Abstract
RA receptors (RARs) have been thought to function through a binary repressor-activator mechanism: in the absence of ligand, they function as transcriptional repressors, and, in the presence of ligand, they function as transcriptional activators. This prevailing model of RAR mechanism has been derived mostly from in vitro studies and has not been widely tested in developmental contexts. Here, we investigate whether zebrafish RARs function as transcriptional activators or repressors during early embryonic anterior-posterior patterning. Ectopic expression of wild-type zebrafish RARs does not disrupt embryonic patterning and does not sensitize embryos to RA treatment, indicating that RAR availability is not limiting in the embryo. In contrast, ectopic expression of hyperactive zebrafish RARs induces expression of a RA-responsive reporter transgene as well as ectopic expression of endogenous RA-responsive target genes. However, ectopic expression of dominant negative zebrafish RARs fails to induce embryonic phenotypes that are consistent with loss of RA signaling, despite their ability to function as transcriptional repressors in heterologous cell culture assays. Together, our studies suggest that zebrafish RAR function is context-dependent and that, during early patterning, zebrafish RARs function primarily as transcriptional activators and may only have minimal ability to act as transcriptional repressors. Thus, it seems that the binary model for RAR function does not apply to all in vivo scenarios. Taking into account studies of RA signaling in tunicates and tetrapods, we propose a parsimonious model of the evolution of RAR function during chordate anterior-posterior patterning.
Keywords: zebrafish, retinoic acid receptors, transcription
Introduction
Retinoic acid (RA) signaling regulates a diverse array of developmental processes. One of its well-known roles is in determining anterior-posterior (A-P) positional identity during early development, through regulation of Hox gene expression (Marletaz et al., 2006). In addition to the early role of RA signaling during A-P patterning, it has later roles in growth, homeostasis and patterning of numerous organs (Capdevila and Izpisua Belmonte, 2001; Kastner et al., 1997; Lohnes et al., 1994; Mendelsohn et al., 1994; Niederreither and Dolle, 2008). RA signaling requires RA receptors (RARs), members of the nuclear hormone family of receptors, of which there are 3 paralogs (alpha, beta and gamma) in mammals (Aranda and Pascual, 2001; Bastien and Rochette-Egly, 2004). Zebrafish have 4 RARs: species-specific paralogs of the RARalphas (RARaa and ab) and gammas (RARga and gb), but no beta ortholog (Hale et al., 2006; Waxman and Yelon, 2007). In both mice and zebrafish, there is a high degree of functional redundancy between RARs in early development, although there are some requirements for individual receptors (Linville et al., 2008; Lohnes et al., 1994; Mendelsohn et al., 1994).
Prior studies have proposed a binary ligand-dependent transcriptional repressor-activator paradigm for RAR function (Bastien and Rochette-Egly, 2004; Chen and Evans, 1995; Hauksdottir et al., 2003; Perissi et al., 1999; Torchia et al., 1998; Xu et al., 1999). In the absence of ligand (all-trans RA), the RARs are thought to interact with transcriptional repressors. Upon the binding of RA to RARs, the receptors then exchange the transcriptional repressors for transcriptional activation machinery (Bastien and Rochette-Egly, 2004; Chen and Evans, 1995; Hauksdottir et al., 2003; Perissi et al., 1999; Torchia et al., 1998; Xu et al., 1999). However, this binary model is primarily derived from studies in cell culture and has not been tested in most developmental contexts. So far, there are only two in vivo developmental contexts in which RARs have been shown to be required as transcriptional repressors: Xenopus midbrain-hindbrain boundary (MHB) formation and mouse skeletal growth (Koide et al., 2001; Williams et al., 2009). In Xenopus embryos, depletion of RARa results in loss of the MHB, similar to the effect of adding RA (Koide et al., 2001). This phenotype can be rescued by expression of a dominant negative RARa, further reinforcing the role of RARa as a repressor in this context, consistent with the binary model. More recently, it has been demonstrated that the shortened limbs in mouse RAR knockouts are due to a requirement for RAR transcriptional repressor activity in skeletal growth plates (Williams et al., 2009). These phenotypes are reminiscent of transgenic mice that overexpress a constitutively active RAR in the limb, supporting a repressive role of RA signaling in the limb (Cash et al., 1997). Moreover, these in vivo studies echo in vitro studies in which RAR-mediated transcriptional repression is required for chondrogenic mesenchyme differentiation in murine primary cultures (Weston et al., 2002; Weston et al., 2000).
Despite these characterized roles as transcriptional repressors, many of the developmental phenotypes caused by loss of RARs have been interpreted as representing failure to activate RAR target genes, rather than representing failure to repress RAR target genes (Lohnes et al., 1994; Mendelsohn et al., 1994). Furthermore, transgenic mice expressing dominant negative RARs only exhibit a subset of phenotypes associated with the loss of RARs, even though transgene expression should be fairly broad (Damm et al., 1993; Iulianella and Lohnes, 2002). Likewise, studies in zebrafish embryos seem inconsistent with a requirement for RARs acting as transcriptional repressors. For instance, depletion of RARs does not seem to inhibit MHB formation (Linville et al., 2008), in contrast to what has been shown for Xenopus (Koide et al., 2001). Instead, expression of a zebrafish dnRAR together with injection of anti-RAR morpholinos exacerbates other phenotypes caused by RA deficiency (Stafford and Prince, 2002). Therefore, studies in both mouse and zebrafish hint that RARs may not be required to function as transcriptional repressors or via the binary model in all developmental contexts.
Here, we more rigorously examine the mechanism of transcriptional regulation by RARs during early A-P patterning of the zebrafish embryo using three different assays: novel transgenic reporters of RA signaling, endogenous target genes downstream of RA signaling, and embryonic phenotypes dependent on RA signaling. We show that ectopic expression of wild-type zebrafish RARs neither blocks nor activates expression of RA-responsive targets in the early embryo, nor does it sensitize the embryo to addition of RA. In contrast, ectopic expression of hyperactive VP16-tagged zebrafish RARs is able to cause phenotypes similar to those caused by increased RA signaling. However, dominant negative zebrafish RARs (dnRARs) are not able to act as transcriptional repressors in the early embryo, even though they can repress transcription in heterologous cell culture assays, suggesting that zebrafish RARs have minimal repressive ability during early A-P patterning. Altogether, these results suggest a model in which the ability of zebrafish RARs to function as transcriptional repressors or activators is context-dependent. Specifically, during early A-P patterning, the zebrafish RARs function primarily as transcriptional co-activators to regulate target gene expression.
Materials and Methods
Construction of VP16 chimeras, Engrailed repressor chimeras, and dominant negative RARs
Sequences for wild-type zebrafish RARs have been reported previously (Waxman and Yelon, 2007). For full-length RAR-VP16 chimeric proteins (generally referred to as RAR-vps), the VP16 activator domain was fused directly to the N or C terminus, with the only modifications being the deletion of the start or termination codons of the RARs. For ΔA domain chimeras with RARab, amino acids 1-47 were deleted and the VP16 activator or Engrailed repressor (Enr) domains were fused to amino acid 48 (H). For the ΔA domain chimera with RARga, amino acids 1-41 were deleted and the VP16 activator domain was fused to amino acid 42 (T). For ΔF domain chimeras with RARab, the VP16 activator or Enr domains were placed after amino acid 410 (E). For the ΔF domain fusion with RARga, the VP16 activator domain was placed after amino acid 406 (E). A dominant negative (dn) RARab was made by truncating the protein at amino acid 398 (P), and a dnRARga was made by truncating the protein at amino acid 394 (G), as reported by Damm et al. (1993) for murine RARa and RARg. All fusion and truncation constructs were generated using PCR and confirmed by sequencing. Primer sequences are available upon request. pSP72 human dnRARa, a gift of C. Sagerström, has been reported previously (Roy and Sagerström, 2004). Zebrafish dnRARaa, a gift of T. Schilling, was reported previously (Stafford et al., 2006). All untagged RARs used in experiments were subcloned into pCS2p+, except dnRARaa, which was in pCS2. For addition of myc tags, RARs were subcloned into pCS2+MT (Rupp et al., 1994). Endogenous UTRs were omitted from all constructs in an effort to normalize levels of ectopic expression for all RARs.
Embryo injections
Embryos were injected at the 1 cell stage for all experiments, except Western blots and wholemount immunofluorescence, for which embryos were injected as late as the 4 cell stage. Capped mRNA was made using the Message Machine Kit (Ambion). For all mRNAs, we titrated the amount of mRNA injected and sought to inject the lowest possible dose that produced the maximal phenotypic response consistent with altered RA signaling. For constructs that seemed to cause no overt phenotype, we therefore needed to use the highest dose possible (200 pg), which borders on levels that are typically toxic. Following titration, 100 pg of mRNA was injected for all constructs used, unless otherwise indicated. For dnrar and rarabΔF-enr chimeras, we injected 200 pg of mRNA. For human dnrara, we injected 80 pg of mRNA. For zebrafish rarab-ΔAenr, only 50 pg of mRNA was injected because higher amounts cause aberrant gastrulation movements and cyclopia, phenotypes which are not typical for loss of RA signaling. For all other mRNAs, these non-specific phenotypes, in addition to kinking of the tail, were seen at doses higher than 200 pg.
In situ hybridization
In situ hybridization probes for dhrs3a (ZDB-GENE-040801-217), hoxb5b (ZDB-GENE-000823-6), opl/zic1 (ZDB-GENE-000208-4), eng2a (ZDB-GENE-980526-167), krox20/egr2b (ZDB-GENE-980526-283), and myod (ZDB-GENE-980526-561) have been reported previously.
Construction of 12X RARE reporter transgenes and transgenic lines
The 12X RARE reporters (12XRARE-ef1a:gfp and 12XRARE-tk:gfp) feature the concatenation of 12 direct repeat 5 (DR5) retinoic acid response element (RARE) sites (binding sites for the RARs). RARE sites were modeled after those found in the Hox gene promoters and varied in their 5 nucleotide spacer sequence (Bastien and Rochette-Egly, 2004). Six 5’ RAREs were placed in the reverse orientation and six 3’ RAREs were placed in the forward orientation (Figure 1A). All RARE sites were placed upstream of either an elongation factor-1 alpha (ef1a) or thymidine kinase (tk) minimal promoter within a vector containing egfp flanked by adeno-associated viral inverted terminal repeat elements and I-SceI sites (gift of D. Prober; Prober et al., 2006). Sequences of reporter transgenes are available upon request.
