel cell polyomavirus (MCPyV) was discovered and found to be integrated into the host genome in ~80% of MCC tumors (2). This association of MCPyV with MCC has been confirmed by multiple groups worldwide (3). MCPyV infection, defined by serology or detection of viral DNA, is prevalent in both healthy persons and MCC patients (4). However, in MCC patients, MCPyV acquires oncogenic potential via rare integration and T-antigen (T-Ag) truncation mutations (5, 6). These large T-Ag (LT) truncation mutants bind and inactivate the retinoblastoma tumor suppressor, but no longer induce lytic viral replication that would be lethal to a cancer cell (7). The small T-Ag (ST) shares the N-terminus with LT and plays an important role in activating the AKT-mTOR signaling pathway (8, 9). Importantly, MCPyV T-Ag oncoproteins are persistently expressed in MCCs and are required for growth (6, 9).
The cellular immune system appears to be critical in preventing and controlling MCC. Patients who are chronically immune suppressed by HIV, chronic lymphocytic leukemia or medications after solid organ transplant have a 3–30 fold increased risk of MCC and represent approximately 10% of MCC patients (10). There are numerous cases of spontaneous MCC regression, suggesting immune-mediated cancer control (11, 12). Furthermore, intratumoral infiltration of CD8 lymphocytes is an independent predictor of improved survival in MCC (13). The high prevalence of antibodies to MCPyV T-Ag (14) and VP1 capsid proteins (15–17) in MCC patients is suggestive of virus-specific helper CD4 T-cell responses.
Despite the virus-dependence of most MCC tumors and the high seroprevalence of MCPyV in the general population (15–17), there have been no reports of MCPyV-specific T-cell reactivity in MCC patients or control subjects. We hypothesized that MCPyV proteins are immunogenic and are recognized by T lymphocytes in MCC tumors and peripheral blood of patients and control subjects. Here we show that MCPyV-specific CD8 and CD4 T cells can localize to MCC tumors and report 26 novel MCPyV T-cell epitopes, some of which may serve as targets for immunotherapy in the future.
Materials and Methods
Human subjects and clinical samples
This study was approved by the Fred Hutchinson Cancer Research Center IRB and performed in accordance with Helsinki principles. Subjects (Table S1) gave informed consent. PBMC obtained from heparinized blood samples were cryopreserved. Tumor samples obtained from medically necessary biopsies or surgeries were transported in T-cell medium (TCM) (18).
Synthetic viral peptides
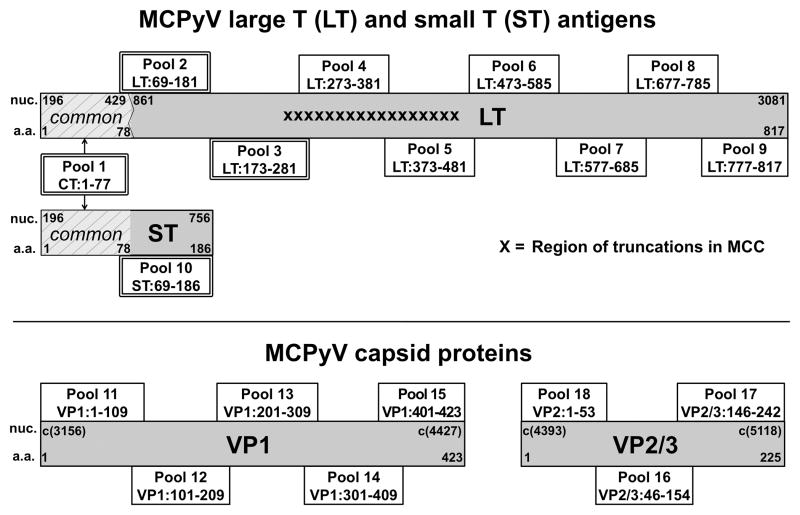
428 peptides (13-mers, overlapping by 9 amino acids) were synthesized (Sigma Aldrich; Table S2) from MCPyV reference sequence Genbank #EU375803 and strain variant #EU375804. Peptide pools (~25 peptides/pool) were organized by viral domains (Figure 1). Additional truncated MCPyV peptides and polyomavirus-derived homologous peptides (predicted by CLUSTAL 2.0.12 sequence alignment software) were synthesized (Table S3).
Figure 1. MCPyV proteome and synthetic peptide pools.
MCPyV proteins (gray boxes) are annotated with nucleotide (nuc) and amino acid (a.a.) numbers. Peptides spanned the entire MCPyV proteome and were grouped into pools (boxes with a.a. range indicated) based on protein domains. Double outlined boxes represent persistently expressed LT, ST, and CT (shared N-terminus, light-gray slashed lines) domains. Jagged line at nucleotide 429 represents the genomic splice site. c(#) indicates the corresponding complementary DNA nucleotide position number.
Tumor infiltrating lymphocytes (TIL) culture
Minced MCC tissue was plated in 48-well plates in TCM plus 1.6 μg/ml PHA (Remel), 32 units/ml nIL-2 (Hemagen Diagnostics), and allogeneic irradiated PBMC. nIL-2 in fresh TCM was added every second day for 14–20 days (18). When performed, a second TIL expansion used anti-CD3 mAb OKT3 (Ortho), recombinant IL-2 (Chiron Corporation), and feeder cells (19). Cultures expanded with anti-CD3 mAb are comprised entirely of CD3-Qdot655 (Invitrogen) positive cells (Figure S1).
T-cell cloning
Subject w347 primary tumor TIL were cloned by limiting dilution and subsequently expanded (20). Subject w347 metastatic tumor was collagenase (Sigma)-digested and plated as single cell suspensions in TCM plus PHA and nIL-2 for limiting dilution cloning. Subject w447 bulk TIL were tetramer-sorted and expanded (19).
