Abstract
Although the rhesus macaque (Macaca mulatta) is commonly used for biomedical research and becoming a preferred model for translational medicine, quantification of genome-wide variation has been slow to follow the publication of the genome in 2007. Here we report the properties of 4040 single nucleotide polymorphisms discovered and validated in Chinese and Indian rhesus macaques from captive breeding colonies in the United States. Frequency-matched measures of linkage disequilibrium were much greater in the Indian sample. Although the majority of polymorphisms were shared between the two populations, rare alleles were over twice as common in the Chinese sample. Indian rhesus had higher rates of heterozygosity, as well as previously undetected substructure, potentially due to admixture from Burma in wild populations and demographic events post-captivity.
Keywords: Macaca mulatta, nonhuman primate, SNP, Linkage Disequilibrium
1. Introduction
Genomic characterization of the rhesus macaque (Macaca mulatta) has been underway since a complete genome sequence was published in 2007 [1], and is now progressing to a massively parallel scale. The National Center for Research Resources (NCRR) at the NIH currently sponsors six rhesus macaque Working Groups (WGs), one of which, the Genetics and Genomics WG, has assumed the stated goal of instituting uniform SNP-based genetic characterization protocols for parentage testing, ancestry determination and population genetic assessments [2]. In addition, several research groups are in the process of quantifying genomic variation in captive populations of rhesus macaques [3–5]. The data presented here were collected as part of a larger project to discover, quantify and validate single nucleotide polymorphisms (SNPs) in regional populations of rhesus macaques included in captive breeding populations in the US.
The rhesus macaque is an underused model in biomedicine [6], given that most captive colonies maintain extended multi-generational pedigrees, collect detailed phenotypic data and curate extensive veterinary records, making it suitable for many studies requiring both familial data and/or quantified phenotypic or genetic variance and a complex mammalian system. Approximately six million mice are involved in biomedical research in the United States [7] while the number of nonhuman primates is less than 1/100th of that number [8]. Rhesus macaques specifically comprise even less than that proportion, although they are the most widely used non-human primate model for biomedical research.
Rhesus macaques have been shown to be an effective and adaptable research model, with applications for multi-factorial diseases, including endometriosis, type 2 diabetes and asthma [6] as well as for determining the genetic basis of behavior and gene by environment interactions [9]. Rhesus macaques have demonstrated success as a translational model, especially in testing delivery methods for gene therapies, for example, Duchenne muscular dystrophy [10], artherosclerosis [11], muscular degeneration [12] and L-Dopa-induced dyskinesia [13]. In addition, recent translational studies have demonstrated the efficacy of microbicide cells containing anti-retroviral drugs in mucosally challenged macaques both vaginally [14] and rectally [15], demonstrating the impact of macaque research on the development of cost-effective strategies to prevent HIV transmission in humans.
The most common area of research utilizing rhesus macaques is microbiology, including HIV/AIDS [16]. Rhesus macaques have been heavily used in HIV drug and vaccine initiatives, especially the STEP vaccine trial, which used a non-replicating recombinant adenovirus 5 (rAD5) vector to stimulate T-cells. Phase 2b trials sponsored by Merck and the National Institutes of Health (NIH) were terminated when it was determined that the vaccine did not provide protection against infection, nor did it reduce viral loads post-infection [17]. Haigwood [18] noted that even in the non-human primate trials, viral load was reduced by several orders of magnitude in animals infected with the chimeric human/simian virus SHIV-89.6P but not when infected with the more virulent SIVmac239 and the progression to human trials was overly optimistic. The difference in immune response underlines the point that successful application of animal models will only come from a more sophisticated understanding of host genetics. Unlike Drosophila, C. elegans or the mouse models, there is no central curated resource for genomic and phenotypic information (flybase.org, wormbase.org and www.informatics.jax.org, respectively) on the rhesus macaque model. The development of such a resource would encourage collaboration between biomedical and genetic research, allowing greater integration of phenotypic, immunological and genomic information.
In this study we focus on curating genomic information and report SNP discovery and validation in rhesus macaques. As we develop rhesus macaques as a research resource we will expand available information to include identification and quantification of copy number variants, location of population-specific genomic rearrangements, and other genome-level factors known to influence phenotype. The greater availability of these data will make rhesus macaques an even more attractive research model for genetic epidemiology, multi-factorial disease and translational medicine.
2. Methods
Our method of SNP discovery is described in detail in Malhi, et al. [19]. A DNA sample from a female rhesus macaque of western Chinese origin was submitted to 454 Life Sciences (Roche Diagnostics, Branford, CT) for large-scale parallel pyrosequencing, producing a total of 339,967 reads with an average read length of 104 bp. The reads were aligned against the published rhesus genome version 1.1 [1], known to be derived from an Indian-origin animal. This alignment identified approximately 23,000 prospective polymorphisms.