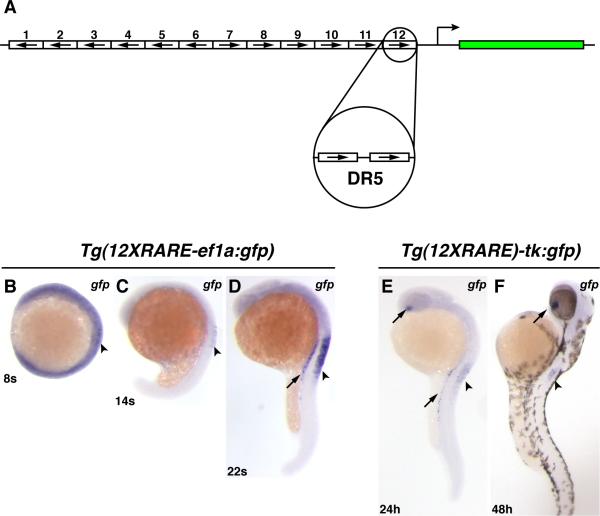
Figure 1. Zebrafish RARE transgenic reporter lines.
(A) Schematic representation of the 12XRARE reporters. Arrows indicate direction of each DR5 RARE site. (B) At 8 somites, the expression of gfp from Tg(12XRARE-ef1a:gfp) can be seen at low levels in the anterior spinal cord (arrowhead). At this stage, embryos require a much longer exposure to visualize gfp expression than at subsequent stages. (C) At 14 somites, gfp expression is more easily detected in the spinal cord (arrowhead). (D) By 22 somites, the reporter is expressed at higher levels in the pronephros (arrow) and spinal cord (arrowhead). Shortly afterward, the reporter is also expressed in the ventral eye (not shown), similar to Tg(12XRARE-tk:gfp). (E) Expression of Tg(12XRARE-tk:gfp) does not initiate until closer to 24 hpf, when it is seen in the ventral eye (upper arrow), pronephros (lower arrow), and the anterior spinal cord (arrowhead). (F) Expression of Tg(12XRARE-tk:gfp) in the eye (arrow) and spinal cord (arrowhead) is maintained past 48 hpf. All images are lateral views, with anterior up and dorsal to the right.
Transgenic lines were made as previously reported (Prober et al., 2006). Briefly, ~100pg of RARE reporter transgene plasmid was digested with I-SceI and injected into embryos at the 1 cell stage. These embryos were raised to adulthood, and their progeny were screened for fluorescence.
RA treatment and pharmacological reagents
For analysis of responsiveness of the Tg(12XRARE-ef1a:gfp) and Tg(12XRARE-tk:gfp) lines, treatment with pharmacological reagents began at 24 or 30 hpf, respectively, and was continuous for 6 hours. For treatment with pharmacological antagonists of RA signaling, we used doses that we have previously found to cause phenotypes consistent with loss of RA signaling when treatments were initiated prior to gastrulation (Waxman et al., 2008). These phenotypes include loss of forelimb (pectoral fin), enlarged heart, and hindbrain defects. For BMS189453 (BMS; Bristol-Meyers Squibb) and DEAB (Sigma), working concentrations of 1μM were used. For Ro41-5253 (Biomol.com), a RARα specific antagonist, a working concentration of 0.5 μM was used.
For analysis of embryos injected with zebrafish RARs, embryos were treated continuously from 40% epiboly with 0.05 μM RA. For analysis of Tg(12XRARE-ef1a:gfp) embryos at the tailbud stage, embryos were treated continuously beginning at 40% epiboly with 0.2 μM RA. For all other experiments involving early RA treatment, embryos were treated for 1 hour with 0.5 μM or 0.2 μM RA beginning at 40% epiboly. For AM580 treatments (Biomol.com), a RARα specific agonist, a working concentration of 0.1 μM was used. For all other pharmacological reagents (see Supplemental Material), working concentrations of 1 μM were used.
Cell culture and luciferase assays
For luciferase reporter assays, the 12XRARE-ef1a and 12XRARE-tk promoters were placed into the NotI site of the pGL3-basic vector (Promega). HEK 293 cells were grown in DMEM supplemented with 10% fetal bovine serum and 1% Pen/Strep under standard conditions. All assays were performed in 96 well dishes. Approximately 10,000 cells per well were transfected with a total of 0.1μg DNA using SiPort NEO (Ambion). For all experiments, cells were transfected with 0.025μg of RAR and β-gal plasmids. Either 0.025 or 0.05ug of RARE reporter plasmid and 0.025μg of renilla luciferase plasmid were also included to bring the total amount of DNA transfected to 0.1μg. Similar results were obtained with either amount of RARE reporter plasmid. Luminescence was read after 3 days using the Dual-Glo system (Promega). Levels of firefly luciferase were standardized relative to renilla luciferase. For RA treatments, the media was removed one day after transfection and replaced with media containing either 1 μM RA or a DMSO control, and luminescence was read at day 3.
Western blotting and wholemount immunofluorescence
Western blots and wholemount immunofluorescence were performed essentially as described by Mintzer et al. (2001). To check the expression of the tagged proteins, 100 pg of each of the tagged mRNAs was injected at the 1-4 cell stage. Embryos were then lysed or fixed, respectively, for Western blots and wholemount immunofluorescence at 70-90% epiboly. For both analyses, the monoclonal 9e10 anti-myc antibody (Covance) was used as the primary antibody. For a loading control, a mouse anti-α-tubulin antibody (Sigma) was used. For Western blotting, a HRP-conjugated anti-mouse (Sigma) was used as the secondary antibody. For wholemounts, a FITC-conjugated anti-mouse IgG1 (Southern Biotech) was used as the secondary antibody.
Results
Characterization of transgenic reporters of RA signaling
In order to understand the mechanisms of RA signaling during early developmental processes, it is important to examine the ability of RARs to regulate transcription through RARE target sites within the embryo. We therefore generated transgenes containing a synthetic RARE reporter, in which 12 RAREs are concatenated (Fig. 1A). We henceforth refer to the two novel synthetic RARE reporter constructs used to make transgenic zebrafish lines as Tg(12XRARE-ef1a:gfp) and Tg(12XRARE-tk:gfp), reflecting the promoter used in each construct. We analyzed 4 different Tg(12XRARE-ef1a:gfp) lines and 2 different Tg(12XRARE-tk:gfp) lines. All recovered lines exhibited a Mendelian mode of inheritance indicating that they likely represent insertions at single loci (JSW and DY, unpublished data). For this study, we focus on two representative lines Tg(12XRARE-ef1a:gfp)sk71 and Tg(12XRARE-tk:gfp)sk73.
Expression of Tg(12XRARE-ef1a:gfp) is detectable by in situ hybridization for gfp at about 8 somites (13 hours post-fertilization (hpf); Fig. 1B). Despite the early detectability of gfp RNA, which is maintained in the spinal cord through 14 somites (Fig. 1C), GFP fluorescence is not detectable until about 20-22 hpf. Fluorescence becomes progressively stronger through 30 hpf (Fig. 2C), but is no longer clearly visible by about 3 days post-fertilization (dpf). Expression of Tg(12XRARE-tk:gfp) does not initiate until closer to 24 hpf (Fig. 1E), but it is maintained through 5 dpf (Figs. 1F and 2D; JSW and DY, unpublished data). Therefore, the transgenic lines differ slightly in the temporal initiation and duration of expression. None of our transgenic lines initiated gfp expression prior to gastrulation, similar to what has been reported previously for another transgene (Perz-Edwards et al., 2001), suggesting that this may be a common characteristic of the responsiveness of synthetic RARE reporters in zebrafish.
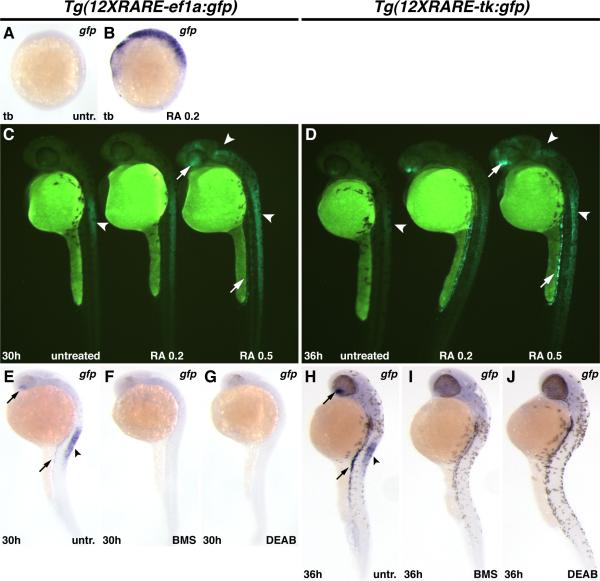
Figure 2. Zebrafish RARE transgenic reporter lines require and are responsive to RA signaling.
(A) Untreated Tg(12XRARE-ef1a:gfp) embryo. (B) Treatment of Tg(12XRARE-ef1a:gfp) embryos with 0.2 μM RA induces reporter expression. (C, D) Tg(12XRARE-ef1a:gfp) and Tg(12XRARE-tk:gfp) embryos respond to treatment with RA for 6 hours beginning at 24 hpf (C) or 30 hpf (D). Untreated sibling embryos (left) exhibit expression in the anterior spinal cord (arrowhead), the ventral eye, and pronephros. Treatment with 0.2 μM RA (center) causes a more modest response than does treatment with 0.5 μM RA (right). Increased fluorescence is induced in the anterior brain/eye (upper arrow), midbrain and spinal cord (arrowheads) and pronephros (lower arrow). (E, H) Untreated Tg(12XRARE-ef1a:gfp) (E) and Tg(12XRARE-tk:gfp) (H) embryos, respectively. Treatment with RA signaling antagonists BMS (F, I) and DEAB (G, J) inhibit expression of Tg(12XRARE-ef1a:gfp) and Tg(12XRARE-tk:gfp). Embryos were treated with RA signaling antagonists for 6 hours beginning at 24 hpf (E-G) or 30 hpf (H-J). All images are lateral views, with anterior up and dorsal to the right.