ELISPOT
Cultured assays: PBMC plated in TCM at 3×105 cells/well in 96-well flat bottom plates were stimulated on day 1 with MCPyV peptide pools (1 μg/ml of each peptide), individual peptides (1 μg/ml), CMV, EBV, influenza peptides (CEF, 2 μg/ml; Cellular Technology Ltd) as positive control or DMSO (<0.2%) and or TCM as negative control and cultured for 10 days. Fresh TCM with 20 U/mL IL-2 (Chiron Corporation) and 20 ng/ml IL-7 (R&D Systems) was added on days 3, 6 and 9. On day 11, cells were plated in 96-well multiscreen IP plate (Millipore) precoated with anti-IFN-γ capture antibody (1-D1K, Mabtech). Mitogens corresponding to the prior stimulation cycle were added on day 12 and the plates developed on day 13 (21).
Direct assays: 5×105 cells/well PBMC plated in TCM were stimulated with relevant peptides. After 24 hours, plates were developed (21), scanned using an ELISPOT reader (C.T.L.) and counted using ImmunoSpot 5.0 Software (C.T.L.). Stringent ELISPOT interpretation criteria were used to determine T-cell reactivity (SUPPLEMENTARY methods).
Intracellular cytokine (ICC) and flow cytometry
Responder cells stimulated with peptide in the presence of anti-CD28 and anti-CD49d mAb, and brefeldin A (22) were incubated with CFSE-(Invitrogen) labeled antigen presenting cells (APCs) for 12–18 hours (23). Controls included DMSO and PMA-Ionomycin. Cells subsequently stained with LIVE/DEAD-Violet (Invitrogen), followed by mAbs CD4-PerCP and CD8-APC (BD Biosciences) were permeabilized and stained with anti-IFN-γ-PE (BD Biosciences) (23). Data acquired with FacsCanto-II cytometer (Becton Dickinson) and BD-FACSDiva software (v6.1.1) were analyzed using FlowJo (v9.1). Data are reported as percentage viable cells of phenotypic interest identified as CFSE-negative responder cells in the lymphocyte forward/side scatter region.
Cytokine analysis
LCLs (2×105 cells/well) and responders (2×105 cells/well) were co-cultured with peptides (1 μg/ml) in a 48-well plate in 1 ml TCM. Supernatants collected at 24 and 48 hours were analyzed for IFN-γ, IL-10 or IL-5 using fluorescent microbead-based flow cytometric ELISA (FHCRC, Seattle).
Plasmids, transfection and imumunoblot
Cos7 cells were transfected with plasmid-encoding MCPyV LT and/or HLA-A*2402 cDNA using Fugene6 transfection reagent (Roche) manufacturer-recommended protocol (SUPPLEMENTARY methods). Protein product was confirmed by immunoblotting for MCPyV LT-specific mAb CM2B4 (Santa Cruz) (24). Expression of HLA-A*2402 was confirmed by flow cytometry with A23/A24 mAb (One Lambda) followed by PE-labeled IgG secondary antibody (Invitrogen) at 48 hours. Controls included HLA-A*2402-positive and negative LCLs. Transfected Cos7 or control cells were used as APCs in ICC.
Cytotoxicity assay
Target cell lysis was determined by a 4-hour 51Chromium release assay in quadruplicates (25). Negative controls included LT-transfected Cos7 plus effectors and Cos7 transfected with LT and HLA*A2402 without effectors. Positive control included detergent-lysed target cells.
Tetramer staining
PE- and APC-conjugated complexes of HLA-A*2402 and peptide LT-92-101 (EWWRSGGFSF, CPC Scientific) or irrelevant peptide (NY-ESO-158-166, (26)), were synthesized at the Immune Monitoring Laboratory (FHCRC, Seattle). Cells were stained with tetramer for 60 minutes at 37°C, followed by anti-CD8-APC mAb (BD Biosciences) for 30 minutes at 4°C, then washed with FACS buffer, fixed and analyzed.
MCPyV DNA sequencing and detection
DNA extraction, amplification and sequencing are detailed in SUPPLEMENTARY methods (3, 27). Sequence data were submitted to GenBank for subjects w447 (accession number JF912157) and w347 (accession number JF912158).
HLA typing
HLA-typing genotypes from blood-derived DNA were determined using sequence based typing of exons 2 and 3 (28) (FHCRC, Seattle) or with commercially available (OneLambda; Qiagen) sequence-specific primer PCR kits (PSBC, Seattle; Hematologisk Laboratorium, Denmark).
Immunohistochemistry (IHC)
MCPyV T-Ag staining was performed using CM2B4 antibody (Santa Cruz) (14, 24). Anti-CD8 (Novocastra) and anti-CD4 (Cell Marque) antibodies were used at 1:200 and 1:25 dilution respectively.
Statistical analyses
Analyses were performed using two-tailed Fisher’s exact test in Stata v11.0 (StataCorp). A p-value <0.05 was considered significant.
Results
To study the T-cell response to MCPyV proteins, we isolated lymphocytes from MCC tumor tissues and blood donated by MCC patients and control subjects (Table S1). Fresh tumor specimens were obtained from ten MCC patients requiring diagnostic or therapeutic surgery. TIL were successfully cultured from each tumor in sufficient numbers to test T-cell reactivity to regions of T-Ag that are persistently expressed in MCC tumors (peptide pools 1–3 and 10; Figure 1 and Table S2). MCPyV T-Ag oncoproteins were present in 24 of 33 tumors (72%) as evaluated by IHC. T-cell responses to MCPyV T-Ag as assessed by IFN-γ secretion were found in two of six T-Ag protein-expressing tumors. T-Ag-specific T cells were not detected among the four tumors that did not express viral oncoproteins.