Malhi et al. [19] described the discovery of approximately 23,000 candidate SNPs distributed throughout the rhesus macaque genome. Our goal was to select and validate markers distributed approximately 1 megabase apart from this pool of candidates. However, the median distance between adjacent candidate polymorphisms is only 65 kilobases (mean=125 kilobases ±223 kb), indicating that the majority of candidate markers were far too close together for the construction of an equidistantly spaced SNP map and thus the actual number of suitable markers for such a map was much smaller than 23,000. Accordingly, 8342 of these candidate SNPs were selected for validation by identifying the most proximal polymorphism on each chromosome and polymorphisms spaced approximately 1 megabase apart across the entire sequence. When probes for the polymorphisms were not designable on the Illumina GoldenGate™ platform, failed to amplify during the genotyping reaction or did not show any segregating polymorphisms in the genotyped individuals, the nearest verifiable polymorphism, either upstream or downstream, was included instead.
Quality-screening of the candidate SNPs is described in Satkoski et al. [4]. Polymorphic locations with pyrofragment Phred scores less than 20 and only a single overlapping fragment were discarded. For the remaining putative polymorphisms, the chromosome and nucleotide position of each fragment containing a candidate SNP within the rhesus genome was confirmed with the genome BLAST [20] function of the National Center for Biotechnology Information website (NCBI, www.ncbi.nlm.nih.gov). Fragments that were confirmed as single copy and produced a high-quality (+98%) match to the rhesus genome were selected for further analysis. Fifty-two of the 8342 candidate markers selected for validation produced no BLAST matches and 3494 produced multiple BLAST hits, suggesting that the sequence flanking the polymorphism is repetitive or exists in multiple copies, leaving 4796 SNPs for validation.
We employed Illumina (San Diego, CA) GoldenGate technology to genotype the resulting candidate markers. Of the candidates, 125 could not be incorporated into the Illumina oligo pool (OPA), resulting in 4671 markers submitted for validation. These markers were genotyped on both the BeadXPress (with one 96-plex OPA and four 384-plex OPAs) and the iScan platforms (with two 1536-plex OPAs). The individuals selected for genotyping were, to the best of our knowledge, not first or second degree relatives; sample information is shown in Table 1. These animals were either imported directly from the country of origin (VBS, TSS) or had sufficient colony documentation to support their assignment to the appropriate region (CPRC, UM, ONPRC and CNPRC). In addition, all individuals had been sequenced for a 830-bp section of the mitochondrial genome [21] and 24 nuclear microsatellite loci [22], used to confirm their assignment to a specific geographic region (partial data presented in Satkoski Trask et al. [23], additional data not shown). Approximately 5% of the markers and at least two individuals were chosen at random and duplicated across each OPA and each run as controls. Of the genotyped markers, 365 did not meet the minimum quality (GenTrain) score of 0.4 and were excluded from further analysis. An additional 266 markers exceeded the maximum 5% missing genotype criterion and were also excluded, leaving a total of 4040 validated SNPs.
Table 1.
Geographic origin and haplotype of the Indian and Chinese samples.
| India | |||||
|---|---|---|---|---|---|
| Geographic Origin | mtDNA haplotype | Sample Size | Source | Sex Ratio | |
| Uttar Pradesh, Central | Ind1 | 20 | CPRC | M=16% F=24% U=36% |
|
| Kashmir | Ind1 | 2 | UM | ||
| Central India | Ind2 | 3 | ONPRC | ||
| China | |||||
| Guangdong | ChiE | 1 | VBS | M=24% F=56% U=20% |
|
| Sichuan | ChiW | 11 | VBS, TSS | ||
| Suzhou | ChiW (N=4) ChiE (N=8) |
12 | CNPRC | ||
| Unknown | ChiS | 1 | COV |
CPRC, Caribbean Primate Research Center; UM, University of Miami; ONPRC, Oregon National Primate Research Center; VBS, Valley Biosystems; TSS, Three Springs Scientific; CNPRC, California National Primate Research Center; COV, Covance Inc., Alice TX.
Minor allele frequencies (MAF) and observed heterozygosities were calculated with PLINK 1.06 (http://pngu.mgh.harvard.edu/purcell/plink, [24]). Principal component analysis (PCA) was performed with the adegenet 1.2–8 package for R [25] to identify genetic structure within the data independently of a priori assignment to a particular geographic origin or breeding center. To identify which markers were located within genes, SNPs were localized relative to known genes using the RefSeq Genes track for the MGSC Merged 1.0/rheMac2 assembly of the rhesus macaque genome in the UCSC Genome Browser (http://genome.ucsc.edu [26, 27]) through the Galaxy web interface [28, 29].
Once validated, information on each polymorphism was submitted to the dbSNP online database (http://www.ncbi.nlm.nih.gov/projects/SNP). Information on each SNP described herein, including chromosome and nucleotide position, dbSNP ss#, flanking sequence and MAF values from the Indian and Chinese samples can be found at http://primate.bioinformatics.ucdavis.edu, a custom UCSC Genome Browser instance.