The spatial expression of the reporter appeared similar in all isolated transgenic lines, with major sites of expression apparent in the anterior spinal cord, the ventral eye, and the pronephros by 24 hpf (Figs. 1D-F and 2E, H; JSW and DY, data not shown). The only minor difference detected was that Tg(12XRARE-tk:gfp) lines had low levels of expression in the anterior notochord (JSW and DY, data not shown). This difference was also found in previous zebrafish RARE transgenic reporter lines, which also compared the use of ef1a and tk promoters (Perz-Edwards et al., 2001). In contrast to some of the previously reported lines (Perz-Edwards et al., 2001), we did not observe expression in other mesodermal derivatives, such as the somites, heart, and pharyngeal arches, or in the dorsal retina.
To determine if our transgenic lines are responsive to RA signaling, we treated transgenic embryos with RA. Exposure of Tg(12XRARE-ef1a:gfp) embryos to RA from 40% epiboly (5 hpf) until tailbud stage (tb; 10 hpf) induced ectopic transgene expression (Figs. 2A, B). However, Tg(12XRARE-tk:gfp) embryos did not respond to early treatment with RA (JSW and DY, unpublished data). Despite their difference in early responsiveness, both reporter lines responded to treatment with RA beginning at later stages (Figs. 2C, D).
We next tested if reporter expression requires RA signaling by treating embryos with the RA signaling antagonists BMS 189453 (BMS), a pan-RAR inhibitor, and DEAB, an inhibitor of RA synthesis (Russo et al., 1988; Schulze et al., 2001). Treatment with either of these RA signaling antagonists eliminated reporter expression (Figs. 2E-J). Other antagonists and agonists of RA signaling were also able to affect reporter expression, while pharmacological reagents specific to other nuclear hormone receptors did not affect reporter expression (Fig. S1). Therefore, although the transgenic reporters do not seem to respond at all sites of endogenous RA signaling, both reporter lines are capable of responding to RA and can be used as tools to better understand RA signaling.
Hyperactive zebrafish RARs differentially affect developmental processes
We next used these tools to aid us in analyzing the mechanisms of zebrafish RAR function within the context of the embryo. First, we examined the effects of overexpressing two of the wild-type (WT) zebrafish RARs (RARab and RARga) in Tg(12XRARE-ef1a:gfp) embryos. Prior studies in cell culture have shown that RARs interact with and act as transcriptional repressors in the absence of RA (Chen and Evans, 1995; Hauksdottir et al., 2003; Perissi et al., 1999), suggesting that embryonic ectopic expression of WT RARs might cause phenotypes consistent with loss of RA signaling. However, embryos injected with rarab or rarga mRNA appeared normal at 48 hpf and beyond, and expression of the transgene and endogenous target genes was not affected (Figs. 3B-D; JSW and DY, unpublished data). Furthermore, embryos injected with either of these mRNAs and treated with RA were not more severely posteriorized and did not activate the transgene more than uninjected embryos treated with RA (Figs. 3E-G), as might have been expected from cell culture experiments (Bastien and Rochette-Egly, 2004; Chen and Evans, 1995; Hauksdottir et al., 2003; Perissi et al., 1999; Torchia et al., 1998; Xu et al., 1999). The inability of WT zebrafish RARs to repress transcription or sensitize embryos to RA treatment does not seem to be due to failure of protein expression, as tagged versions of these proteins can be robustly expressed in embryos (Fig. S2). Therefore, availability of RARs within the zebrafish embryo is not a limiting factor in the regulation of transcription in response to RA signaling; more likely, RA availability, along with the availability of RAR-interacting transcriptional modifiers, is the major limitation.
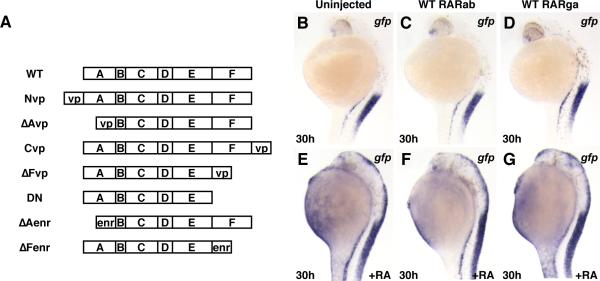
Figure 3. Ectopic expression of zebrafish RARs does not sensitize embryos to RA signaling.
(A) Schematic composition of RAR constructs used in experiments. (B) Uninjected Tg(12XRARE-ef1a:gfp) embryo. (C, D) Tg(12XRARE-ef1a:gfp) embryos injected with zebrafish rarab (C) or rarga (D) mRNA appear normal, and reporter expression is unaffected. (E) Tg(12XRARE-ef1a:gfp) embryo continuously treated with 0.05 μM RA. (F, G) Tg(12XRARE-ef1a:gfp) embryo injected with zebrafish rarab (F) or rarga (G) mRNA and treated with 0.05 μM RA are not more affected than uninjected embryos. All images are lateral views, with anterior up and dorsal to the right.
The binary repressor-activator model predicts that interactions with other transcriptional regulators underlie the regulatory roles of RARs. If interactions of RARs with transcriptional activators modify their functions, then RAR-VP16 fusion proteins should affect the abilities of RARs to control the transcription of RA-responsive targets. We first tested zebrafish versions of previously reported “constitutively active” RARs, in which the VP16 domain was fused directly to the N-terminus of an RAR (Blumberg et al., 1997; Koide et al., 2001). Because A domains of nuclear hormone receptors can modify transcriptional ability (Bastien and Rochette-Egly, 2004; Mark et al., 2006; Nagpal et al., 1992), we were not sure whether this zebrafish version of the previously reported VP-RAR fusion (here termed a Nvp construct; Fig. 3A) would be an optimal indicator of activator activity. Therefore, we also made fusion proteins in which the VP16 domain was fused directly to the C-terminus, to the B domain, or to the E domain of each RAR (here termed Cvp, ΔAvp, and ΔFvp constructs, respectively; Fig. 3A). We found that all of the RAR-vp constructs were able to induce ectopic reporter expression by 8 somites, although induction by the RARga-Cvp was very weak (Figs. 4A-E, K-M; and Figs. 5A1-I1). Ectopic expression was maintained through 24 hpf (Fig. S3 and Fig. S4). Comparing trends of transgene activation by the different fusion proteins, the RAR-ΔAvp fusion proteins (RARab-ΔAvp and RARga-ΔAvp) produced the strongest activation, although RARab-ΔAvp activation is only marginally greater than the other RARab-vps (Figs. 4A-E, K-M and Figs. 5A1-I1). Overall, the RARab-vp constructs induced stronger activation of the reporter than the RARga-vp constructs (Figs. 4A-E, K-M, Figs. 5A1-I1, Fig. S3 and Fig. S4). Therefore, in the majority of cases, VP16-tagged RARs can activate ectopic reporter expression.
Figure 4. RAR-vps require RA to function.
All images depict Tg(12XRARE-ef1a:gfp) embryos. (A, F) Uninjected control embryos. (B-E, K-M) Injection with (B) rarab-Nvp, (C) rarab-ΔAvp, (D) rarab-Cvp, (E) rarab-ΔFvp, (K) rarga-Nvp, (L) rarga-ΔAvp, or (M) rarga-ΔFvp mRNA induces ectopic reporter expression. (F-P) Treatment with 1μM DEAB beginning at 40% epiboly blocks reporter expression in injected embryos. All images are dorsal views with anterior up. (Q) In HEK 293 cells transfected with the pGL3-basic:12XRARE-tk reporter, co-transfection with RAR-vp constructs caused similar trends of reporter activation to those found in zebrafish embryos. Arbitrary Units (AU) represents the ratio of firefly luciferase to renilla luciferase. Each column represents the average AU plus standard deviation (bars) of 3 replicates for a condition from one representative experiment. Equivalent trends were observed using the pGL3-basic:12XRAR-ef1a reporter; additionally, DEAB treatment of cells transfected with RAR-vp constructs reduced reporter activation (JSW and DY, data not shown).
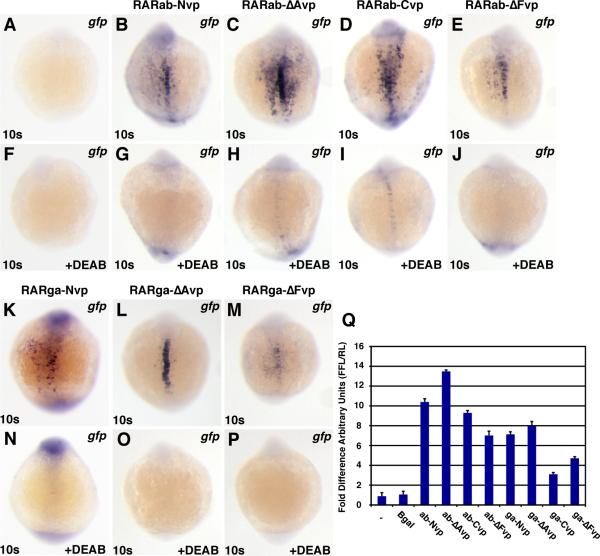
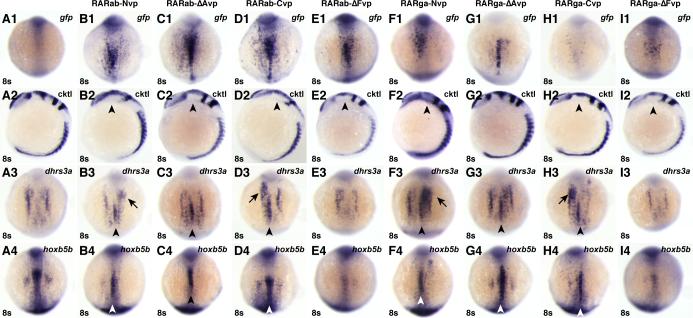
Figure 5. RAR-vps can induce ectopic expression of RA target genes.