Identification of the MCPyV large T-Ag epitope recognized by CD8+ TIL
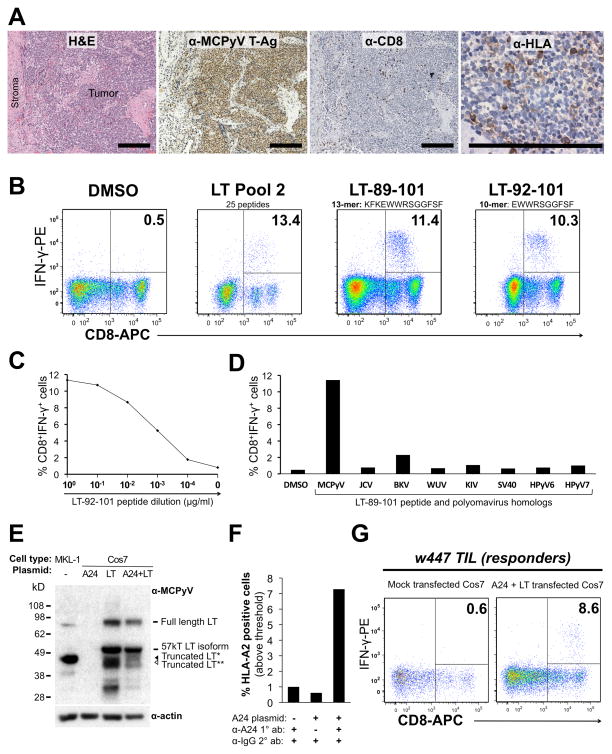
Subject w447 presented at age 67 with a 1.5 cm primary MCC lesion on his left hip. The patient underwent wide local excision of the tumor, a portion of which was used for obtaining TIL. IHC revealed CD8+ cells within an MCPyV T-Ag protein-positive tumor that had low HLA class I expression (Figure 2A). After fourteen days of expansion in the presence of PHA and nIL-2, TIL were screened by ICC assay for IFN-γ production to peptides from regions of T-Ag that are persistently expressed in MCC tumors (pools 1, 2, 3, and 10). A discrete population of IFN-γ secreting CD8 cells was only reactive to peptide pool 2 (Figure 2B). Upon challenge with single peptides from pool 2, these TIL responded to a single immunogenic peptide, LT-89-101 (KFKEWWRSGGFSF) (Figure 2B). Truncation analysis identified a 10 amino acid section, LT-92-101 (EWWRSGGFSF) that generated strong CD8 IFN-γ responses that persisted in peptide titrations to as low as 1 ng/ml (Figure 2B–C). Notably, engagement of TCR signals may downregulate CD8 expression (29), explaining slightly low CD8 signal among IFN-γ producing cells. Sequencing of DNA amplified from this patient’s tumor confirmed the predicted MCPyV large T-Ag amino acid sequence (amino acids 92–101, data not shown).
Figure 2. CD8+ TIL isolated from an MCC patient specifically recognize an MCPyV-derived peptide and endogenously processed large T-Ag (LT).
(A) Serial sections from w447 MCC tumor. From left: H&E stain; T-Ag expression (CM2B4 antibody); CD8 lymphocyte staining; HLA-I staining (EMR8-5 clone). Scale bar = 200μm. (B) CD8 IFN-γ response to LT-derived peptides in an ICC assay. From left: TIL stimulated with DMSO; pool 2; 13-mer peptide LT-89-101 from pool 2; and minimal 10-mer epitope LT-92-101. Autologous PBMC were used as APCs. (C) TIL have high functional avidity for peptide LT-92-101. (D) TIL recognize MCPyV LT-89-101 but not homologous peptides from indicated polyomaviruses. Y-axis represents flow cytometry data gated as in 2B. (E) Western blot primary bands after probe with anti-MCPyV LT (CM2B4) or anti-alpha-actin loading control in MKL-1 (MCPyV positive MCC cell line) and Cos7 cells transfected as indicated. (*) and (**) indicate distinct truncation isoforms. (F) Flow cytometry analysis of HLA-A24 expression on Cos7 cells transfected as indicated. Data represent percent cells above PE-fluorescence intensity threshold. (G) TIL IFN-γ responses to Cos7 cells transfected as indicated. Numbers shown in plots are the percentage of CD8-gated TIL.
We next determined whether TIL from this subject cross-reacted to peptide homologs of MCPyV LT-92-101 derived from other human polyomaviruses. Cross-reactivity with other polyomaviruses could indicate T-cell priming by other pathogens prior to tumor infiltration. Conversely, if the TIL response was MCPyV – specific, this would suggest that MCPyV was the relevant immunogen. To address this, we tested 13-mer peptides homologous to MCPyV LT-92-101 (Table S3) and did not detect cross-reactivity among TIL to these peptides (Figure 2D).
HLA restriction and endogenous processing of MCPyV large T-Ag
We sought to determine the restricting HLA allele from among this patient’s genotype (A*03G1, A*24G1, B*1501, B*3502, Cw*0304 and Cw*04G1). Two published HLA-peptide binding algorithms (30) predicted strong binding of the immunogenic peptide (MCPyV LT 92–101) to HLA-A*2402. This in silico prediction was confirmed using cells matched to subject w447 only at HLA-A*2402. While allogeneic partially-matched PBMC induced IFN-γ secretion in CD8 TIL from subject w447, peptide-loaded PBMC that were not matched at any HLA locus induced minimal IFN-γ secretion (Figure S2).
To study MCPyV oncoprotein processing into short peptides, we co-transfected full-length MCPyV LT and HLA-A*2402 into the primate cell line, Cos7 (Figure 2E–F). Transfected cells were exposed to subject w447 TIL and T-cell activation was measured by IFN-γ production. Mock-transfected cells, or cells transfected with A*2402 alone, did not activate subject w447 TIL. However, co-transfection of LT and A*2402 led to readily detectable activation of CD8 TIL by IFN-γ ICC (Figure 2G). Furthermore, we observed specific lysis of Cos7 cells that expressed MCPyV LT and HLA-A*2402 by a TIL-derived CD8 T cell clone (Figure S3). These MCPyV-specific CD8 cells also have excellent cytolytic potential against peptide-pulsed autologous cells (91% and 100% lysis of PBMCs pulsed with peptide LT.92-101 at 1μg/ml and 5μg/ml respectively, Figure S3). We conclude that, 1) MCPyV LT can be processed into appropriate peptides, 2) these peptides can access the HLA class-I antigen presentation pathway for proper loading, and 3) MCPyV-specific CD8 TIL can recognize and kill cells expressing LT and the appropriate HLA.
