Abstract
7-Dehydrocholesterol reductase (DHCR7) catalyzes the final step in cholesterol synthesis. The enzyme utilizes NADPH as a source of electrons and has been reported to require NADPH-cytochrome P450 reductase (POR) as its redox partner. To test this hypothesis, microsomes were prepared from the livers of mice in which hepatic cytochrome P450 reductase expression was extinguished during maturation. These microsomes contained negligible levels of POR but had 2.5-fold greater DHCR7 activity than did microsomes from wild-type mice. Consistent with this greater activity, immunoblot analysis of DHCR7 expression indicated that DHCR7 protein levels were elevated two-fold in POR-null microsomes. Addition of POR to these microsomes provided no stimulation of DHCR7 activity, confirming the lack of a role for POR in DHCR7 activity. Because the original observation that POR was necessary for DHCR7 activity was based, in part, on antibody inhibition studies with POR antibody, the ability of an antibody to the full-length POR protein to inhibit DHCR7 activity and cytochrome c reductase activity was tested; the antibody had no effect on DHCR7 activity but decreased cytochrome c reductase activity (a POR-catalyzed reaction) by 50%. Immunoblot analysis further demonstrated no cross-reactivity between POR and DHCR7 with antibodies to either protein. We conclude that cytochrome P450 reductase is not involved in 7-dehydrocholesterol reductase activity.
Keywords: 7-Dehydrocholesterol reductase, NADPH-cytochrome P450 reductase, cholesterol synthesis, Smith-Lemli-Opitz Syndrome, liver microsomes
1. Introduction
7-Dehydrocholesterol reductase (DHCR7) is a 54 kDa microsomal enzyme that catalyzes the final step in cholesterol synthesis, the reduction of the sterol C(7–8) double bond, using NADPH as an electron source. Mutations in the DHCR7 gene that impair or inactivate the enzyme result in Smith-Lemli-Opitz Syndrome (SLOS), a disease characterized by developmental and neurological deficits due to cholesterol deficiency [1]. Human and rat DHCR7 cDNAs were cloned in 1998 and 1999, respectively [2,3], and the enzymes were expressed in yeast and shown to be active in microsomal preparations. A subsequent study with rat liver microsomes [4] presented several lines of evidence that DHCR7 required NADPH-cytochrome P450 reductase (POR) for activity; this included a marked inhibition of DHCR7 activity in microsomes by addition of antibody to POR, as well as a reconstitution of DHCR7 activity by addition of purified POR to protease-treated microsomes and to partially purified DHCR7 preparations. Although NADPH-cytochrome P450 reductase is more widely recognized for its role in the reduction of cytochrome P450 in the oxidation of xenobiotics and steroids, POR is required for cholesterol synthesis, where it is the requisite electron donor to lanosterol demethylase (CYP51A1) [5] and the principal, but not exclusive, electron donor to squalene monooxygenase [6]. POR is ubiquitously expressed in mammalian cells and also in yeast.
DHCR7 has not yet been purified to homogeneity or expressed in a host lacking POR (e.g., E. coli), so the requirement for POR for activity has not been unequivocally established. Moreover, the absence of a recognizable NADPH binding domain in this protein, or the closely related reductase, Δ14-sterol reductase (TM7SF2) has further clouded the issue. The recent availability of hepatic POR-null mice [7] provided an opportunity to examine the requirement for this electron transfer protein in mammalian microsomes that lack this reductase. Our results argue strongly that POR is not required for DHCR7 activity.
2. Material and methods
7-Dehydrocholesterol reductase activity was measured by following the conversion of ergosterol to brassicasterol as described [8] in liver microsomes prepared from wild-type and hepatic POR-null mice [7]. Incubations (500 µl) contained 100 µg of microsomal protein and 60 µM ergosterol solubilized in 5% methyl-β-cyclodextrin (Sigma-Aldrich, St. Louis, MO) and were carried out for 60 min at 37°C. Lipids were saponified (1M ethanolic NaOH) for 1 h at 90°, extracted twice with hexane, derivatized, and identified and quantified by GC/MS at the UK Mass Spectrometry facility, focusing on mass ions 363 (ergosterol), 380 (brassicasterol), and 394 (stigmasterol, internal standard). PORmediated cytochrome c reductase activity was measured on an HP8453 UV/Vis spectrophotometer at 550 nm at room temperature. Sterols were obtained from the following suppliers: 7-dehydrocholesterol (Sigma-Aldrich); ergosterol, brassicasterol, 7α-hydroxycholesterol, and stigmasterol (Steraloids, Inc., Newport, RI). Cytochrome c was from Sigma-Aldrich; purified rat cytochrome P450 reductase was purchased from ENZO Life Sciences, Plymouth Meeting, PA.