(A1-I1) Injection of the rar-vp mRNAs differentially activates reporter expression. (A2-I2) Injection of the rar-vp mRNAs, except for rarga-ΔAvp, eliminates the MHB. Cocktail (cktl) of probes includes opl/zic1 (telencephalon, diencephalic-midbrain boundary), eng2a (MHB), krox20/egr2b (rhombomeres 3 and 5), and myod (somites and adaxial cells). Arrowheads indicate loss of enr2 expression in the MHB. (A3-I3, A4-I4) Injection of the rarNvp, rarΔAvp, and rarCvp mRNAs can induce ectopic expression of the RA signaling target genes dhrs3a and hoxb5b, while injection of the rarΔFvp mRNAs inhibits their expression. Arrowheads indicate ectopic dhrs3a or hoxb5b expression in the notochord. Arrows indicate ectopic dhrs3a expression more anteriorly. (A1-A4) Control uninjected embryos. (B1-B4) rarabNvp mRNA-injected embryos. (C1-C4) rarabΔAvp mRNA-injected embryos. (D1-D4) rarabCvp mRNA-injected embryos. (E1-E4) rarabΔFvp mRNA-injected embryos. (F1-F4) rargaNvp mRNA-injected embryos. (G1-G4) rargaΔAvp mRNA-injected embryos. (H1-H4) rargaCvp mRNA-injected embryos. (I1-I4) rargaΔFvp mRNA-injected embryos. All images except A2-I2 are dorsal views with anterior up. Images in A2-I2 are lateral views with dorsal to the right.
The reporter expression induced by VP16 fusion proteins was largely restricted to areas adjacent to the expression of raldh2, which encodes the major RA producing enzyme in the embryo (Begemann et al., 2001; Grandel et al., 2002). Ectopic ectodermal and notochord expression was primarily restricted to the trunk region and the ventral eye (Figs. 4A-E, K, L, Fig. S3, and Fig. S4). This regionalized enhancement of expression suggested that the VP16-tagged RARs may be acting as ligand-dependent hyperactive proteins, rather than as truly constitutively active proteins. To confirm this, we treated Tg(12XRARE-ef1a:gfp) embryos injected with the rar-vp mRNAs with DEAB beginning at 40% epiboly. DEAB treatment was able to eliminate ectopic activation of the reporter (Fig. 4A-P). This dependence on the synthesis of RA suggests that the VP16 fusion proteins are ligand-dependent, so we now refer to these tools as hyperactive RARs.
We also noticed that the phenotypes induced by hyperactive RARs were reminiscent of but different than the effects of RA treatment. RA causes a concentration-dependent posteriorization of the anterior central nervous system (CNS) (Hernandez et al., 2007). Higher concentrations of RA can posteriorize embryos and eliminate anterior structures, while lower concentrations lead to reduction of anterior structures (Fig. S5 and Figs. S6B,C). Examining anterior CNS markers at 8 somites revealed that the full-length RARab-vp proteins were the most potent, eliminating both the MHB and anterior hindbrain (Figs. 5B2, D2), compared to just the MHB for the RARab-ΔAvp and -ΔFvp (Figs. 5C2, E2). Interestingly, of the RARga-vps, the RARga-ΔAvp did not affect the anterior CNS (Fig. 5G2), while the 3 others all eliminated the MHB (Fig. 5F2, H2, I2). Despite strong effects on the MHB and hindbrain, the most anterior CNS was relatively unaffected by hyperactive RARs (Fig. 5A2-I2 and S5D-K). This lack of effect on the most anterior CNS is in contrast to the phenotype caused by RA treatment (Fig. S6C). Interestingly, the strong effects of the full length RARab-vps (Nvp and Cvp) on the CNS (Figs. 5B2 and D2) contrast with the trends observed for activation of the transgenic RARE reporter (Figs. 5B1 and D1), for which RARab-ΔAvp was the most potent activator (Fig. 5C1). While the RARga-ΔAvp was the best of the tagged RARga constructs at activating the transgenic reporter (Fig. 5G1), it did not affect the CNS (Fig. 5G2). Together, these data show that ectopic expression of hyperactive RARs does not completely recapitulate the effects of RA treatment on the anterior CNS, which is likely due to the ligand-dependency of the hyperactive RARs. Furthermore, when comparing the effects of particular RARs, there is not a correlation between the effects of hyperactive RARs on CNS markers and on the transgenic reporter, given that the RAR-vp constructs that were best at eliminating CNS structures were not the most potent activators of transgene expression.
We next looked at the ability of the hyperactive RARs to affect endogenous targets of RA signaling. We examined two positively regulated target genes, dhrs3a and hoxb5b, which we have recently shown to be downstream and potentially direct targets of RA signaling (Feng et al., 2010; Waxman et al., 2008; Figs. S4D-I). In contrast to the generally more potent activation of the transgenic RARE reporter by RARab-vps, the abilities of the RARab-vps and RARga-vps to induce ectopic expression of endogenous target genes were quite similar (Figs. 5A3-6I3, 6A4-6I4). RAR-vps (Nvp, ΔAvp, and Cvp for both RARab and RARga) induced ectopic expression of the endogenous targets (Figs. 5B3-D3, F3-H3, B4-D4, F4-H4). Like with transgene expression, ectopic expression of target genes was usually restricted to the trunk region of the notochord or to regions adjacent to sites of endogenous expression (Figs. 5B3-D3, F3-H3, B4-D4, F4-H4). In contrast, the RAR-ΔFvp fusion proteins (for both RARab and RARga) did not induce ectopic expression of dhrs3a and hoxb5b, but instead seemed to slightly reduce their endogenous expression (Figs. 5E3, E4, I3, I4).
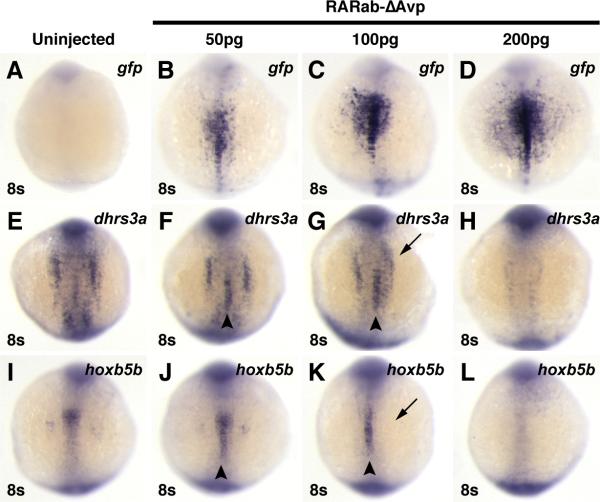
Figure 6. Injection of high doses of hyperactive RAR mRNA can inhibit target gene expression.
Injection of increasing concentrations of rarabΔAvp mRNA increasingly activates reporter expression (A-D), but inhibits endogenous target gene expression (E-L). (A, E, I) Uninjected control embryos. (B, F, J) In embryos injected with 50 pg of rarabΔAvp mRNA, modest levels of the reporter are induced (B), as well as ectopic expression of dhrs3a and hoxb5b (arrowheads in F and J). (C, G, K) In embryos injected with 100 pg of rarabΔAvp mRNA, higher levels of the reporter are induced (C). Although ectopic activation of dhrs3a and hoxb5b is induced (arrowheads in G and K), their endogenous expression is also inhibited (arrows in G and K). (D, H, L) In embryos injected with 200 pg of rarabΔAvp mRNA, the highest levels of the reporter are induced (D), while endogenous dhrs3a and hoxb5b expression are inhibited and ectopic expression is not induced (arrowheads in H and L). All images are dorsal views with anterior up.
For the RAR-vps (Nvp, ΔAvp, and Cvp) that did induce ectopic expression of the target genes, regions of endogenous expression were sometimes lost (Figs. 5B4, C4, F4, H4). This suggested that increasing the amount of RAR-vp expression may also inhibit endogenous target gene expression. Through titrating the amounts of rarab-ΔAvp mRNA injected, we found that the lowest concentrations did not affect normal expression and induced ectopic target gene expression in the notochord, while the highest concentrations inhibited all expression of target genes (Figs. 6E-L). In contrast to its effects on target genes, increasing the amount of rarab-ΔAvp mRNA injected induced stronger reporter expression (Figs. 6A-D). Altogether, in contrast to the differential abilities of RAR-vps to activate the RARE transgene, there was little difference between the abilities of zebrafish RARab-vps and RARga-vps to induce ectopic expression of endogenous target genes. However, the inhibition of target genes by both the RAR-ΔFvps and higher levels of RARab-ΔAvp emphasize that a proper level of regulation of RAR function is needed for correct expression of target genes in endogenous locations, which we postulate is dictated through the availability of co-factors.
Incorporating all of the experiments with RAR-vps, our results suggest that previously described RAR-vps function as hyperactive RARs. Importantly, while these function as transcriptional activators of synthetic RARE reporters, they can also inhibit expression of endogenous targets. This context-dependence of RAR-vp function could have significance to the interpretation of studies incorporating these fusion proteins (Blumberg et al., 1997; Koide et al., 2001). Finally, while we do observe differences between the effects of RARab and RARga constructs on synthetic reporters, these may be a bit misleading, as RARab and RARga constructs have similar abilities to affect endogenous target gene expression in the context of the embryo.
Context-dependent function of dominant negative zebrafish RARs
Having found that hyperactive zebrafish RARs can activate transcription in the early embryo, we next wanted to test the ability of the zebrafish RARs to act as transcriptional repressors. We did this by truncating the majority of the RAR F domain, in the same manner as for previously reported dominant negative RARs (dnRARs) (Fig. 3; Damm et al., 1993; Koide et al., 2001). These truncated proteins are thought to act as dominant transcriptional repressors due to an inability to release the transcriptional repressive machinery in the presence of RA (Damm et al., 1993; Koide et al., 2001). In Xenopus, injection of these constructs has been shown to cause patterning defects of the anterior CNS consistent with loss of RA signaling (Blumberg et al., 1997; Koide et al., 2001; Kolm et al., 1997; Sharpe and Goldstone, 1997). To confirm that the zebrafish dnRARs can function similarly to the previously studied human and Xenopus dnRARs, we first tested their ability to function in HEK 293 cells.