MCPyV-specific CD8 T cells are enriched in the tumor as compared to blood
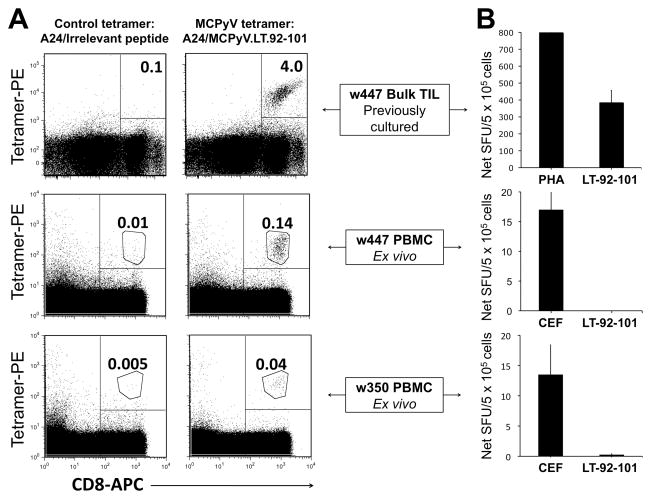
We synthesized a fluorescent HLA-peptide tetramer (A24:MCPyV.LT.92-101). This MCPyV-specific tetramer, and a control HLA-A*2402 tetramer, were used to stain subject w447 PBMC and his TIL that had been expanded in an unbiased fashion with PHA and IL-2. Our data is consistent with marked enrichment of MCPyV-specific CD8 T cells among TIL (4.0%) as compared to blood (0.14%) (Figure 3A, top and middle panels).
Figure 3. Detection and functional status of MCPyV-specific CD8 T cells in tumor and blood of MCC patients.
(A) Subject w447 TIL (top), w447 PBMC (middle) and w350 PBMC (bottom) specifically stain with an MCPyV-specific tetramer, but not with control tetramer. Specific gating for cell clusters was used in the bottom four panels as indicated. (B) IFN-γ secretion by TIL and PBMC in response to LT-92-101 and positive controls as assessed by direct ELISPOT assay.
Tetramer-positive T cells identified in PBMC in two MCC patients fail to produce IFN-γ
PBMC from five MCC patients (all of whom were positive for an HLA-A*24xx genotype) were stained with the MCPyV tetramer. We observed a well-defined population of tetramer-bright, CD8-positive cells in PBMC from both the index patient, w447, and a second patient, w350 (Figure 3A, middle and bottom panels), but not in other HLA-A24-positive or HLA-A24-negative subjects (Figure S4). Both of these patients had T-Ag expressing tumors. In contrast to expanded TIL from subject w447, physiologically-relevant direct ELISPOT functional analyses of PBMC from subjects w447 and w350 revealed no IFN-γ production in response to antigen (Figure 3B). These observations were further supported by ICC assays examining IFN-γ production in response to peptide among tetramer-specific cells in subject w447. While 20% of tetramer-positive TIL secreted IFN-γ, only 2% of tetramer-positive PBMC produced IFN-γ (Figure S5). The PBMC analyzed for IFN-γ were drawn during an early asymptomatic time-point after recurrence (subject w447) or one month post tumor removal (w350).
CD4+ T cells with the same MCPyV-specificity isolated from an MCC primary tumor and a subsequent metastasis
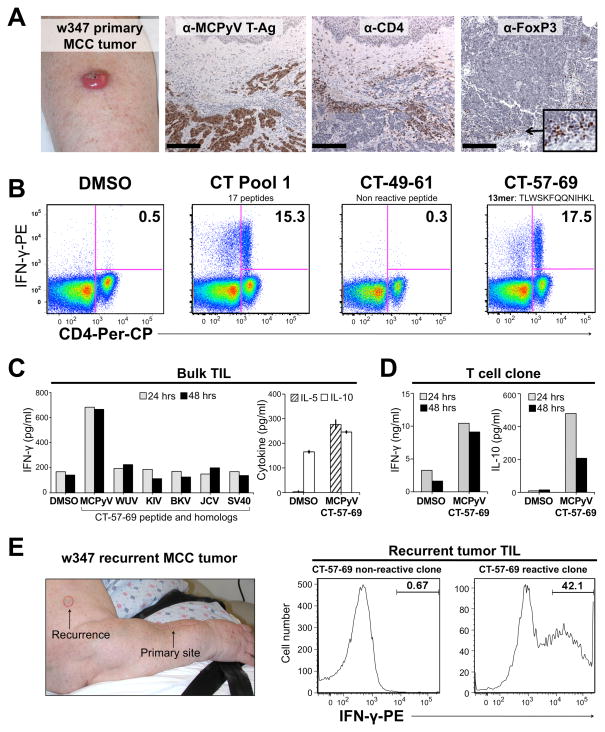
Subject w347 was 78 years old when she presented with a 3.5 cm MCC on the right forearm. IHC revealed CD4+ and FoxP3+ cells within an MCPyV T-Ag protein-positive tumor (Figure 4A). TIL were expanded in the absence of exogenous antigen and later screened for IFN-γ production to peptide pools 1, 2, 3, and 10 using the ICC assay. CD4 reactivity was observed only to peptide pool 1 (Figure 4B). Pool breakdown showed reactivity only to peptide CT-57-69 (TLWSKFQQNIHKL) (Figure 4B). Of note, strong antigenic triggers may downregulate CD4 cell surface expression (31), among IFN-γ producing cells. Sequencing of MCPyV DNA amplified from tumor w347 confirmed the predicted T-Ag sequence (amino acids 57–69, data not shown).
Figure 4. Characterization and cloning of MCPyV-specific CD4 TIL from an MCC patient.
(A) MCC tumor from subject w347. From Left: Primary tumor arising on forearm; MCPyV T-Ag expression (CM2B4); CD4 and FoxP3 (236A/E7 clone) staining. Scale bar = 200μm. (B) Identification of IFN-γ CD4 T-cell responses to CT domain peptides using ICC. From left: TIL stimulated with DMSO; CT pool 1; representative non-reactive pool 1 peptide; reactive pool 1 peptide CT-57-69. PBMC were used as APCs. Gating as in Figure 2. (C) TIL cytokine response to MCPyV peptide or its homologs as measured by ELISA. Left: Bulk CD4 TIL IFN-γ response to CT-57-69 or homologous peptides from the indicated polyomaviruses. Right: Bulk CD4 TIL secretes IL-5 and IL-10. Error bars represent standard error between triplicates. (D) CD4 clone isolated from primary tumor TIL secretes IFN-γ and IL-10 in response to CT-57-69. Data represent single measurements at two time points. (E) CD4 TIL from recurrent lesion recognize the same peptide as TIL from primary lesion. From left: Recurrent w347 MCC (red outline); representative non-reactive T-cell clone; T-cell clone reactive to peptide CT-57-69.