Linkage disequilibrium (LD) was measured as r2, the correlation coefficient between the allele frequencies of the two markers [30], and calculated with Haploview [31]. Only pairwise LD calculations with non-zero T-int values were considered; T-int is a statistic used by the HapMap project that measures the completeness of information provided by a set of markers in a genomic region [31]. For both the Indian and Chinese samples, markers were sorted into MAF bins of 0.1, 0.2, 0.3, 0.4 and 0.5. The r2 values were filtered to include only markers in the same
MAF bin to create frequency-matched pairs [32]. By comparing the positions of the markers in the frequency-matched pairs to the MGSC Merged 1.0/rheMac2 version of the rhesus genome on the UCSC genome browser (http://genome.ucsc.edu), it was determined that all were located outside of known genes. Hernandez et al. [33] used SNPs located in ENCODE regions to estimate that LD in rhesus macaques decayed completely by 50 kb; we considered all frequency-matched pairs regardless of distance to ensure that we captured the point at which LD reached zero.
3. Results
Of the 8342 markers chosen for validation, 4040 (40%) met all the QC and genotype standards. These SNPs are distributed throughout the genome, located on all 20 autosomes and the X chromosome. We have validated one SNP approximately every 723 kilobases (±142 kb) on the autosomes, and one SNP approximately every 1.9 megabases on the X chromosome. No SNPs were discovered on the Y chromosome due to the lack of a published Y chromosome sequence. The number and spacing of SNPs on each chromosome is shown in Table 1. Of the 4040 validated SNPs, 80 (2%) were located in 76 different genes.
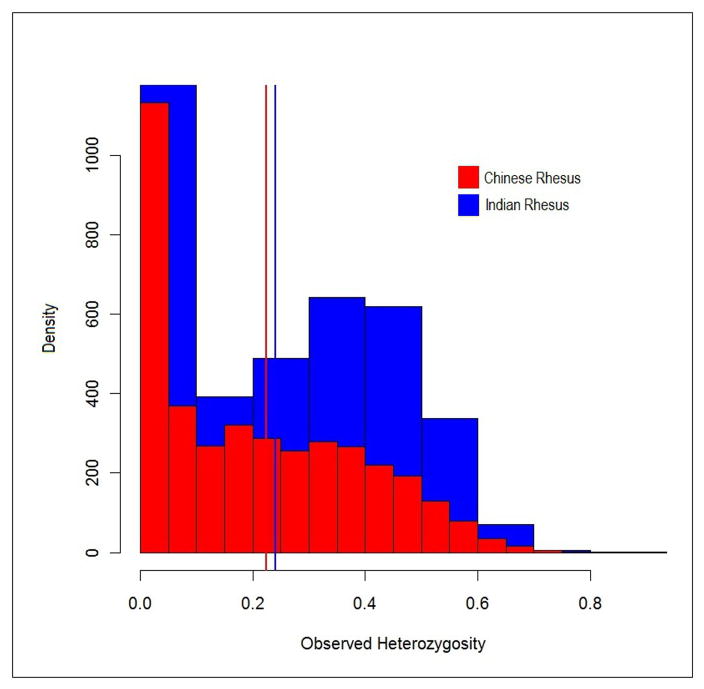
Of the 4040 markers, 2760 (68%) were polymorphic in both the Indian and Chinese samples. Thirteen percent of the markers were polymorphic only in the Chinese sample, compared to 19% of the markers polymorphic only in the Indian sample. The MAF distributions for both the Indian and Chinese samples are shown in Figure 1. The average MAF for the Indian sample was 0.19, compared with 0.17 in the Chinese sample. This difference is statistically significant (p<0.000001, two-sample t-Test, unequal variances). Not only are the average MAF value and proportion of population-specific SNPs higher in the Indian sample, but, as shown in Figure 1, many more markers are of high frequency in the Indian sample with a corresponding low frequency in the Chinese sample than the reverse. Of the 3023 markers polymorphic in Indian animals, 308 (10.2%) were below 5% minor allele frequency, compared with 767 (23.0%) of the 3336 markers polymorphic in Chinese individuals. Of the markers polymorphic in both populations, only 65 (2.4%) had a minor allele frequency below 5%. Differences in observed heterozygosity are shown in Figure 2. The average heterozygosity for the Chinese sample was 0.22, while the average heterozygosity in the Indian sample was 0.24. This difference was statistically significant (p<0.0001, two-sample t-Test for equal variances).
Figure 1.
Distribution of minor allele frequencies in the Chinese and Indian samples. Mean minor allele frequency is significantly different (p<0.01).
Figure 2.
Histograms of observed heterozygosity in the Chinese and Indian samples. The mean heterozygosities for the two samples are statistically significantly different (p<0.01).
The r2 values for the two samples are shown plotted against distance in Figure 3. Six hundred and twenty-five marker pairs met the stated criteria in the Indian sample, compared with 724 marker pairs in the Chinese sample. We fit logarithmic, exponential and linear models to the data and found that the logarithmic model had the greatest predictive power. Using the appropriate equation for each population, we calculated the slope and x intercept of the logarithmic regression line, which allowed us to estimate the value of r2 at 10 kb (x=10) as well as the point at which LD dissipates completely (x intercept). For the Indian sample, r2 at 10 kb was 0.54, compared to 0.30 for the Chinese sample. LD was predicted to reach 0 at 582.55 kb and 1.81 Mb, respectively.
Figure 3.
The decay of linkage disequalibrium in the Indian and Chinese samples. R2 values are calculated for frequency-matched markers located on the same chromosome. The x intercept of each line marks the complete disintegration of linkage disequilibrium.