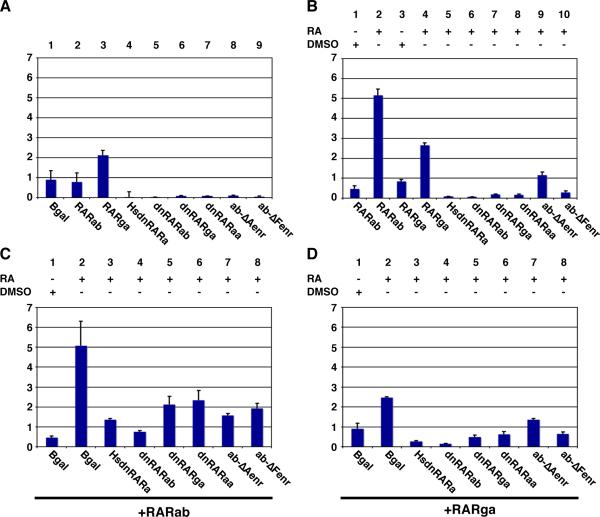
HEK 293 cells appear to exhibit a low level of endogenous RA signaling that can activate basal levels of the RARE reporters (Fig. 7A – column 1); this activation is not seen when the RARE sites have been deleted from the reporter (JSW and DY, unpublished data). Additionally, transfection of the RA-dependent RAR-vps results in reporter activation, with similar trends of activation to those observed in Tg(12XRARE-ef1a:gfp) embryos (Fig. 4Q). In contrast to the benign effects of injecting WT rarab and rarga mRNA into zebrafish embryos, both zebrafish RARs were able to stimulate reporter expression in HEK 293 cells upon treatment with RA, although activation by RARga was consistently less than activation by RARab (Fig. 7B – columns 1-4). Interestingly, without RA treatment, RARga caused more of an activation of the reporter than did RARab (Fig. 7A – columns 1 and 3). This difference parallels what has been reported for the human RARa and RARg (Farboud et al., 2003; Hauksdottir et al., 2003), suggesting that, in this context, zebrafish RARa and RARg proteins have characteristic differences that are conserved with other vertebrates.
Figure 7. Dominant negative RARs can act as transcriptional repressors in HEK 293 cells.
(A) Transfection of zebrafish RARab does not affect the basal activation of the reporter (column 2), but transfection of zebrafish RARga causes a modest activation (column 3). Transfection of the dominant negative and Enr fusion proteins inhibits basal activation of the reporter (columns 4-9). (B) RA treatment of cells transfected with RARab or RARga activates the reporter (columns 2 and 4). RA treatment of HEK 293 cells transfected with any of the dominant negative proteins or with RARabΔFenr does not activate the reporter (columns 5-8 and 10), while RARabΔAenr modestly activates the reporter (column 9). (C) The dominant negative and Enr fusion proteins can inhibit the activation of the reporter that is induced by RA treatment of cells transfected with zebrafish RARab. (D) The dominant negative and Enr fusion proteins can inhibit the activation of the reporter that is induced by RA treatment of cells transfected with zebrafish RARga. All bar graphs reflect activation of the pGL3-basic:12XRAR-ef1a reporter in terms of fold difference of AU and are presented as in Figure 4.
Having established that the zebrafish RARs are able to activate the reporter in HEK 293 cells in the presence of RA, we then tested the ability of zebrafish dnRARs to function as dominant transcriptional repressors. Alone, the zebrafish and human dnRARs inhibited basal reporter activation (Fig. 7A – columns 4-6) and were not activated by RA treatment (Fig. 7B – columns 5-7). In the presence of zebrafish RARs (ab and ga), we found that both zebrafish dnRARs (ab and ga), like the human dnRARa, were able to inhibit RAR function (Fig. 7C – columns 1-5, and Fig. 7D – columns 1-5).
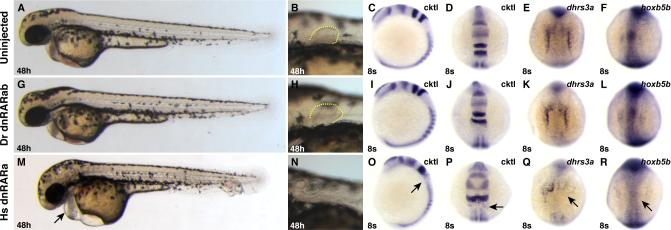
Because the zebrafish dnRARs could function as dominant transcriptional repressors in this heterologous signaling assay, we next tested if they could act similarly in the zebrafish embryo. Previous studies have shown that injection of human dnrara mRNA into zebrafish embryos can affect expression of hox genes (Roy and Sagerström, 2004) and that heat-shock induction of a human dnrara transgene can result in loss of pectoral fins, abnormal hindbrain patterning and heart enlargement (Waxman et al., 2008), all of which are characteristic of loss of RA signaling. Consistent with those reports, we found that injection of human dnrara mRNA caused loss of the pectoral fin, expansion of the anterior hindbrain, loss of rhombomere 5, and loss of dhrs3a and hoxb5b expression (Figs. 8A-F, M-R). However, zebrafish embryos injected with zebrafish dnRARab or dnRARga appeared normal, and expression of endogenous target genes was not affected (Figs. 8G-L and Figs. S5B,G; JSW and DY, data not shown). Therefore, it appears that, despite their ability to function in HEK 293 cells, the zebrafish dnRARab and dnRARga cannot function as dominant transcriptional repressors in the context of the early zebrafish embryo.
Figure 8. Zebrafish dnRARab does not act as a transcriptional repressor in zebrafish embryos.
(A-F) Uninjected control embryos. (G-L) Embryos injected with zebrafish dnrarab mRNA appear equivalent to uninjected control embryos. (M-R) Injection of human dnrara mRNA causes phenotypes consistent with loss of RA signaling, including enlarged hearts (arrow in M), loss of pectoral fin (N), loss of rhombomere 5 (arrows in O and P), and loss of endogenous target gene expression (arrows in Q and R). Phenotypes induces by injection of the human dnrara mRNA are similar to, if not stronger than, those induced when the human dnRARa is expressed from a heat-shock inducible transgene (Waxman et al., 2008). Cktl of probes is the same as in Figure 6. (A-C, G-I, M-O) Images are lateral views with dorsal to the right. (D-F, J-L, P-R) Images are dorsal views, anterior up.
Since others have reported the use of a zebrafish dnRARaa (the paralog of RARab) as a repressor (Stafford and Prince, 2002), we were hopeful that differences between zebrafish RARaa and RARab could explain the contextual ability to act as a transcriptional repressor. However, we found that dnraraa mRNA, like dnrarab mRNA, did not cause phenotypes consistent with strong loss of RA signaling (JSW and DY, data not shown), including repression of target genes (Fig. S7C,H), even though both dnRARaa and dnRARab can function as transcriptional repressors in cell culture (Fig. 7A-D). Therefore, we cannot completely rule out that there may be some ability of zebrafish RARs to function as transcriptional repressors. However, it is likely that the effects of the zebrafish dnRARaa are minimal, since it needed to be co-injected with RAR-specific morpholinos to see a loss of RA signaling phenotype in the previously reported experiments (Stafford and Prince, 2002).
Due to the high conservation of human and zebrafish RARs, we were surprised to find that the zebrafish dnRARs, unlike human dnRARa, did not act as dominant transcriptional repressors in the embryo. It is possible that low levels of expression of exogenous dnRARs might not be able to interfere with endogenous RAR activity. Although no tools are currently available to monitor endogenous zebrafish RAR protein levels, we do not think that the lack of function of zebrafish dnRARs reflects their poor expression, since tagged versions of zebrafish dnRARs are expressed ubiquitously and robustly in injected embryos, at levels that we assume to be in excess of the endogenous RARs (Fig. S2). Therefore, we next tried to increase the repressive capabilities of zebrafish RARab by replacing either its A or F domain with the Engrailed repressor (Enr) domain (RARab-ΔAenr and -ΔFenr; Fig. 3A). We replaced the A and F domains because replacing them with the VP16 domain in either zebrafish RARab or RARga had produced some of the more potent hyperactive proteins. In HEK 293 cells, both RARab-enr proteins were able to inhibit basal activity of the reporter (Fig. 7A – columns 8 and 9), but RARab-ΔAenr modestly activated the RARE reporter in the presence of RA (Fig. 7B – column 9). Interestingly, this modest activation by RARab-ΔAenr is reminiscent of the activity of the PML-RAR fusion protein implicated in acute myelogenous leukemia (AML), after which this fusion protein was modeled, which can activate RA targets in the presence of high RA concentrations (Soprano et al., 2004). Like the zebrafish dnRARs, the zebrafish RARab-enr proteins were able to inhibit zebrafish RAR activation of the RARE reporter in HEK 293 cells (Fig. 7C, D; columns 7 and 8). However, as with the zebrafish dnRARs, the zebrafish RARab-enr constructs did not affect expression of endogenous target genes or alter embryonic phenotypes (Fig. S7D, E , I, J; JSW and DY, data not shown). Since even the addition of an Enr domain does not allow zebrafish RARs to act as dominant transcriptional repressors in the embryo, it seems likely that the zebrafish RARs are not required to act as transcriptional repressors in this context.
Discussion
Together, our results indicate that zebrafish RARs function as context-dependent transcriptional activators. Ectopic expression of WT zebrafish RARs does not affect development nor sensitize the embryo to RA treatment. Hyperactive RARs can activate RA-responsive targets, although different VP16 fusions exhibit differing abilities to function depending on the context analyzed. To our surprise, we did not find that zebrafish versions of dominant negative RARs were able to act as transcriptional repressors in the early embryo, even though a human dnRAR can produce phenotypes consistent with loss of RA signaling. Together, our studies suggest that, during early A-P patterning of the zebrafish embryo, RARs function primarily as transcriptional activators rather than via a binary repressor-activator mechanism. Thus, it seems that the binary model for RAR function does not apply to all in vivo scenarios.
Our analysis of zebrafish RA-responsive transgenes demonstrates that RARE sites alone cannot recapitulate the expression patterns of all RA-responsive target genes, such as some anterior hox genes or dhrs3a (Marletaz et al., 2006; Waxman et al., 2008). The post-gastrulation initiation of RARE reporter transgene expression in zebrafish, observed in our studies and by Perz-Edwards and colleagues (2001), contrasts with the expression of two similar mouse transgenes, which begins in pregastrula embryos and more closely recapitulates the expression of known target genes (Balkan et al., 1992; Perz-Edwards et al., 2001; Rossant et al., 1991). This contrast suggests the intriguing notion that there are species-specific differences in RARE responsiveness. However, we cannot rule out the possibility that these differences simply reflect position-dependent influences upon differentially integrated transgenes.