To determine if this CD4 TIL response was cross-reactive, we tested 13-mer homolog peptides (Table S3) from other human polyomaviruses. No specific reactivity to the homologous peptides was noted (Figure 4C, left panel). To investigate the nature of the CD4 response to MCPyV CT-57–69, we investigated the cytokine secretion profile of the expanded CD4 TIL population. Interestingly, we observed the secretion of both Th1 and Th2 type cytokines, namely IFN-γ, IL-5, and IL-10 (Figure 4C), although the latter cytokine was only modestly increased compared to background. Moreover, 2 of 58 tested T-cell clones isolated by limiting dilution from the primary TIL also secreted IFN-γ and 1 of 1 clones tested secreted IL-10 in response to peptide (Figure 4D).
Two months after primary tumor excision, subject w347 developed a skin recurrence of MCC (Figure 4E, left panel). After clonal microculture of the digested tumor tissue, 1 of 53 clones tested produced a robust IFN-γ response to the same peptide (CT-57-69) as in the primary tumor (Figure 4E, right panel).
T-cell responses to MCPyV in the peripheral blood
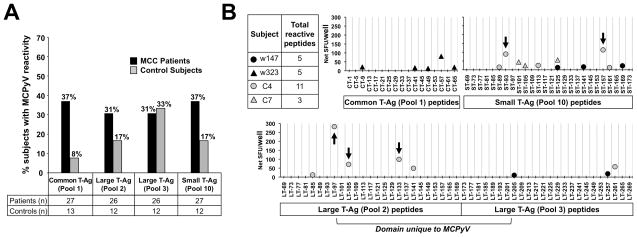
PBMC reactivity was screened against the MCC-associated T-Ag oncoprotein domains (pools 1, 2, 3 and 10) in MCC patients and control subjects. We observed a trend toward a higher frequency of T-cell responses in patients than control subjects for pool 1 (37% of patient PBMC samples vs. 8% of control subject samples), pool 2 (31% vs. 17%) and pool 10 (37% vs. 17%) (Figure 5A). The magnitude of pool-level responses for each subject in Figure 5A is included as Figure S6. We detected PBMC responses against T-Ag in 12 of 19 patients with MCPyV-positive tumors. In contrast, we found T-Ag-specific T-cell reactivity in blood from 1 of 6 MCC patients with MCPyV-negative tumors (p=0.07).
Figure 5. Prevalence of PBMC reactivity to MCPyV oncoproteins and determination of immunoreactive epitopes.
(A) PBMC T-cell reactivity (as described in ELISPOT interpretation criteria in SUPPLEMENTARY methods) to specified MCPyV peptide pools amongst MCC patients (black) and control subjects (gray). Although there is a trend, data did not reach statistical significance. (B) Identification of immunoreactive epitopes derived from previously reactive T-Ag peptide pools by cultured IFN-γ ELISPOT in MCC patients (black) and control subjects (gray). In subject C4, five peptides (arrows) were identified to be immunogenic to CD4+ T cells.
Twenty-four additional immunogenic MCPyV epitopes were identified. Breakdown of the reactive pools to single reactive peptides is shown in Figure 5B for two MCC patients and two control subjects. A T-cell response to a single peptide (ST-125-137) was common to both subject C7 and w147. Responder T-cell phenotype was determined for five immunoreactive T-Ag-derived peptides in control subject C4 using a cultured ICC assay. In each case, the reactive cells were CD4-positive (data not shown). To explore whether these CD4 T-cell responses were likely to be MCPyV specific, we compared the sequence of the reactive MCPyV 13-mers to the homologous regions from other known polyomaviruses (Table S3). Among these five peptides, three were from an MCPyV-unique protein domain and one was sequence-divergent when compared to homologous peptides, suggesting MCPyV-specificity in most cases.
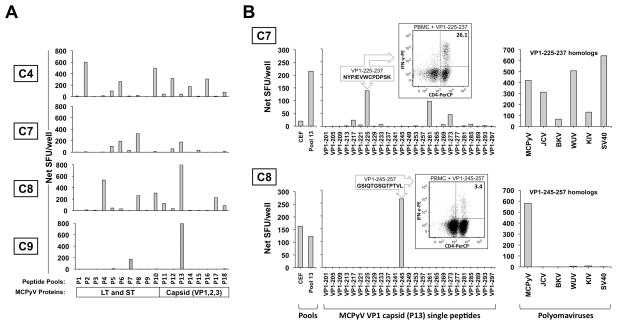
T-cell responses to the entire MCPyV proteome (18 peptide pools) were also assessed in four control subjects. PBMC from each subject demonstrated strong reactivity to at least one portion of the MCPyV proteome (Figure 6A). Of note, PBMC from all four control subjects reacted to peptides in pool 13 (derived from the capsid protein VP1). Single-peptide level responses were deconvoluted for two subjects (C7 and C8). Subject C7 demonstrated robust T-cell responses to multiple peptides within pool 13 (Figure 6B, top panel). The peptide with the strongest response (VP1-225-237) was shown to be CD4-positive (Figure 6B, top panel, inset). This particular peptide shares amino acid identity at five of thirteen positions with all of the homologous peptides derived from five other polyomaviruses (Table S3). All six of these related peptides elicited T-cell reactivity from this subject’s PBMC (Figure 6B, top panel, right). In subject C8, VP1-245-257 was the only reactive peptide within pool 13 and it stimulated CD4 cells (Figure 6B, bottom panel and inset). This immunoreactivity was MCPyV-specific as homologous peptides from other polyomaviruses were non-reactive (Figure 6B, bottom panel, right). Aggregate results for all 26 peptides described in this study are shown in Table S4.
Figure 6. MCPyV proteome-wide screen of T-cell responses and characterization of select epitopes.