Polyclonal antibodies to an N-terminal peptide (Santa Cruz Biotechnology, Santa Cruz, CA) or an internal segment (Sigma-Aldrich) of human 7DHCR, to full-length rat POR (ENZO Life Sciences), and a monoclonal antibody to β-actin (Sigma-Aldrich) were used to detect these proteins on 8% SDS-polyacrylamide gel immunoblots on nitrocellulose (Bio-Rad, Hercules, CA) using fluorescent-labeled secondary antibodies (GE Healthcare, Pittsburgh, PA). Image intensity was quantified on a GE Typhoon 9400 Imager.
3. Results
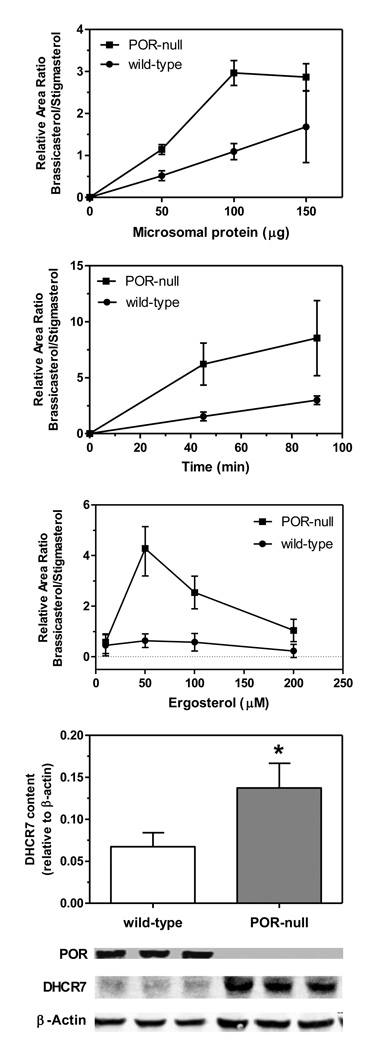
As shown in Fig. 1, 7-dehydrocholesterol reductase activity, measured as the conversion of ergosterol to brassicasterol [8], was prominent in microsomes prepared from the livers of mice in which cytochrome P450 reductase expression is extinguished during maturation [7]. This activity was consistently 2–3-fold greater than that found in microsomes from the corresponding control (wild-type) mice when evaluated on the basis of microsomal protein concentration, time-course, or substrate concentration. As noted by Shefer et al. [8], the substrate ergosterol was inhibitory to activity, with maximal activity at ~50 µM. DHCR7 protein content in microsomes correlated with activity, and was approximately two-fold higher in POR-null microsomes; POR protein was undetectable in these preparations.
Fig. 1.
Activity of DHCR7 in wild-type and hepatic POR-null mouse liver microsomes. Microsomal protein content (n=2), time (n=2), and substrate concentration (n=3) were varied and brassicasterol formation from ergosterol was monitored. Standard conditions were 100 µg of microsomal protein and 60 µM ergosterol for 60 min at 37°. DHCR7 protein content was measured by immunoblotting, with 20 µg of microsomal protein per lane, n=3. All data presented as mean ± range; *, significantly different from wild-type, p<0.05, Student’s t test.