The results of the PCA analyses are illustrated in Figures 4 and 5. PC1 represents 24.7% of the total sample variance, and PC2 and 3 represent 3.2% and 2.9%, respectively. As shown in Figure 4, the Chinese and Indian sample sorted cleanly, with the exception of individual 22375. This individual was classified as Indian and determined to have the Ind2 mitochondrial haplotype that is relatively rare (5%) in Indian rhesus macaques yet fixed in Burmese [21] and Bangladesh [34] rhesus macaques. Given that there appears to be structure in the Indian sample along the PC2 axis, we plotted PC2 against PC3 in Figure 5. These individuals form at least three, possibly four, distinct clusters unrelated to mitochondrial haplotype or geography: one cluster consisting solely of Ind1 animals from the CPRC, one cluster containing CPRC animals and Ind2 individual 22375 and one containing both Ind1 and Ind2 animals from the other sample sources.
Figure 4.
Principal component analysis of Indian and Chinese rhesus macaques. Principal component (PC) 1 separates Indian from Chinese individuals with the exception of sample Orhs22375.
Figure 5.
Differentiation within the Indian samples shown by PC2 and PC3 in the principal component analysis.
4. Discussion
Of the SNPs chosen for validation, 42% were rejected due to their location within duplicated or repetitive regions, as reported by a BLAST search against the rhesus macaque reference genome. The human genome is composed of approximately 50% repetitive sequences [35], and the amount of repetitive DNA in the rhesus macaque is comparable [1] although the proportion of this sequence attributable to segmental duplication (2.3%) is substantially lower than the comparable values for humans and chimpanzees. Therefore, the percentage of markers rejected due to their location in a repetitive or duplicated region is reasonable. This high rate of failure for marker validation seems to be an innate quality of the primate genome [35] rather than an issue with the validation method, and should be taken into account when researchers seek to estimate the number of usable markers resulting from any SNP discovery effort.
Previously undetected flanking polymorphisms in the Indian and Chinese samples or repetitive DNA sequence not reflected in the published genome are potential explanations for the 365 markers (an additional 4% of the markers originally selected for validation) that failed to meet the GenTrain quality threshold during Illumina GoldenGate genotyping. A much smaller proportion of markers chosen for validation (1%) failed during the probe-design process. A probe design failure (as opposed to a polymorphic site that is designable but flagged with a warning) is likely due to low sequence complexity in the flanking region (Illumina Technical Note: Designing Custom GoldenGate® Assays). These failures highlight the importance of genomic completeness and adequate genome annotation during SNP map construction. The available annotation of the rhesus macaque genome has not been updated since its release in 2006 [36]. Although, as demonstrated here, it is possible to identify sequence regions inappropriate for SNP genotyping through genotype failure or low-quality scores, bioinformatic screening of markers prior to wet lab validation is much faster and massively more cost effective.
The largest collection of rhesus macaque SNPs was previously published by Hernandez et al. [33]. Their sample included nine individuals from China (seven from Suzhou, one from Kunming and one from Guangdong) and thirty-eight individuals of Indian origin, the ancestry of most of which could not be attributed to a specific geographic region. Hernandez et al. [33] sequenced five ENCODE regions in 166 non-overlapping windows, resulting in the discovery of 1,476 SNPs. In contrast, our study includes 50 individuals, with Chinese and Indian individuals equally represented. Simulations of LD using simulated data have suggested that sufficient sample size is an important consideration when designing a study of linkage disequilibrium, since insufficient population representation can fail to capture recombination events and overestimate haplotype block size [37]. Fifty individuals (or 100 chromosomes) are generally considered to be the minimum viable sample size for estimating LD around common alleles, a number confirmed by studies of LD in human populations [38].
Hernandez et al. [33] did not report the individual heterozygosity or MAF values of their reported markers but noted that very few of the SNPs were shared across populations. Only 33% of their markers, compared to 68% in the present study, were shared between both populations, while 61% and 39% were found only in the Chinese and Indian samples, respectively, compared to 13% and 19% in our study. A comparison with the baboon genome (to infer the ancestral state) determined that the Chinese population contained an excess of rare markers under the assumption of constant population size, while the Indian sample contained many SNPs of intermediate and high frequency. A much greater proportion of the markers examined in our study were polymorphic in both the Indian and Chinese samples, and the proportion of markers that were population specific much smaller. This difference is probably largely due to a bias towards the discovery of high MAF, shared SNPs inherent in our discovery method [23]: the comparison of just two individuals, one Indian and one Chinese, led to preferential discovery of high frequency polymorphisms shared between the two populations. Low frequency polymorphisms had a lower probability of discovery, while discovery of SNPs polymorphic only in the Indian or Chinese populations required the sequenced individual to be a heterozygote. This phenomenon is probably exacerbated by the fact that the Indian genome used for alignment represented only a single strand and thus, could not be heterozygotic. Another factor that potentially contributes to this difference is the larger, more geographically variable composition of our Chinese sample. A third likely explanation is the difference in marker location: while the markers described in Hernandez et al.’s study were located entirely in ENCODE regions, only 2% of the markers in our study are located in coding regions and probably less subject to the action of selection on the Indian and Chinese rhesus macaque populations. One result of this study did reproduce the results of Hernandez et al., specifically, the paucity of intermediate and high-frequency alleles in China, relative to the Indian sample. As illustrated in Figure 1, while markers with low MAF in the Indian sample also tended to have a low MAF in the Chinese sample, the converse was not true. Rare alleles (MAF equal to or less than 5%) were over twice as common in the Chinese sample, relative to the Indian sample. This pattern is consistent with a genetic bottleneck in Indian (but not Chinese) rhesus macaques that eliminated some of the rare alleles in their common ancestors, as speculated by Hernandez et al. [33].