(A) PBMC from four control subjects demonstrated reactivity to both MCPyV T-Ag and structural proteins as measured by cultured IFN-γ ELISPOT. (B) Characterization of T-cell responses to pool 13 peptides derived from VP1 capsid protein in control subject C7 (top panel) and C8 (bottom panel). Left: Stimulation with CEF positive control, pool 13, or single 13-mer peptides from pool 13. Inset: ICC/flow cytometry analysis identifies IFN-γ+CD4+ T cells upon peptide stimulation. Right: PBMC challenged with relevant peptide or homologous peptides from indicated polyomaviruses.
Discussion
The strong association of MCC with both a polyomavirus and T-cell dysfunction suggests that dissection of the specificity, trafficking, and effector functions of MCPyV-specific T cells is medically significant. We demonstrate that circulating and tumor infiltrating T cells can recognize specific peptides from MCPyV T-Ag oncoproteins and capsid structural proteins, and that both CD8 and CD4 MCPyV-specific T-cells are locally enriched in at least some MCC tumors. Twenty-six novel MCPyV T-cell epitopes have been identified. We confirmed that the T-Ag oncoprotein can be processed and presented to stimulate cell lysis by virus-specific CD8 cells isolated from a patient’s tumor. We developed an MCPyV tetramer that should serve as the first of several tools for isolating and determining the function of MCPyV-specific CD8 T cells directly ex vivo. These findings advance our understanding of the association of immune suppression with MCC, and help enable rational therapies for MCC that augment virus-specific T-cell activity.
Although not previously described, several lines of evidence suggest that MCPyV-specific T cells should be present in patients and control subjects. Serologic data indicate MCPyV infection is commonly acquired during childhood (16, 32, 33). The presence of MCPyV-specific antibodies in 53–88% of healthy persons (15–17) implies the presence of MCPyV-specific CD4 T-cell help (indeed, all MCPyV-responsive T cells identified in our PBMC-based assays were CD4+). Similar to these findings in MCPyV, T-cell responses have been described (34–37) in other cancer-associated viruses such as HPV and HCV but are typically of low abundance (38, 39). Cultured ELISPOT or ICC are usually required for their detection (35, 40), but occasionally their abundance is sufficient to be detected ex vivo by tetramers (41).
Our data document significant enrichment of MCPyV-specific T cells in an MCC tumor compared to blood. In subject w447 (with a small, slow-growing tumor) we detected a marked enrichment of antigen-specific CD8 cells in TIL compared to blood using tetramer staining. Since primary tumor tissue was sparse, TIL were expanded using a non-specific mitogen (PHA+IL-2) cocktail that uniformly expands the peripheral T cell repertoire (42). We therefore believe the relative frequency of antigen-specific TIL remains stable after expansion. Regardless, this finding indicates that local immune evasion mechanisms in this tumor did not block the trafficking and enrichment of virus-specific T cells. However, the tumor may persist despite these antigen – specific CD8+ T cells due in part to low HLA-I expression. Conversely, chronic exposure to antigen may mediate an exhausted phenotype of antigen-specific CD8+ T cells with poor effector function and sustained expression of inhibitory receptors, such as PD-1 and CTLA-4 (43). Interestingly, in subject w323 (with a large, rapidly-growing tumor) we detected MCPyV T-Ag-specific T cells only in the blood, but not in TIL derived from her T-Ag expressing tumor. In the latter case, there are several possible explanations regarding antigen-specific T cells: 1) they were present in the blood but failed to home to the tumor; 2) they homed to the tumor, but were undetectable because they failed to produce IFN-γ; or 3) they were present in the tumor, but failed to expand in culture.
It is possible that local MCPyV-specific T cells actually promoted tumor growth in some cases. Chronic antigen exposure, a common feature of MCC tumors, has been reported to induce IL-10 and IFN-γ secreting CD4 T cells that have a regulatory function (44). In subject w347, we observed MCPyV-specific IFN-γ and IL-10 cytokine secretion by CD4 T cells isolated from the primary tumor. Notably, we also detected FoxP3+ cells within the tumor microenvironment, which may be similar to the antigen-specific, IL-10 secreting, FoxP3+ T-regulatory cell subset found to be immunosuppressive in metastatic melanoma (45). Further investigation is required to determine the role of T-regulatory cells in MCC.
There are several limitations to this study. PBMC availability prevented proteome-wide screens for some subjects, therefore we focused on the oncogenic portions of T-Ag persistently expressed in tumors. Cultured ELISPOT may sometimes generate false positive reactivity via in vitro priming (46). In order to eliminate such artifactual results, we employed a stringent ELISPOT interpretation criteria (SUPPLEMENTARY methods). Finally, it was not possible to determine the extent of prior exposure to MCPyV for all studied subjects because no ‘gold standard’ serologic or viral shedding test exists. The single most relevant indicator of MCPyV status is viral oncoprotein expression in the tumor, which was assessed for 33 of 38 patients.
In conclusion, this study is the first report of T-cell immune responses to the Merkel cell polyomavirus. The necessary and persistent expression of T-Ags to drive MCC proliferation makes these oncoproteins especially promising targets for measuring and manipulating tumor-specific immune responses. As has been accomplished in other cancers (20, 47, 48), the identified MCPyV T-cell epitopes may be used to design peptide-specific vaccines, to generate virus-specific T cells for adoptive immunotherapy, or to track tumor-associated antigen-specific T-cell responses.
Supplementary Material
Translational relevance.
Merkel cell carcinoma (MCC) is an aggressive neuroendocrine skin cancer (46% 5-year mortality) with limited treatment options for progressive disease. Current evidence suggests that the recently discovered Merkel cell polyomavirus (MPCyV) is causally associated with most MCCs. Reported MCC dependence on viral oncoproteins and our findings that virus-specific CD8 and CD4 T cells are present in MCC patients suggest that immunotherapeutic targeting of MCPyV and reversal of immune evasion mechanisms are promising options for patients with this disease.
The MCPyV epitopes in this report provide tools to 1) isolate both antigen- and tumor-specific T lymphocytes from blood and tumors of MCC patients 2) characterize immune evasion mechanisms 3) develop tumor-specific therapies such as peptide vaccines or adoptive immunotherapy and 4) track T-cell responses during tumor progression or clinical trials.