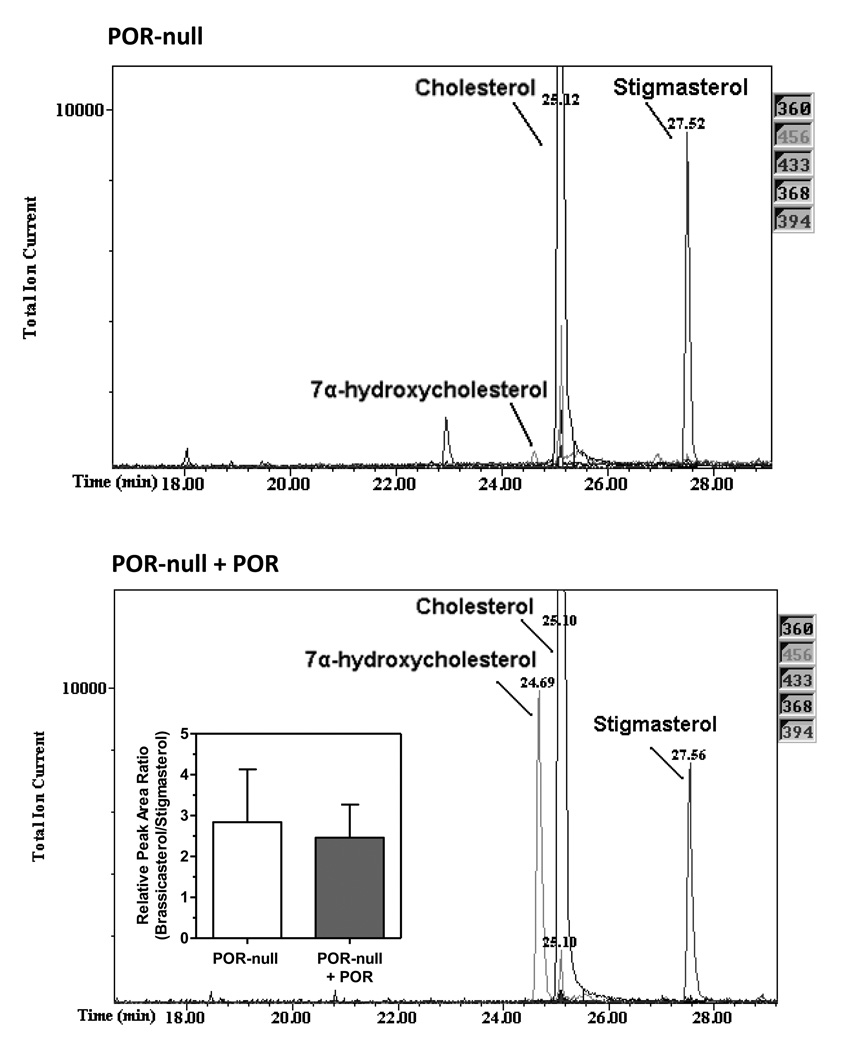
To determine if POR was stimulatory to DHCR7 activity, purified rat POR was added to POR-null microsomes and DHCR7 and cholesterol 7α-hydroxylase activity was monitored by GC/MS. As shown in Fig. 2, DHCR7 activity did not change in the presence of 50 nM POR (inset). This addition of POR restored cholesterol 7α-hydroxylase activity catalyzed by CYP7A1, a POR-dependent microsomal enzyme, indicating that the added reductase was functional in this system.
Fig. 2.
Addition of POR to POR-null microsomes does not stimulate DHCR7 activity. Two µg of purified rat POR (0.026 nmol) was added to POR-null microsomes (50 nM final concentration) and DHCR7 and cholesterol 7α-hydroxylase activity was measured. The inset bar graph shows DHCR7 activity (mean ± range, n=2); the chromatograms depict the 456 m/e− ion abundance eluting at 24.7 min characteristic for 7α-hydroxycholesterol in POR-null and POR-supplemented POR-null microsomes.
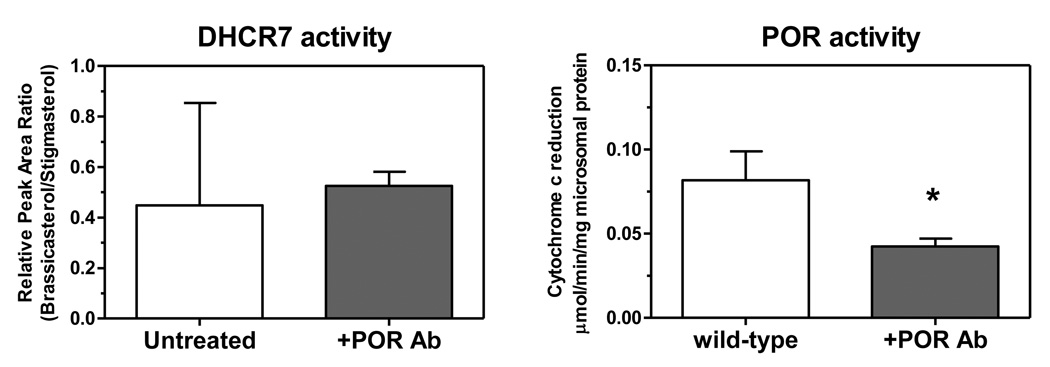
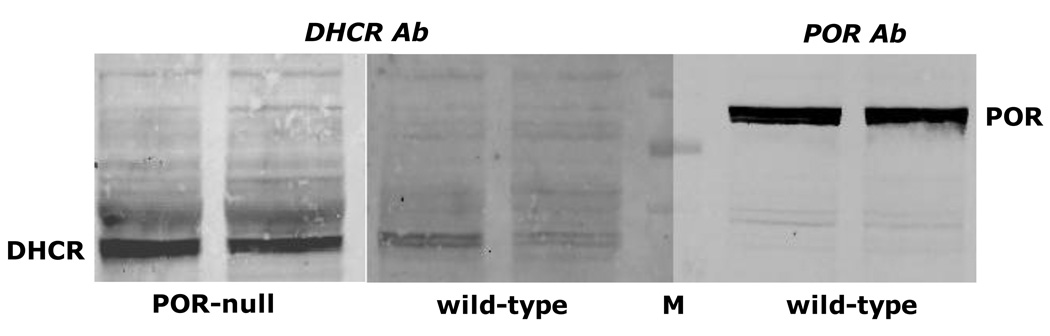
To address the possibility that antibody to POR is inhibitory to DHCR7 (as found by Nishino and Ishibashi [4]), a commercial polyclonal antibody to the full-length POR protein was added to wild-type microsomes and DHCR7 activity was measured. As shown in Fig. 3A, POR antibody had no effect on the conversion of ergosterol to brassicasterol. This antibody was effective in inhibiting a POR-catalyzed reaction, the reduction of cytochrome c, by about 50%, as shown in Fig. 3B. POR antibody did not cross-react with DHCR7, as evidenced by the immunoblot of Fig. 4, and polyclonal antibodies to an internal segment or the N-terminal segment (data not shown) of DHCR7 did not detect POR in wild-type microsomes, arguing that these two enzymes do not share common epitopes.
Fig. 3.
Antibody to POR inhibits POR but not DHCR7. Polyclonal antibody to POR was added to liver microsomes from wild-type mice (1 µg/µg microsomal protein) and DHCR7 activity (left) and cytochrome c reduction (right) was measured, mean ± range, n=2; *, p<0.05, Student’s t test.