As shown in Figure 3, and in agreement with Hernandez et al. [33], linkage disequilibrium dissipates much more rapidly in Indian rhesus macaques compared to their Chinese counterparts. The linkage distance in the Chinese sample was over twice that of the Indian sample, although both were much greater than predicted by Hernandez et al. [33], making it difficult to assess the impact of admixture of the Indian sample with rhesus macaques dispersing from the east carrying the Ind2 mtDNA haplogroup. The LD values reported by Hernandez et al. [33], r2 at 10 kb of 0.15 and 0.52 for Chinese and Indian rhesus macaques, respectively, were somewhat different from those estimated in the present study (r2 at 10 kb of 0.30 and 0.54, respectively), although more so for the Chinese animals. This difference may be due to the fact that only nine Chinese individuals were included in the former study, compared to 38 Indian individuals, raising the possibility that Chinese recombinants were identified at a lower rate. Also probably contributing to the difference is the fact that LD in the Hernandez et al. [33] study was calculated from SNPs in ENCODE regions, while the frequency-matched LD values presented here eliminated all comparisons of markers located within the same gene and contained only LD measurements from SNPs in non-coding regions.
Consistent between the two studies was the substantial difference between LD in the Indian and Chinese rhesus macaque samples. The longer linkage distance observed in Indian rhesus could potentially be due to nuclear genetic admixture with Burmese rhesus, via the Bramaputra River [21]. Additionally, a selective sweep, or local reduction in genetic variation, can be caused by the rapid fixation of a beneficial mutation, resulting in high LD around the site of this mutation [39]. If the selective sweep happened quickly, local variation will diminish to zero, followed by the re-accumulation of variation through novel mutation and recombination, leading to an overabundance of rare alleles. In human populations, researchers have identified incomplete selective sweeps around the genes for lactase (LCT) and glucose-6-phosphate dehydrogenase (G6PD), displaying haplotypes that appear to be selection, but have not yet reached 100% frequency. This process produces a pattern of locally identical haplotypes segregating at high frequencies, with the other haplotypes displaying normal variability [40]. Preliminary analysis of LD in geographically and phenotypically variable rhesus macaques have identified regions potentially under selection in Indian, but not Chinese populations, which could contribute to the large difference in linkage distance [41]. Although the measurements of LD presented here and in Hernandez et al. [33] provide insight into the differing evolutionary histories of the Indian and Chinese rhesus populations, a true linkage map will not be possible until these SNPs can be genotyped in extended families [42].
Although the differentiation between the Indian and Chinese samples was quite strong and consistent with previously reported results [4, 23], two results unique to the present study are the greater heterozygosity in Indian than in Chinese rhesus macaques and the presence of population structure of unknown source in the Indian sample (Figures 4 and 5). Previous studies of microsatellite (STR) markers [43], mtDNA sequence [22] and SNPs in the 3’ ends of rhesus macaque genes [3] and ENCODE regions [33] have reported higher levels of heterozygosity in Chinese than in Indian rhesus macaques. While the majority of the Indian-origin animals sampled by Hernandez et al. [33] came from the Yerkes National Primate Research Center, most of the Indian-origin animals in the present study were selected from the Caribbean Primate Research Center (CPRC). Figure 4 shows that the individuals form several distinct clusters along PC2, and this continuum is also visible along PC3, with the CPRC individuals forming two groups, exclusive of the Indian-origin animals from the University of Miami (UM) and the Oregon National Primate Research Center (ONPRC). In contrast, the individuals from the latter two centers are far less differentiated than the individuals in the UM and ONPRC colonies. While individual 22375 appeared to be of Chinese origin, it clusters with other individuals of Indian origin in Figure 4, consistent with the hypothesis that the IND2 haplotype originated in Burma with these individuals serving as a source of novel alleles in the Indian population. We have previously reported the presence of a historic signal of Chinese admixture among individuals of haplotype IND2 from ONPRC [44] using a smaller set of 829 SNPs.