Restricting the comparison to different zebrafish RA-responsive transgenes, it seems likely that differences in expression result from locus-specific influences. For example, the more restricted expression of our reporters, in contrast to the more expansive expression of previously reported transgenes (Perz-Edwards et al., 2001), may reflect the ability of our longer transgenes to better insulate themselves against outside influences. In support of strong context-dependent influences on minimal RARE sites, a recent study has examined the zebrafish cyp26a1 promoter, which is RA-responsive due to two RARE sites, but whose expression is also known to be controlled by other signaling pathways (Hu et al., 2008). Transgenes driven by the cyp26a1 promoter have spatial and temporal expression patterns closely resembling the early expression of the endogenous gene, which differ significantly from the expression patterns of the synthetic RARE reporter transgenes presented here and previously (Perz-Edwards et al., 2001). This further suggests that other transcription factors, acting either within or outside the transgene sequences, heavily influence the temporal and spatial responsiveness of genes with RARE sites within the zebrafish embryo.
The context-dependent nature of influences on RARE sites in the zebrafish embryo fits with the notion that the function of zebrafish RARs may be highly context-dependent. 3 of the 4 zebrafish RARs (RARaa, RARab and RARgb) are expressed ubiquitously prior to gastrulation, while later RARgb is expressed ubiquitously at low levels (Waxman and Yelon, 2007). Thus, the locations where RA signaling occurs are likely to be dictated primarily by the availability of RA and other collaborating factors, rather than by the RARs themselves. We found multiple distinctions between the abilities of the RARvp fusion proteins in three different functional assays: RARE reporter transgene expression, endogenous target gene expression, and CNS patterning phenotypes. In general, RARab-vps were able to more strongly induce expression from the transgenic reporter compared to the RARga-vps, although there were also subtle differences in activation ability among the RARab-vps and RARga-vps. In contrast to the differences found with the RARE reporter transgenes, the RARab-vps and RARga-vps similarly induced the ectopic expression of endogenous target genes. The endogenous promoters may represent a more permissive context, which buffers differences between specific RARs within the embryo. Because the expression of endogenous target genes can be repressed by an overload of RAR-vp expression or the lack of an F domain, coordinated input from other factors must also be necessary for proper regulation of target gene expression.
Most of the hyperactive zebrafish RARs can eliminate the MHB, similar to the reported effects of increased RA signaling in Xenopus (Blumberg et al., 1997; Koide et al., 2001). However, recent studies in zebrafish have not suggested a conserved role for RARs to act as repressors in the same context, although it cannot be ruled out that this could be due to an inability to adequately deplete the zebrafish RARs (Linville et al., 2008). Interestingly, we have found that the individual zebrafish RAR-vps have differing potency in affecting the anterior CNS, although these differences did not correlate with their ability to activate the RARE reporter transgene. These differences in ability to affect the CNS seem to be additional indicators of context-specific modifiers at the transcriptional level in the embryo. Together, the distinctions between the assays revealed by the hyperactive RARs indicate that transgenic reporter activation can at times be misleading relative to the effect on endogenous targets. Thus, multiple convergent assays should be performed to achieve a complete assessment of RAR function in a given in vivo context.
In contrast to the abilities of the hyperactive RARs, zebrafish dnRARs are not capable of acting as transcriptional repressors in the early zebrafish embryo, suggesting that zebrafish RARs may have minimal endogenous requirement as transcriptional repressors. To our surprise, even replacing the RARab A- or F-domains with an Enr domain does not allow the chimeric proteins to function as transcriptional repressors in the zebrafish embryo. We have found that fusing the Enr domain to a zebrafish retinoid X receptor (a related nuclear hormone receptor and heterodimeric partner of the RARs) can specifically affect its function and strongly abrogate normal development when injected into zebrafish embryos (JSW and DY, unpublished observations), indicating that the Enr domain can alter transcriptional function of members of the nuclear hormone family of receptors in zebrafish. Furthermore, both the zebrafish dnRAR and RAR-enr proteins are able to function as transcriptional repressors in heterologous cell culture signaling assays. It is possible that the ineffectiveness of the ectopically expressed dnRAR and RAR-enr proteins in zebrafish embryos is due to technical limitations, such as a lack of significant overexpression above endogenous protein levels or variability between expression levels of different proteins. However, we think these scenarios are unlikely, based on the observed robust expression of tagged versions of these proteins (Fig. S2).
What we have found more perplexing is that a human dnRARa can cause phenotypes resembling loss of RA signaling in the zebrafish embryo, suggesting it is able to function as a transcriptional repressor (Roy and Sagerström, 2004; Waxman et al., 2008) in this context. We presume that there are structural differences between human and zebrafish RARs that are responsible for their context-dependent functional differences, but it is not yet clear where these differences reside. The human and zebrafish dnRARs used are 78% identical and 85% similar, with most of the differences in the variable A domain and hinge region/D domain. Initial assessments of human-zebrafish chimeric dominant negative proteins, made by fusing the human RARa A-C domains to the D-ΔF domains of the zebrafish RARab dominant negative protein and vice versa, were not informative, since both chimeras were able to act as weak dominant negative proteins relative to the human dnRARa (Fig. S8). Therefore, it appears that aspects of both halves of the human protein are required to confer ability to act as a repressor within the context of the early zebrafish embryo. Future studies employing additional chimeras will be necessary to discern which residues are important for the context-specific differences. Altogether, the inability of the zebrafish dnRARs to function in the zebrafish embryo, in contrast to the ability of the human dnRARa, emphasizes the inherent differences of these homologous proteins and the limitations of a binary model for RAR function in some developmental contexts.
Together, our data suggest there is significant context dependence for the responsiveness of RA signaling at the transcriptional level. Putting these results into a broader perspective, such context dependence is not surprising in light of mechanisms for transcriptional regulation of developmental genes in other settings. Elegant studies of gene regulatory networks in organisms such as Drosophila, sea urchins, and frogs have demonstrated that precise tissue-specific regulation of gene expression often involves multiple cis-regulatory modules harboring multiple transcriptional elements that correspond to transcription factors with context-dependent functions (Levine, 2010; Levine and Davidson, 2005; Wilczynski and Furlong, 2010). Future studies will be aimed at elucidating the precise function of RARs in a variety of contexts and the nature of their interactions with partners in cis-regulatory modules containing RAREs.
The acquisition of RA signaling and its control of hox gene expression has been associated with development of the chordate body plan (Marletaz et al., 2006). Although RA signaling has been shown to regulate hox1 expression in amphioxus (Schubert et al., 2005), it has been shown recently that the thalacian urochordates have lost all of the major components important for RA signaling (RAR, Cyp26 and Raldh) and do not require it for A-P patterning (Canestro and Postlethwait, 2007). While ascidian urochordates have retained the RA signaling machinery, it appears that they may not require RA signaling for A-P patterning either (Canestro and Postlethwait, 2007). It is clear that RA signaling is required for A-P patterning in zebrafish (Begemann et al., 2001; Grandel et al., 2002; Maves and Kimmel, 2005); however, our results suggest that the regulation of RA target genes may have a strong dependence on additional transcriptional regulators in zebrafish embryos. Based on studies of RA-responsive genes and the more expansive expression of RARE reporter transgenes in mice (Balkan et al., 1992; Oosterveen et al., 2003; Rossant et al., 1991; Sharpe et al., 1998), we also hypothesize that RA signaling in tetrapods may function more independently at the transcriptional level.
Assimilating the observations from basal chordates, zebrafish, and tetrapods, we propose two possible models for the evolution of RAR function. In the first model, the ancestral role of RA signaling and RARs during early A-P patterning could have been subordinate to or redundant with other necessary transcription factors. If the role of RA signaling were minimal, acting only in concert with other transcription factors in the common chordate ancestor, it would be easier for these other factors to compensate for loss of RA signaling in the urochordates and maintain proper embryonic A-P patterning. In contrast, tetrapod RARs could have become better able to interact with regulators of transcriptional machinery and therefore less dependent on interactions with other cis-regulators, allowing RA signaling to take on a more singular role in regulation of A-P patterning. Thus, RARs may have evolved from being transcription factors with minimal singular competence during A-P patterning of the chordate body plan to having a more pronounced individual role in tetrapods. Alternatively, in the second model, the manner in which the zebrafish RARs function could be a derived characteristic of teleosts, or even zebrafish specifically. In this scenario, there would be little or no functional difference between RARs from amphioxus and mammals, while teleost RARs have lost the ability to act as repressors in some contexts. Other explanations would therefore be needed to explain why and how proper A-P patterning is maintained in the absence of RA signaling in urochordates. Future studies comparing RARs and the regulation of RA signaling target genes in chordates will allow for an enhanced understanding of the evolution of RA signaling and its role in A-P patterning of the chordate body plan.
Supplementary Material
Supplemental Figure Legends
Supplemental Figure 1. RAR-specific pharmacological reagents affect expression of Tg(12XRARE-ef1a:gfp), while pharmacological reagents specific for other nuclear hormone receptors do not. (A) Untreated control embryo. (B) Ro41-5253, a RARα-specific antagonist, inhibits reporter expression. (C) AM580, a RARα-specific agonist, activates ectopic reporter expression. Arrows (ventral eye) and arrowheads (anterior spinal cord) in B and C indicate where restricted expression is normally found in an untreated embryo (A). (D) The pan-RXR agonist methoprene acid (Biomol.com) does not affect reporter expression. Agonists of (E) PPARα (GW7647), (F) PPARδ (GW0742), and (G) PPARγ (GW1929) do not activate the reporter. The pan-PPAR antagonist LY171883(H) and the PPARα specific antagonist GW6471 (I) do not inhibit reporter expression. All PPAR-related reagents were purchased from Sigma. All images are lateral views, with anterior up and dorsal to the right.
Supplemental Figure 2. Myc-tagged zebrafish RARs are expressed in zebrafish embryos. (A) Western blot of myc-tagged Hs dnRAR and zebrafish WT RARs and dnRARs at 70-90% epiboly in zebrafish embryos that were injected with the indicated mRNAs. The decreasing relative expression from left to right reflects the timing of execution for this representative experiment, in which constructs on the left were injected earlier than those on the right. Approximate sizes of the myc-tagged proteins are: Hs myc-dnRARa, 55kDa; Dr myc-RARab, 61kDa; Dr myc-dnRARab, 55kDa; Dr myc-RARga, 67kDa; Dr myc-dnRARga, 52kDa. The approximate size of α-tubulin is 50kDa. (B-D) Wholemount immunofluorescence at 70-90% epiboly of zebrafish embryos that were injected with mRNAs for myc-tagged human and zebrafish RARs. Exogenous protein expression was analyzed at mid- to late-gastrulation stages because this is when RA signaling is required to pattern the early zebrafish embryo (Hernandez et al., 2007; Hernandez et al., 2004; Linville et al., 2008; Maves and Kimmel, 2005; Stafford et al., 2006).