Fig. 4.
Antibodies to DHCR7 and POR do not cross-react. Liver microsomes (20 µg of protein) from two wild-type and two hepatic POR-null mice were fractionated by SDS-polyacrylamide gel electrophoresis and electroblotted to nitrocellulose for immunodetection with polyclonal antibodies to an internal segment of DHCR7 or full-length POR. M, molecular weight markers at 95, 72, and 52 kDa.
4. Discussion
The present results argue strongly against a role for cytochrome P450 reductase in 7-dehydrocholesterol reductase activity. The two-fold increase in DHCR7 activity in liver microsomes from hepatic POR-null mice matched the two-fold increase in immunoreactive DHCR7 protein, even while POR protein levels were reduced below detectable levels. This increase in DHCR7 activity and protein is consistent with the 1.3 to 1.9-fold increase in DHCR7 gene expression that results from the loss of POR expression in this tissue [9,10]. The lack of increase in DHCR7 activity when purified POR was added to POR-null microsomes further argues that POR does not serve as an alternative or supplemental electron donor to DHCR7.
The selective suppression of cytochrome P450 reductase expression in mouse liver results in a number of metabolic changes in this tissue, most notably the loss of cytochrome P450-mediated drug metabolism [7,11]. Although some POR can be detected immunologically [6], in mature POR-null mouse liver it is less than 5% of wild-type levels and may be derived from nonparenchymal cells. In the microsomes used in these studies POR protein was not evident (Fig. 1) and no NADPH (POR)-dependent CYP2E1-catalyzed activity could be detected [12], making it unlikely that any residual cytochrome P450 reductase was responsible for the DHCR7 activity found here. Indeed, the loss of POR expression severely impacts the activity of all known POR-dependent enzymes [7,11].