The early history of the CPRC colony is described in detail by Carpenter [45], Buettner-Janusch et al. [46] and Johnsen [47]. The population was initiated in 1938/1939 with the release of 409 individuals, 14 gibbons and three Macaca nemestrina on the island of Cayo Santiago, off the southeastern coast of Puerto Rico. The population in March of 1940 was approximately 350 animals, but this had dropped to 150 prior to 1956 but grew to 791 by 1968. In 1970, when the colony on Cayo Santiago became part of the CPRC, the population was reduced to 333 animals. A genetic study of the transferrin locus by Buettner-Janusch et al. [46] conducted both before and after the 1970 population reduction found that although the allele frequencies had not changed significantly, the total number of transferrin phenotypes had fallen from 15 to 12. Olivier et al. [48] confirmed this result with low Fst values among social groups calculated from the serum protein transferrin and the isozymes carbonic anhydrase II and 6-phosphogluconate dehydrogenase. In contrast, Duggleby [49] found that the red blood cell phenotypes I, J, K, L, P and Q were significantly heterogeneous over social groups. These early protein polymorphism studies suggest, as do these results, that although the complicated demographic history of the CPRC has not significantly impacted variation of selectively important loci, the impact on neutral loci, or loci under weak selection, could be profound. Although Indian rhesus within US breeding colonies are generally considered to be far more genetically homogeneous than their Chinese counterparts (who are often recently imported or the offspring of imported animals), the results of our study call this into question and demonstrate cryptic population structure in Indian rhesus macaques; the importance of this structure on the phenotypic and immunological variance within the US captive Indian-origin rhesus population is difficult to gauge at this point.
4. Conclusions
Forty-two percent of SNPs selected for validation were rejected due to their location in a duplicated or repetitive region, confirming estimates that the amount of repetitive DNA in the rhesus macaque genome is comparable to that in humans. Unlike previously published reports of SNP variability in rhesus macaques [33], the majority of polymorphisms were shared between the Indian and Chinese samples. Rare alleles were over twice as common in Chinese rhesus. Linkage disequilibrium was much stronger in the Indian sample relative to the Chinese sample, potentially due to a combination of admixture with rhesus macaques from Burma and differential selection on Indian populations. Although a paucity of rare alleles in the Indian sample is consistent with the hypothesis of a bottleneck in this population, high heterozygosity and the presence of previously undetected substructure indicates that Indian-origin rhesus macaques in US breeding centers may contain cryptic genetic variation. The results of this study underline the importance of quantifying genomic variation present in biomedical research models, especially as the rhesus macaque increases in popularity as a translational model.
Supplementary Material
Table 2.
The number of markers on each chromosome and the average distance between markers.
| Rhesus Chromosome | Number of SNPs | Mean Gap (Kb) | Median Gap (Kb) |
|---|---|---|---|
| 1 | 344 | 664.57 | 511.01 |
| 2 | 315 | 600.81 | 494.68 |
| 3 | 288 | 681.46 | 538.90 |
| 4 | 258 | 647.70 | 509.50 |
| 5 | 280 | 651.23 | 542.89 |
| 6 | 272 | 648.67 | 522.97 |
| 7 | 273 | 618.72 | 543.87 |
| 8 | 214 | 688.63 | 593.58 |
| 9 | 201 | 662.12 | 484.88 |
| 10 | 138 | 685.66 | 550.93 |
| 11 | 207 | 652.38 | 543.18 |
| 12 | 154 | 677.20 | 551.21 |
| 13 | 214 | 636.28 | 497.95 |
| 14 | 151 | 881.63 | 633.93 |
| 15 | 158 | 695.24 | 625.23 |
| 16 | 101 | 784.60 | 789.43 |
| 17 | 152 | 618.62 | 531.11 |
| 18 | 81 | 894.22 | 758.78 |
| 19 | 53 | 1194.82 | 769.40 |
| 20 | 101 | 871.52 | 571.50 |
| X | 82 | 1862.98 | 1371.66 |
| Grand Mean | Grand Median | ||
| 777.10 | 543.87 |
Highlights.
We validated 4040 SNPs in a geographically variable sample of rhesus macaques.
Chinese rhesus had a greater proportion of rare alleles than Indian-origin rhesus.
Linkage disequilibrium was much greater in the Indian sample.
The Indian sample was more heterozygous.
The Indian sample possessed previously undetected population substructure.