Acknowledgments
We thank Stephanie Pietromonaco, Miranda Schmidt, Renee Thibodeau, Sherry Lee, Yuki Yoshimatsu, Jianhong Cao, Sine Hadrup, Julie Randolph-Habecker’s lab and FHCRC Specimen Processing for their contributions towards this study.
Financial Support:
NIH-RC2-CA147820-01, NIH-F30ES019463-01, FHCRC Molecular Diagnostics Pilot Award, NIH-K02-AR50993, ACS-RSG-08-115-01-CCE, NIH-K24-CA139052-0, Poncin scholarship, David & Rosalind Bloom Endowment for MCC Research, UW MCC Patient Gift Fund.
Footnotes
Disclosure of Potential Conflicts of Interest: None
References
- 1.Albores-Saavedra J, Batich K, Chable-Montero F, Sagy N, Schwartz AM, Henson DE. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: a population based study. J Cutan Pathol. 2009 doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 2.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker JC, Houben R, Ugurel S, Trefzer U, Pföhler C, Schrama D. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol. 2009;129:248–250. doi: 10.1038/jid.2008.198. [DOI] [PubMed] [Google Scholar]
- 4.Foulongne V, Kluger N, Dereure O, Mercier G, Molès JP, Guillot B, Segondy M. Merkel cell polyomavirus in cutaneous swabs. Emerging Infect Dis. 2010;16:685–687. doi: 10.3201/eid1604.091278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houben R, Schrama D, Alb M, Pföhler C, Trefzer U, Ugurel S, Becker JC. Comparable expression and phosphorylation of the retinoblastoma protein in Merkel cell polyoma virus-positive and negative Merkel cell carcinoma. Int J Cancer. 2010;126:796–798. doi: 10.1002/ijc.24790. [DOI] [PubMed] [Google Scholar]
- 7.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, Gaubatz S, Becker JC, Schrama D. An intact retinoblastoma protein binding site in merkel cell polyomavirus large T antigen is required for promoting growth of merkel cell carcinoma cells. Int J Cancer. 2011 doi: 10.1002/ijc.26076. ePub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Sablina AA, Hahn WC. SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev. 2008;27:137–146. doi: 10.1007/s10555-008-9116-0. [DOI] [PubMed] [Google Scholar]
- 9.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. The Journal of clinical investigation. 2011 doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Penas PF, Nghiem P. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J Am Acad Dermatol. 2008;58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubo H, Matsushita S, Fukushige T, Kanzaki T, Kanekura T. Spontaneous regression of recurrent and metastatic Merkel cell carcinoma. J Dermatol. 2007;34:773–777. doi: 10.1111/j.1346-8138.2007.00382.x. [DOI] [PubMed] [Google Scholar]
- 12.Wooff JC, Trites JR, Walsh NMG, Bullock MJ. Complete Spontaneous Regression of Metastatic Merkel Cell Carcinoma: A Case Report and Review of the Literature. Am J Dermatopathol. 2010 doi: 10.1097/DAD.0b013e3181cd3158. [DOI] [PubMed] [Google Scholar]
- 13.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, Schrama D, Simonson WT, Lemos BD, Byrd DR, Koelle DM, Galloway DA, Leonard JH, Madeleine MM, Argenyi ZB, Disis ML, Becker JC, Cleary MA, Nghiem P. Transcriptome-Wide Studies of Merkel Cell Carcinoma and Validation of Intratumoral CD8+ Lymphocyte Invasion As an Independent Predictor of Survival. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulson KG, Carter JJ, Johnson LG, Cahill KW, Iyer JG, Schrama D, Becker JC, Madeleine MM, Nghiem P, Galloway DA. Antibodies to Merkel Cell Polyomavirus T Antigen Oncoproteins Reflect Tumor Burden in Merkel Cell Carcinoma Patients. Cancer research. 2010 doi: 10.1158/0008-5472.CAN-10-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 2009;5:e1000578. doi: 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst. 2009;101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koelle DM, Corey L, Burke RL, Eisenberg RJ, Cohen GH, Pichyangkura R, Triezenberg SJ. Antigenic specificity of human CD4+ T cell clones recovered from recurrent genital HSV-2 lesions. Journal of Virology. 1994;68:2803–2810. doi: 10.1128/jvi.68.5.2803-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koelle DM, Chen H, Gavin MA, Wald A, Kwok WW, Corey L. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. Journal of Immunology. 2001;166:4049–4058. doi: 10.4049/jimmunol.166.6.4049. [DOI] [PubMed] [Google Scholar]
- 20.Yee C, Gilbert MJ, Riddell SR, Brichard VG, Fefer A, Thompson JA, Boon T, Greenberg PD. Isolation of tyrosinase-specific CD8+ and CD4+ T cell clones from the peripheral blood of melanoma patients following in vitro stimulation with recombinant vaccinia virus. J Immunol. 1996;157:4079–4086. [PubMed] [Google Scholar]
- 21.Hosken N, McGowan P, Meier A, Koelle DM, Sleath P, Wagener F, Elliott M, Grabstein K, Posavad C, Corey L. Diversity of the CD8+ T-cell response to herpes simplex virus type 2 proteins among persons with genital herpes. J Virol. 2006;80:5509–5515. doi: 10.1128/JVI.02659-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Posavad CM, Wald A, Hosken N, Huang ML, Koelle DM, Ashley RL, Corey L. T cell immunity to herpes simplex viruses in seronegative subjects: silent infection or acquired immunity? J Immunol. 2003;170:4380–4388. doi: 10.4049/jimmunol.170.8.4380. [DOI] [PubMed] [Google Scholar]
- 23.Jing L, Chong TM, McClurkan CL, Huang J, Story BT, Koelle DM. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J Immunol. 2005;175:7550–7559. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, Nalesnik MA, Kant JA, Weiss LM, Moore PS, Chang Y. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koelle DM, Posavad CM, Barnum GR, Johnson ML, Frank JM, Corey L. Clearance of HSV-2 from recurrent genital lesions correlates with infiltration of HSV-specific cytotoxic T lymphocytes. Journal of Clinical Investigation. 1998;101:1500–1508. doi: 10.1172/JCI1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi H. Identification of HLA-A24-Restricted CTL Epitope from Cancer-Testis Antigen, NY-ESO-1, and Induction of a Specific Antitumor Immune Response. Clinical Cancer Research. 2004;10:890–896. doi: 10.1158/1078-0432.ccr-1086-3. [DOI] [PubMed] [Google Scholar]