The loss of POR expression also disables cholesterol and bile acid synthesis, as both pathways are dependent upon microsomal cytochromes P450. While this lowers plasma cholesterol and triglyceride levels it paradoxically increases hepatic triglyceride levels [7,11], perhaps due at least in part to the loss of bile acid-mediated stimulation of FXR [13]. Cholesterol synthesis is blocked at CYP51 (lanosterol demethylase), resulting in the accumulation of 24-dihydrolanosterol [6]. As might be expected, the loss of cholesterol synthesis modestly upregulates gene expression in the cholesterolgenic pathway [9,10], although enzyme protein and activity levels have not been systematically evaluated in these livers. Indeed, we noted [6] that HMG-CoA reductase and squalene monooxygenase enzyme levels were decreased by half in POR-null mouse livers, which we attributed to secondary post-translational down-regulation of these enzymes in response to elevated levels of 24-dihydrolanosterol; lanosterol and 24-dihydrolanosterol have been shown to enhance the degradation of HMG-CoA reductase [14,15] but not squalene monooxygenase [16]. In contrast to HMG-CoA reductase and squalene monooxygenase, 7-dehydrocholesterol reductase protein levels were increased several-fold in POR-null mouse liver in the present study (Fig. 1), consistent with the modest increase seen in mRNA level for this enzyme [9,10]. Because DHCR7 is downstream of lanosterol demethylase it is not surprising that it is not subject to post-translational suppression by accumulating 24-dihydrolanosterol in POR-null liver. It is clear from this and other recent studies [e.g., 15,16] that the post-translational regulation of cholesterol synthesis is considerably more complex than previously appreciated.
The study by Nishino and Ishibashi [4] noted that antibody to POR inhibited DHCR7 activity. We were unable to replicate this finding with a commercial polyclonal antibody to the full-length reductase, although it was effective in inhibiting a marker activity for POR, the reduction of cytochrome c. The antibody used by Nishino and Ishibashi was raised to the trypsin-released catalytic fragment of rat POR and was inhibitory to the rat POR and DHCR7 enzymes, whereas our antibody was raised to the complete rat enzyme but was tested against mouse DHCR7 and POR. It is possible that species differences may explain the different inhibitory results obtained, as their antibody showed greater effectiveness in inhibition of cytochrome c reductase activity than we obtained with our antibody with mouse liver microsomes. We considered the possibility that POR and DHCR7 share antigenic epitopes, and that inhibition of DHCR7 by antibody to POR was due to direct interaction with DHCR7. However, we could find no evidence of cross-reaction between the POR antibody and DHCR7 protein in microsomes (Fig. 4). In addition, antibody to DHCR7 did not cross react with microsomal POR protein. It remains possible that other antibodies might recognize epitopes shared by these two proteins, and might account for cross inhibition of activity. It also must be considered that the antibody used by Nishino and Ishibashi recognized both proteins due to contamination of the trypsin-released POR antigen with trypsin-released DHCR7 protein.
We are unable to provide an explanation for why partially purified fractions of DHCR7 required the addition of purified, detergent-solubilized POR for activity in the study by Nishino and Ishibashi, but would note that the DHCR7 assay used by this group followed 7-dehydrocholesterol disappearance by monitoring absorbance at 280 nm by HPLC, whereas we directly monitored brassicasterol formation from ergosterol by GC/MS. Substrate disappearance is generally considered a less reliable means to measure enzyme activity, and does not exclude other mechanisms by which the substrate might be removed. We attempted to measure 7-dehydrocholesterol disappearance by GC/MS in wild-type and POR-null microsomes but were unable to detect significant changes in substrate levels by this semi-quantitative technique; similarly, it is not possible to measure cholesterol formation due to the high levels of cholesterol in microsomes. It remains formally possible that DHCR7 uses different redox mechanisms (e.g., direct binding of NADPH vs electron transfer from POR) for different substrates (i.e., 7-dehydrocholesterol vs ergosterol), but we are not aware of any oxidoreductases that discriminate redox sources by substrate. Ultimately this issue can best be resolved by reconstitution studies with the purified native or recombinant DHCR7 enzyme in a defined lipid milieu.
RESEARCH HIGHLIGHTS.
-
✓
7-Dehydrocholesterol reductase does not require cytochrome P450 reductase for activity
-
✓
Cytochrome P450 reductase does not stimulate nor augment 7-dehydrocholesterol reductase activity
-
✓
7-dehydrocholesterol reductase expression is elevated in microsomes from livers that lack cytochrome P450 reductase
Acknowledgment
We thank Drs. Xinxin Ding and Jun Gu for providing the liver tissue used in these studies; generation and maintenance of this mouse line was supported in part by NIH Grant ES-07462. Ms. Lauren Smith carried out the initial studies, and Dr. Jack Goodman provided advice and guidance on the mass spectrometric analysis.