Acknowledgments
The authors acknowledge funding from the following sources: NIH/NCRR R24RR025871 and NIH/NCRR R24RR005090. The authors would like to thank the staff of the University of California (UC), Davis Molecular Anthropology Laboratory for assistance with DNA extraction and preparation. We would also like to acknowledge the staff and faculty of the UC Davis Genome Center, especially Dr. Dawei Lin and the Bioinformatics core, for insight on data analysis, and Dr. Charles Nicolet and the Genome Technologies Core, for valuable discussion of SNP genotyping.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gibbs R R.M.G.S.a.A. Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 2.Kanthaswamy S, Capitanio JP, Dubay CJ, Ferguson B, Folks T, Ha JC, Hotchkiss CE, Johnson ZP, Katze MG, Kean LS, Michael Kubisch H, Lank S, Lyons LA, Miller GM, Nylander J, O’Connor DH, Palermo RE, Smith DG, Vallender EJ, Wiseman RW, Rogers J. Resources for genetic management and genomics research on non-human primates at the National Primate Research Centers (NPRCs) Journal of Medical Primatology. 2009;38:17–23. doi: 10.1111/j.1600-0684.2009.00371.x. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson B, Street SL, Wright H, Pearson C, Jia Y, Thompson SL, Allibone P, Dubay CJ, Spindel E, Norgren RB. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2007;8:43. doi: 10.1186/1471-2164-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satkoski J, Malhi RS, Kanthaswamy S, Tito RY, Malladi V, Smith DG. Pyrosequencing as a method for SNP identification in the rhesus macaque (Macaca mulatta) BMC Genomics. 2008;9:256. doi: 10.1186/1471-2164-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fawcett GL, Raveendran M, Derios DR, Chen D, Yu F, Harris RA, Ren Y, Muzny D, Reid JG, Wheeler DA, Worley KC, Shelton SE, Kalin NH, Milosavljevic A, Gibbs R, Rogers J. Characterization of single-nucleotide variation in Indian-origin Rhesus Macaques (Macaca mulatta) BMC Genomics. 2011;12:311. doi: 10.1186/1471-2164-12-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadfield RM, Pullen JG, Davies KF, Wolfensohn SE, Kemnitz JW, Weeks DE, Bennett ST, Kennedy SH. Toward developing a genome-wide microsatellite marker set for linkage analysis in the rhesus macaque (Macaca mulatta): Identification of 76 polymorphic markers. American Journal of Primatology. 2001;54:223–231. doi: 10.1002/ajp.1032. [DOI] [PubMed] [Google Scholar]
- 7.Hart LA, Dassler A. Mouse in Science: Why Mice? UC Davis Center for Animal Alternatives; [Google Scholar]
- 8.U.S.D.o. Agriculture; A.a.P.H.I. Service. Annual Report Animal Usage by Fiscal Year: Fiscal Year 2009. 2011. [Google Scholar]
- 9.Barr CS, Newman TK, Becker ML, Parker CC, Champoux M, Lesch KP, Goldman D, Suomi SJ, Higley JD. The utility of the non-human primate model for studying gene by environment interactions in behavioral research. Genes, Brain and Behavior. 2003;2:336–340. doi: 10.1046/j.1601-1848.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 10.Rodino-Klapac LR, Janssen PML, Montgomery CL, Coley BD, Chicoine LG, Clark KR, Mendell JR. A translational approach for limb vascular delivery of the micro-distrophin gene without high volume or high pressure for treatment of Duchenne muscular dystrophy. Journal of Translational Medicine. 2007;5:45. doi: 10.1186/1479-5876-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiBlasio-Smith EA, Arai M, Quinet EM, Evans MJ, Kornaga T, Basso MD, Chen L, Feingold I, Halpern AR, Liu QY, Nambi P, Savio D, Wang S, Mounts WM, Isler JA, Slager AM, Burczynski ME, Dorner AJ, LaVallie ER. Discovery and implementation of trascriptional biomarkers of synthetic LXR agonists in peripheral blood cells. Journal of Translational Medicine. 2008;6:59. doi: 10.1186/1479-5876-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kota J, Handy CR, Haidet AM, Montgomery CL, Eagle A, Rodino-Klapac LR, Tucker D, Shilling CJ, Therfall WR, Walker CM, Weisbrode SE, Janssen PML, Clark KR, Sahenk Z, Mendell JR, Kaspar BK. Follistatin gene delivery enhanses muscle growth and strength in nonhuman primates. Science Translational Medicine. 2009;1:6ra15. doi: 10.1126/scitranslmed.3000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed MR, Berthet A, Bychkov E, Porras G, Li Q, Bioulac BH, Carl YT, Bloch B, Kook S, Aubert I, Dovero S, Doudnikoff E, Gurevitch VV, Gurevitch EV, Bezard E. Lentiviral overexpression of GRK6 alleviates L-Dopa-induced dyskinesia in experimental Parkinson’s Disease. Science Translational Medicine. 2010;2:28ra28. doi: 10.1126/scitranslmed.3000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parikh UM, Dobard C, Sharma S, Cong M, Jia H, Martin A, Pau CP, Hanson DL, Guenthner P, Smith J, Kersh E, Garcia-Lerma JC, Novembre FJ, Otten R, Folks T, Heneine W. Complete protection from repeated vaginal SHIV exposures in macaques by a topical gel containing tenofovir alone or with emtricitabine. Journal of Virology. 2009;83:10358–10365. doi: 10.1128/JVI.01073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Lerma JG, Cong M, Mitchell J, Youngpairoj AS, Zheng Q, Masiotra S, Martin A, Kuklenyik Z, Holder A, Lipscomb J, Pau CP, Barr JR, Hanson DL, Otten R, Paxton L, Folks T, Heneine W. Intermittent prophylaxis with oral Truvada protects macaques from rectal SHIV infection. Science Translational Medicine. 2010;2:14ra14. doi: 10.1126/scitranslmed.3000391. [DOI] [PubMed] [Google Scholar]
- 16.Carlsson HE, Schapiro SJ, Hau J. Use of primates in research: a global overview. American Journal of Primatology. 2004;63:225–237. doi: 10.1002/ajp.20054. [DOI] [PubMed] [Google Scholar]