- 27.Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Invest Dermatol. 2009;129:246–248. doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eiz-Vesper B, Deluca DS, Blasczyk R, Horn PA. The nature of recombination in HLA-B*4207. Tissue Antigens. 2007;70:164–168. doi: 10.1111/j.1399-0039.2007.00873.x. [DOI] [PubMed] [Google Scholar]
- 29.Park JH, Adoro S, Lucas PJ, Sarafova SD, Alag AS, Doan LL, Erman B, Liu X, Ellmeier W, Bosselut R, Feigenbaum L, Singer A. ‘Coreceptor tuning’: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat Immunol. 2007;8:1049–1059. doi: 10.1038/ni1512. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, Lund O, Lundegaard C, Nielsen M, Ponomarenko J, Sette A, Zhu Z, Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Research. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weyand CM, Goronzy J, Fathman CG. Modulation of CD4 by antigenic activation. J Immunol. 1987;138:1351–1354. [PubMed] [Google Scholar]
- 32.Chen T, Hedman L, Mattila PS, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. Serological evidence of Merkel cell polyomavirus primary infections in childhood. J Clin Virol. 2010;50:125–129. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gheuens S, Bord E, Kesari S, Simpson DM, Gandhi RT, Clifford DB, Berger JR, Ngo L, Koralnik IJ. Role of CD4+ and CD8+ T-Cell Responses against JC Virus in the Outcome of Patients with Progressive Multifocal Leukoencephalopathy (PML) and PML with Immune Reconstitution Inflammatory Syndrome. Journal of Virology. 2011;85:7256–7263. doi: 10.1128/JVI.02506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneidawind D, Schmitt A, Wiesneth M, Mertens T, Bunjes D, Freund M, Schmitt M. Polyomavirus BK-specific CD8+ T cell responses in patients after allogeneic stem cell transplant. Leuk Lymphoma. 2010;51:1055–1062. doi: 10.3109/10428191003746323. [DOI] [PubMed] [Google Scholar]
- 36.Chan PKS, Liu SJ, Cheung JLK, Cheung TH, Yeo W, Chong P, Man S. T-cell response to human papillomavirus type 52 L1, E6, and E7 peptides in women with transient infection, cervical intraepithelial neoplasia, and invasive cancer. J Med Virol. 2011;83:1023–1030. doi: 10.1002/jmv.21889. [DOI] [PubMed] [Google Scholar]
- 37.Scott M, Nakagawa M, Moscicki AB. Cell-mediated immune response to human papillomavirus infection. Clin Diagn Lab Immunol. 2001;8:209–220. doi: 10.1128/CDLI.8.2.209-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Jong A, van Poelgeest MIE, van der Hulst JM, Drijfhout JW, Fleuren GJ, Melief CJM, Kenter G, Offringa R, van der Burg SH. Human papillomavirus type 16-positive cervical cancer is associated with impaired CD4+ T-cell immunity against early antigens E2 and E6. Cancer research. 2004;64:5449–5455. doi: 10.1158/0008-5472.CAN-04-0831. [DOI] [PubMed] [Google Scholar]
- 39.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 40.Ramaswami B, Popescu I, Macedo C, Metes D, Bueno M, Zeevi A, Shapiro R, Viscidi R, Randhawa PS. HLA-A01-, -A03-, and -A024-binding nanomeric epitopes in polyomavirus BK large T antigen. Hum Immunol. 2009;70:722–728. doi: 10.1016/j.humimm.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima MA, Marzocchetti A, Autissier P, Tompkins T, Chen Y, Gordon J, Clifford DB, Gandhi RT, Venna N, Berger JR, Koralnik IJ. Frequency and phenotype of JC virus-specific CD8+ T lymphocytes in the peripheral blood of patients with progressive multifocal leukoencephalopathy. J Virol. 2007;81:3361–3368. doi: 10.1128/JVI.01809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moretta A, Pantaleo G, Moretta L, Cerottini JC, Mingari MC. Direct demonstration of the clonogenic potential of every human peripheral blood T cell. Clonal analysis of HLA-DR expression and cytolytic activity. J Exp Med. 1983;157:743–754. doi: 10.1084/jem.157.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frebel H, Richter K, Oxenius A. How chronic viral infections impact on antigen-specific T-cell responses. Eur J Immunol. 2010;40:654–663. doi: 10.1002/eji.200940102. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Liu XS. Development and function of IL-10 IFN- -secreting CD4+ T cells. J Leukoc Biol. 2009;86:1305–1310. doi: 10.1189/jlb.0609406. [DOI] [PubMed] [Google Scholar]
- 45.Vence L, Palucka AK, Fay JW, Ito T, Liu YJ, Banchereau J, Ueno H. Circulating tumor antigen-specific regulatory T cells in patients with metastatic melanoma. Proc Natl Acad Sci USA. 2007;104:20884–20889. doi: 10.1073/pnas.0710557105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janetzki S, Price L, Britten CM, van der Burg SH, Caterini J, Currier JR, Ferrari G, Gouttefangeas C, Hayes P, Kaempgen E, Lennerz V, Nihlmark K, Souza V, Hoos A. Performance of serum-supplemented and serum-free media in IFNgamma Elispot Assays for human T cells. Cancer Immunol Immunother. 2010;59:609–618. doi: 10.1007/s00262-009-0788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, Yee C. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med. 2008;358:2698–2703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burch PA, Croghan GA, Gastineau DA, Jones LA, Kaur JS, Kylstra JW, Richardson RL, Valone FH, Vuk-Pavlović S. Immunotherapy (APC8015, Provenge) targeting prostatic acid phosphatase can induce durable remission of metastatic androgen-independent prostate cancer: a Phase 2 trial. Prostate. 2004;60:197–204. doi: 10.1002/pros.20040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.