Abbreviations
- DHCR7
7-dehydrocholesterol reductase
- POR
NADPH-cytochrome P450 oxidoreductase
- GC/MS
gas chromatography/mass spectrometry
- TM7SF2
Δ14-sterol reductase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Porter FD. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet. 2008;16:535–541. doi: 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- 2.Moebius FF, Fitzky BU, Lee JN, Paik YK, Glossmann H. Molecular cloning and expression of the human delta7-sterol reductase. Proc Natl Acad Sci U S A. 1998;95:1899–1902. doi: 10.1073/pnas.95.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae SH, Lee JN, Fitzky BU, Seong J, Paik YK. Cholesterol biosynthesis from lanosterol. Molecular cloning, tissue distribution, expression, chromosomal localization, and regulation of rat 7-dehydrocholesterol reductase, a Smith-Lemli-Opitz syndrome-related protein. J Biol Chem. 1999;274:14624–14631. doi: 10.1074/jbc.274.21.14624. [DOI] [PubMed] [Google Scholar]
- 4.Nishino H, Ishibashi T. Evidence for requirement of NADPH-cytochrome P450 oxidoreductase in the microsomal NADPH-sterol Delta7-reductase system. Arch Biochem Biophys. 2000;374:293–298. doi: 10.1006/abbi.1999.1602. [DOI] [PubMed] [Google Scholar]
- 5.Trzaskos J, Kawata S, Gaylor JL. Microsomal enzymes of cholesterol biosynthesis. Purification of lanosterol 14 alpha-methyl demethylase cytochrome P-450 from hepatic microsomes. J Biol Chem. 1986;261:14651–14657. [PubMed] [Google Scholar]
- 6.Li L, Porter TD. Hepatic cytochrome P450 reductase-null mice reveal a second microsomal reductase for squalene monooxygenase. Arch Biochem Biophys. 2007;461:76–84. doi: 10.1016/j.abb.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, Ding X. Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem. 2003;278:25895–25901. doi: 10.1074/jbc.M303125200. [DOI] [PubMed] [Google Scholar]
- 8.Shefer S, Salen G, Honda A, Batta AK, Nguyen LB, Tint GS, Ioannou YA, Desnick R. Regulation of rat hepatic 3beta-hydroxysterol delta7-reductase: substrate specificity, competitive and non-competitive inhibition, and phosphorylation/dephosphorylation. J Lipid Res. 1998;39:2471–2476. [PubMed] [Google Scholar]
- 9.Wang XJ, Chamberlain M, Vassieva O, Henderson CJ, Wolf CR. Relationship between hepatic phenotype and changes in gene expression in cytochrome P450 reductase (POR) null mice. Biochem J. 2005;388:857–867. doi: 10.1042/BJ20042087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng Y, DiRusso CC, Reilly AA, Black PN, Ding X. Hepatic gene expression changes in mouse models with liver-specific deletion or global suppression of the NADPH-cytochrome P450 reductase gene. Mechanistic implications for the regulation of microsomal cytochrome P450 and the fatty liver phenotype. J Biol Chem. 2005;280:31686–31698. doi: 10.1074/jbc.M504447200. [DOI] [PubMed] [Google Scholar]
- 11.Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem. 2003;278:13480–13486. doi: 10.1074/jbc.M212087200. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Porter TD. Chlorzoxazone hydroxylation in microsomes and hepatocytes from cytochrome P450 oxidoreductase-null mice. J Biochem Mol Toxicol. 2009;23:357–363. doi: 10.1002/jbt.20299. [DOI] [PubMed] [Google Scholar]
- 13.Porter TD, Banerjee S, Stolarczyk EI, Zou L. Suppression of Cytochrome P450 Reductase (POR) Expression in Hepatoma Cells Replicates the Hepatic Lipidosis Observed in Hepatic POR-Null Mice. Drug Metab Dispos. 2011;39:966–973. doi: 10.1124/dmd.111.038562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange Y, Ory DS, Ye J, Lanier MH, Hsu FF, Steck TL. Effectors of rapid homeostatic responses of endoplasmic reticulum cholesterol and 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 2008;283:1445–1455. doi: 10.1074/jbc.M706967200. [DOI] [PubMed] [Google Scholar]
- 15.Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Gill S, Stevenson J, Kristiana I, Brown AJ. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13:260–273. doi: 10.1016/j.cmet.2011.01.015. [DOI] [PubMed] [Google Scholar]