- 17.Barouch D. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455:613–619. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haigwood NL. Update on animal models for HIV research. European Journal of Immunology. 2009;39:1991–2058. doi: 10.1002/eji.200939576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malhi RS, Sickler B, Lin D, Satkoski J, Tito RY, George D, Kanthaswamy S, Smith DG. MamuSNP: a resource for rhesus macaque (Macaca mulatta) genomics. PLoS ONE. 2007;2:e438. doi: 10.1371/journal.pone.0000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Smith DG, McDonough J. Mitochondrial DNA variation in Chinese and Indian rhesus macaques (Macaca mulatta) American Journal of Primatology. 2005;65:1–25. doi: 10.1002/ajp.20094. [DOI] [PubMed] [Google Scholar]
- 22.Smith DG, George D, Kanthaswamy S, McDonough J. Identification of country of origin and admixture between Indian and Chinese rhesus macaques. International Journal of Primatology. 2006;27:881–898. [Google Scholar]
- 23.Satkoski Trask JA, Malhi RS, Kanthaswamy S, Johnson J, Garnica WT, Malladi V, Smith DG. The effect of SNP discovery method and sample size on estimation of population genetic data for Chinese and Indian rhesus macaques (Macaca mulatta) Primates. 2011;52:129–138. doi: 10.1007/s10329-010-0232-4. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 26.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic Acids Research. 2010;39:1–7. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blankenberg D, Von Kuster G, Corador N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Currrent Protocols in Molecular Biology. 2010:1–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goecks J, Nekrutenko A, Taylor J, Team TG. Galaxy: a comprehensive approach for supporting accessible, reproducible and transparent computational research in the life sciences. Genome Biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedrick P, Kumar S. Mutation and linkage disequilibrium in human mtDNA. European Journal of Human Genetics. 2001;9:969–972. doi: 10.1038/sj.ejhg.5200735. [DOI] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–264. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.Eberle MA, Rieder M, Kruglyak L, Nickerson D. Allele frequency matching between SNPs reveals an excess of linkage disequilibrium in genic regions of the human genome. PLos Genetics. 2006;2:1319. doi: 10.1371/journal.pgen.0020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez RD, Hubisz M, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, Muzny D, Gibbs R. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- 34.Hasan MK. Mitochondrial DNA of rhesus macaques (Macaca mulatta) from Bangladesh. The 23rd Biannual Meeting of the International Primatological Society; Kyoto, Japan. 2010. [Google Scholar]
- 35.Lander ES I.H.G.S. Consortium, Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 36.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pederson JS, Pohl A, Raney J, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, Sultan-Qurraie A, Thomas DJ, Trumbower H, Weber RJ, Weirauch M, Zweig AS, Haussler D, Kent WJ. The UCSC Genome Browser Database: update 2006. Nucleic Acids Research. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination and mutation. American Journal of Human Genetics. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reich D, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark A. Recent and ongoing selection in the human genome. Nature Reviews Genetics. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satkoski Trask JA, Garnica WT, Malhi RS, Kanthaswamy S, Smith DG. High-throughput single-nucleotide polymorphism discovery and the search for candidate genes for long-term SIVmac nonprogression in Chinese rhesus macaques (Macaca mulatta) Journal of Medical Primatology. 2011;40:224–232. doi: 10.1111/j.1600-0684.2011.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, Johnson Z, Bergstrom M, Novakowski L, Nair P, Vinson A, Newman D, Heckman G, Cameron J. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 2006;87:30–38. doi: 10.1016/j.ygeno.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Smith DG, McDonough J, George D. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. American Journal of Primatology. 2007;69:182–198. doi: 10.1002/ajp.20337. [DOI] [PubMed] [Google Scholar]
- 44.Kanthaswamy S, Satkoski J, Kou A, Malladi V, Glenn Smith D. Detecting signatures of inter-regional and inter-specific hybridization among the Chinese rhesus macaque specific pathogen-free (SPF) population using single nucleotide polymorphic (SNP) markers. Journal of Medical Primatology. 2010;39:252–265. doi: 10.1111/j.1600-0684.2010.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carpenter CR. Breeding colonies of macaques and gibbons on Santiago Island, Puerto Rico. In: Beveridge WIB, editor. Breeding Primates. S. Karger; Basel: 1972. pp. 76–87. [Google Scholar]
- 46.Buettner-Janusch J, Mason GA, Buettner-Janusch V, Sade DS. Genetic studies of serum transferrins of free-ranging rhesus macaques of Cayo Santiago,Macaca mulatta (Zimmerman 1780) American Journal of Physical Anthropology. 1974;41:217–232. doi: 10.1002/ajpa.1330410204. [DOI] [PubMed] [Google Scholar]
- 47.Johnsen DO. History. In: Bennett BT, Abee CR, Hendrickson R, editors. Nonhuman Primates in Biomedical Research. Academic Press; San Diego: 1995. pp. 1–12. [Google Scholar]
- 48.Olivier TJ, Ober C, Buettner-Janusch J, Sade DS. Genetic differentation among matrilines in social groups of rhesus monkeys. Behavioral Ecology and Sociobiology. 1981;8:279–285. [Google Scholar]
- 49.Duggleby C. Blood group antigens and the population genetics of Macaca mulatta on Cayo Santiago: I. Genetic differentiation of social groups. American Journal of Physical Anthropology. 1978;48:35–40. doi: 10.1002/ajpa.1330480107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.