Both analyses indicate that, while there may be differences in precise expression levels of different exogenous RARs, all of the exogenous RARs are expressed broadly and robustly in zebrafish embryos at these stages. Unfortunately, there are no commercially available anti-RAR antibodies that allow evaluation of the endogenous levels of zebrafish RARs. If we presume that endogenous α-tubulin is likely to be present at higher levels than endogenous RARs, the qualitative comparison of exogenous RARs and endogenous α-tubulin suggests that the exogenous RARs are present in excess of their endogenous counterparts.
Supplemental Figure 3. RARabvps and RARgavps differentially activate reporter expression in RARE transgenic reporter lines. (A-D) Ectopic expression of RAR-vps can induce ectopic expression of Tg(12XRARE-ef1a:gfp) at 24 hpf. (E-H) Ectopic expression of RAR-vps can induce ectopic expression of Tg(12XRARE-tk:gfp) at 24 hpf. Induction of expression by RARabvps (center in A-H) is typically stronger than that induced by RARgavps (right in A-H). Uninjected sibling embryos are at the left in A-H. All images are lateral views, with anterior up and dorsal to the right.
Supplemental Figure 4. Hyperactive RAR-vps induce regional activation of the RARE reporter. Tg(12XRARE-ef1a:gfp) embryos overexpressing RAR-vps. (A) Control uninjected embryos and embryos injected with (B) rarab-Nvp, (C) rarab-ΔAvp, (D) rarab-Cvp, (E) rarab-ΔFvp, (F) rarga-Nvp, (G) rargaΔAvp, and (H) rargaΔFvp mRNAs. Reporter expression is enhanced in regions of the embryo near a source of RA. Arrow in C indicates expanded ventral eye expression. Arrowhead in C indicates reporter expression within the trunk. All images are lateral views, with anterior up and dorsal to the right.
Supplemental Figure 5. Phenotypes induced by RA treatment. (A-C) RA treatment eliminates anterior neural gene expression. Cktl of probes is the same as in Figure 6. (A,D,G) Untreated control embryos. (B) Treatment with 0.5 μM RA can strongly posteriorize embryos, resulting in loss of all anterior neural markers. (C) Treatment with 0.2 μM RA can yield moderately posteriorized embryos in which the MHB is lost and the expression of krox20 in rhombomere 3 is reduced. This more modest posteriorization is remniscent of rar-vp mRNA-injected embryos (Figure 6), though the eyes of rar-vp mRNA-injected embryos are never lost. (D-I) RA signaling positively regulates expression of dhrs3a and hoxb5b. (E, H) Treatment with DEAB inhibits dhrs3a and hoxb5b expression. (F, I) Treatment with 0.5 μM RA induces ectopic dhrs3a and hoxb5b expression. Images in A-C are lateral views with dorsal to the right. All other images are dorsal views with anterior up.
Supplemental Figure 6. Ectopic expression of hyperactive RAR-vps does not phenocopy all aspects of RA treatment. (A) Uninjected control embryo. (B) Treatment with 0.5 μM RA causes loss of the anterior CNS (arrow). (C) Treatment with 0.2 μM RA causes dysmorphic, reduced eyes (arrow and dashed yellow outline) and loss of the MHB. Injection of (D) rarab-Nvp, (E) rarab-Cvp, (F) rarga-Nvp, (G) rarga-Cvp, (H) rarab-ΔAvp, (I) rarab-ΔFvp, and (K) rarga-ΔFvp mRNA can eliminate the MHB (arrowheads), but the eyes are not affected (arrows and dashed yellow outline). rarab-Nvp and rarab-Cvp mRNAs are particularly effective at eliminating the MHB. (J) Injection of rarga-ΔAvp mRNA does not affect the MHB (arrowhead).
Supplemental Figure 7. Zebrafish dominant negative RARs and Enr fusion proteins do not inhibit expression of RA-responsive target genes. (A, F) Uninjected control embryos. (B-E, G-J) Injection of mRNAs for zebrafish dnrarga (B, G), dnraraa (C, H), rarab-ΔAenr (D, I) and rarab-ΔFenr (E, J) do not significantly affect expression of RA-responsive target genes. All images are dorsal views with anterior up.
Supplemental Figure 8. Chimeric human/zebrafish dominant negative RARs can inhibit expression of RA-responsive target genes. (A-D) Uninjected control embryos. (E-H) Embryos injected with mRNA encoding a hs-dr RAR fusion protein, in which the human A-C domains are fused to the zebrafish D-ΔF domains. (I-L) Embryos injected with mRNA encoding a dr-hs RAR fusion protein, in which the zebrafish A-C domains are fused to the human D-ΔF domains. Although neither chimeric protein was as effective as the human dnRARa (Fig. 8O-R), either can inhibit expression of RA-responsive genes.
Acknowledgements
We are grateful to Ram Dasgupta and Foster Gonsalves for their generous assistance with establishing luciferase assays. We would also like to thank Holger Knaut and Jennifer Schumacher for comments on the manuscript and members of the Yelon and Torres-Vázquez labs for helpful discussions. This work was supported by NIH R01 HL069594 to DY. JSW was supported by NIH NRSA F32 HL083591 and NIH Pathway to Independence Award K99 HL091126.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- Balkan W, Colbert M, Bock C, Linney E. Transgenic indicator mice for studying activated retinoic acid receptors during development. Proc Natl Acad Sci U S A. 1992;89:3347–51. doi: 10.1073/pnas.89.8.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–94. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Bolado J, Jr., Moreno TA, Kintner C, Evans RM, Papalopulu N. An essential role for retinoid signaling in anteroposterior neural patterning. Development. 1997;124:373–9. doi: 10.1242/dev.124.2.373. [DOI] [PubMed] [Google Scholar]
- Canestro C, Postlethwait JH. Development of a chordate anterior-posterior axis without classical retinoic acid signaling. Dev Biol. 2007;305:522–38. doi: 10.1016/j.ydbio.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Izpisua Belmonte JC. Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Cash DE, Bock CB, Schughart K, Linney E, Underhill TM. Retinoic acid receptor alpha function in vertebrate limb skeletogenesis: a modulator of chondrogenesis. J Cell Biol. 1997;136:445–57. doi: 10.1083/jcb.136.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Damm K, Heyman RA, Umesono K, Evans RM. Functional inhibition of retinoic acid response by dominant negative retinoic acid receptor mutants. Proc Natl Acad Sci U S A. 1993;90:2989–93. doi: 10.1073/pnas.90.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, Hauksdottir H, Wu Y, Privalsky ML. Isotype-restricted corepressor recruitment: a constitutively closed helix 12 conformation in retinoic acid receptors beta and gamma interferes with corepressor recruitment and prevents transcriptional repression. Mol Cell Biol. 2003;23:2844–58. doi: 10.1128/MCB.23.8.2844-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol. 2010;338:1–14. doi: 10.1016/j.ydbio.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Kuchler AM, Schulte-Merker S, Geisler R, Holder N, Wilson SW, Brand M. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–65. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- Hale LA, Tallafuss A, Yan YL, Dudley L, Eisen JS, Postlethwait JH. Characterization of the retinoic acid receptor genes raraa, rarab and rarg during zebrafish development. Gene Expr Patterns. 2006 doi: 10.1016/j.modgep.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Hauksdottir H, Farboud B, Privalsky ML. Retinoic acid receptors beta and gamma do not repress, but instead activate target gene transcription in both the absence and presence of hormone ligand. Mol Endocrinol. 2003;17:373–85. doi: 10.1210/me.2002-0340. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–87. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–20. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- Hu P, Tian M, Bao J, Xing G, Gu X, Gao X, Linney E, Zhao Q. Retinoid regulation of the zebrafish cyp26a1 promoter. Dev Dyn. 2008;237:3798–808. doi: 10.1002/dvdy.21801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iulianella A, Lohnes D. Chimeric analysis of retinoic acid receptor function during cardiac looping. Dev Biol. 2002;247:62–75. doi: 10.1006/dbio.2002.0685. [DOI] [PubMed] [Google Scholar]
- Kastner P, Messaddeq N, Mark M, Wendling O, Grondona JM, Ward S, Ghyselinck N, Chambon P. Vitamin A deficiency and mutations of RXRalpha, RXRbeta and RARalpha lead to early differentiation of embryonic ventricular cardiomyocytes. Development. 1997;124:4749–58. doi: 10.1242/dev.124.23.4749. [DOI] [PubMed] [Google Scholar]
- Koide T, Downes M, Chandraratna RA, Blumberg B, Umesono K. Active repression of RAR signaling is required for head formation. Genes Dev. 2001;15:2111–21. doi: 10.1101/gad.908801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolm PJ, Apekin V, Sive H. Xenopus hindbrain patterning requires retinoid signaling. Dev Biol. 1997;192:1–16. doi: 10.1006/dbio.1997.8754. [DOI] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–63. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102:4936–42. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linville A, Radtke K, Waxman JS, Yelon D, Schilling TF. Combinatorial roles for zebrafish retinoic acid receptors in the hindbrain, limbs and pharyngeal arches. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P, Gansmuller A, Chambon P. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–48. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–80. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- Marletaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci. 2006;2:38–47. doi: 10.7150/ijbs.2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–71. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Mintzer KA, Lee MA, Runke G, Trout J, Whitman M, Mullins MC. Lost-a-fin encodes a type I BMP receptor, Alk8, acting maternally and zygotically in dorsoventral pattern formation. Development. 2001;128:859–69. doi: 10.1242/dev.128.6.859. [DOI] [PubMed] [Google Scholar]
- Nagpal S, Saunders M, Kastner P, Durand B, Nakshatri H, Chambon P. Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell. 1992;70:1007–19. doi: 10.1016/0092-8674(92)90250-g. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–53. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Oosterveen T, Niederreither K, Dolle P, Chambon P, Meijlink F, Deschamps J. Retinoids regulate the anterior expression boundaries of 5' Hoxb genes in posterior hindbrain. Embo J. 2003;22:262–9. doi: 10.1093/emboj/cdg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz-Edwards A, Hardison NL, Linney E. Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol. 2001;229:89–101. doi: 10.1006/dbio.2000.9979. [DOI] [PubMed] [Google Scholar]
- Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci. 2006;26:13400–10. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Zirngibl R, Cado D, Shago M, Giguere V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–44. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- Roy NM, Sagerström CG. An early Fgf signal required for gene expression in the zebrafish hindbrain primordium. Brain Res Dev Brain Res. 2004;148:27–42. doi: 10.1016/j.devbrainres.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–23. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- Russo JE, Hauguitz D, Hilton J. Inhibition of mouse cytosolic aldehyde dehydrogenase by 4-(diethylamino)benzaldehyde. Biochem Pharmacol. 1988;37:1639–42. doi: 10.1016/0006-2952(88)90030-5. [DOI] [PubMed] [Google Scholar]
- Schubert M, Yu JK, Holland ND, Escriva H, Laudet V, Holland LZ. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development. 2005;132:61–73. doi: 10.1242/dev.01554. [DOI] [PubMed] [Google Scholar]
- Schulze GE, Clay RJ, Mezza LE, Bregman CL, Buroker RA, Frantz JD. BMS-189453, a novel retinoid receptor antagonist, is a potent testicular toxin. Toxicol Sci. 2001;59:297–308. doi: 10.1093/toxsci/59.2.297. [DOI] [PubMed] [Google Scholar]
- Sharpe CR, Goldstone K. Retinoid receptors promote primary neurogenesis in Xenopus. Development. 1997;124:515–23. doi: 10.1242/dev.124.2.515. [DOI] [PubMed] [Google Scholar]
- Sharpe J, Nonchev S, Gould A, Whiting J, Krumlauf R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. Embo J. 1998;17:1788–98. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano DR, Qin P, Soprano KJ. Retinoic acid receptors and cancers. Annu Rev Nutr. 2004;24:201–21. doi: 10.1146/annurev.nutr.24.012003.132407. [DOI] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–20. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Stafford D, White RJ, Kinkel MD, Linville A, Schilling TF, Prince VE. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development. 2006;133:949–56. doi: 10.1242/dev.02263. [DOI] [PubMed] [Google Scholar]
- Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–83. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman JS, Yelon D. Comparison of the expression patterns of newly identified zebrafish retinoic acid and retinoid X receptors. Dev Dyn. 2007;236:587–95. doi: 10.1002/dvdy.21049. [DOI] [PubMed] [Google Scholar]
- Weston AD, Chandraratna RA, Torchia J, Underhill TM. Requirement for RAR-mediated gene repression in skeletal progenitor differentiation. J Cell Biol. 2002;158:39–51. doi: 10.1083/jcb.200112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston AD, Rosen V, Chandraratna RA, Underhill TM. Regulation of skeletal progenitor differentiation by the BMP and retinoid signaling pathways. J Cell Biol. 2000;148:679–90. doi: 10.1083/jcb.148.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski B, Furlong EE. Challenges for modeling global gene regulatory networks during development: insights from Drosophila. Dev Biol. 2010;340:161–9. doi: 10.1016/j.ydbio.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Williams JA, Kondo N, Okabe T, Takeshita N, Pilchak DM, Koyama E, Ochiai T, Jensen D, Chu ML, Kane MA, Napoli JL, Enomoto-Iwamoto M, Ghyselinck N, Chambon P, Pacifici M, Iwamoto M. Retinoic acid receptors are required for skeletal growth, matrix homeostasis and growth plate function in postnatal mouse. Dev Biol. 2009;328:315–27. doi: 10.1016/j.ydbio.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–7. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure Legends
Supplemental Figure 1. RAR-specific pharmacological reagents affect expression of Tg(12XRARE-ef1a:gfp), while pharmacological reagents specific for other nuclear hormone receptors do not. (A) Untreated control embryo. (B) Ro41-5253, a RARα-specific antagonist, inhibits reporter expression. (C) AM580, a RARα-specific agonist, activates ectopic reporter expression. Arrows (ventral eye) and arrowheads (anterior spinal cord) in B and C indicate where restricted expression is normally found in an untreated embryo (A). (D) The pan-RXR agonist methoprene acid (Biomol.com) does not affect reporter expression. Agonists of (E) PPARα (GW7647), (F) PPARδ (GW0742), and (G) PPARγ (GW1929) do not activate the reporter. The pan-PPAR antagonist LY171883(H) and the PPARα specific antagonist GW6471 (I) do not inhibit reporter expression. All PPAR-related reagents were purchased from Sigma. All images are lateral views, with anterior up and dorsal to the right.
Supplemental Figure 2. Myc-tagged zebrafish RARs are expressed in zebrafish embryos. (A) Western blot of myc-tagged Hs dnRAR and zebrafish WT RARs and dnRARs at 70-90% epiboly in zebrafish embryos that were injected with the indicated mRNAs. The decreasing relative expression from left to right reflects the timing of execution for this representative experiment, in which constructs on the left were injected earlier than those on the right. Approximate sizes of the myc-tagged proteins are: Hs myc-dnRARa, 55kDa; Dr myc-RARab, 61kDa; Dr myc-dnRARab, 55kDa; Dr myc-RARga, 67kDa; Dr myc-dnRARga, 52kDa. The approximate size of α-tubulin is 50kDa. (B-D) Wholemount immunofluorescence at 70-90% epiboly of zebrafish embryos that were injected with mRNAs for myc-tagged human and zebrafish RARs. Exogenous protein expression was analyzed at mid- to late-gastrulation stages because this is when RA signaling is required to pattern the early zebrafish embryo (Hernandez et al., 2007; Hernandez et al., 2004; Linville et al., 2008; Maves and Kimmel, 2005; Stafford et al., 2006).
Both analyses indicate that, while there may be differences in precise expression levels of different exogenous RARs, all of the exogenous RARs are expressed broadly and robustly in zebrafish embryos at these stages. Unfortunately, there are no commercially available anti-RAR antibodies that allow evaluation of the endogenous levels of zebrafish RARs. If we presume that endogenous α-tubulin is likely to be present at higher levels than endogenous RARs, the qualitative comparison of exogenous RARs and endogenous α-tubulin suggests that the exogenous RARs are present in excess of their endogenous counterparts.
Supplemental Figure 3. RARabvps and RARgavps differentially activate reporter expression in RARE transgenic reporter lines. (A-D) Ectopic expression of RAR-vps can induce ectopic expression of Tg(12XRARE-ef1a:gfp) at 24 hpf. (E-H) Ectopic expression of RAR-vps can induce ectopic expression of Tg(12XRARE-tk:gfp) at 24 hpf. Induction of expression by RARabvps (center in A-H) is typically stronger than that induced by RARgavps (right in A-H). Uninjected sibling embryos are at the left in A-H. All images are lateral views, with anterior up and dorsal to the right.
Supplemental Figure 4. Hyperactive RAR-vps induce regional activation of the RARE reporter. Tg(12XRARE-ef1a:gfp) embryos overexpressing RAR-vps. (A) Control uninjected embryos and embryos injected with (B) rarab-Nvp, (C) rarab-ΔAvp, (D) rarab-Cvp, (E) rarab-ΔFvp, (F) rarga-Nvp, (G) rargaΔAvp, and (H) rargaΔFvp mRNAs. Reporter expression is enhanced in regions of the embryo near a source of RA. Arrow in C indicates expanded ventral eye expression. Arrowhead in C indicates reporter expression within the trunk. All images are lateral views, with anterior up and dorsal to the right.
Supplemental Figure 5. Phenotypes induced by RA treatment. (A-C) RA treatment eliminates anterior neural gene expression. Cktl of probes is the same as in Figure 6. (A,D,G) Untreated control embryos. (B) Treatment with 0.5 μM RA can strongly posteriorize embryos, resulting in loss of all anterior neural markers. (C) Treatment with 0.2 μM RA can yield moderately posteriorized embryos in which the MHB is lost and the expression of krox20 in rhombomere 3 is reduced. This more modest posteriorization is remniscent of rar-vp mRNA-injected embryos (Figure 6), though the eyes of rar-vp mRNA-injected embryos are never lost. (D-I) RA signaling positively regulates expression of dhrs3a and hoxb5b. (E, H) Treatment with DEAB inhibits dhrs3a and hoxb5b expression. (F, I) Treatment with 0.5 μM RA induces ectopic dhrs3a and hoxb5b expression. Images in A-C are lateral views with dorsal to the right. All other images are dorsal views with anterior up.
Supplemental Figure 6. Ectopic expression of hyperactive RAR-vps does not phenocopy all aspects of RA treatment. (A) Uninjected control embryo. (B) Treatment with 0.5 μM RA causes loss of the anterior CNS (arrow). (C) Treatment with 0.2 μM RA causes dysmorphic, reduced eyes (arrow and dashed yellow outline) and loss of the MHB. Injection of (D) rarab-Nvp, (E) rarab-Cvp, (F) rarga-Nvp, (G) rarga-Cvp, (H) rarab-ΔAvp, (I) rarab-ΔFvp, and (K) rarga-ΔFvp mRNA can eliminate the MHB (arrowheads), but the eyes are not affected (arrows and dashed yellow outline). rarab-Nvp and rarab-Cvp mRNAs are particularly effective at eliminating the MHB. (J) Injection of rarga-ΔAvp mRNA does not affect the MHB (arrowhead).
Supplemental Figure 7. Zebrafish dominant negative RARs and Enr fusion proteins do not inhibit expression of RA-responsive target genes. (A, F) Uninjected control embryos. (B-E, G-J) Injection of mRNAs for zebrafish dnrarga (B, G), dnraraa (C, H), rarab-ΔAenr (D, I) and rarab-ΔFenr (E, J) do not significantly affect expression of RA-responsive target genes. All images are dorsal views with anterior up.
Supplemental Figure 8. Chimeric human/zebrafish dominant negative RARs can inhibit expression of RA-responsive target genes. (A-D) Uninjected control embryos. (E-H) Embryos injected with mRNA encoding a hs-dr RAR fusion protein, in which the human A-C domains are fused to the zebrafish D-ΔF domains. (I-L) Embryos injected with mRNA encoding a dr-hs RAR fusion protein, in which the zebrafish A-C domains are fused to the human D-ΔF domains. Although neither chimeric protein was as effective as the human dnRARa (Fig. 8O-R), either can inhibit expression of RA-responsive genes.