Abstract
Natural killer (NK) cells play an important role in innate immunity, especially in the response to viral infections, such as hepatitis C virus (HCV). Killer cell immunoglobulin-like receptors (KIRs) are the primary receptors of NK cells that mediate innate immunity. KIRs are also involved in acquired immunity, because some KIRs are expressed on the surface of certain subsets of T cells. In this study, the frequency of KIR genes, HLA-C allotypes, and combinations of KIR genes with their HLA-C ligands were evaluated in two different groups of the Korean population: controls and patients with chronic HCV infection. The study population consisted of 147 Korean patients with chronic HCV infection. The frequency of KIR2DS2 in patients with chronic HCV infection was 9.5% which was significantly lower than 19.5% of the control (P < 0.01). However, there were no significant differences in the frequency of other KIR genes, HLA-C allotypes or different combinations of KIR genes with their HLA-C ligands. This study can contribute to the further prospective study with a larger scale, suggesting the assumption that KIR2DS2 might aid in HCV clearance by enhancing both the innate and acquired immune responses of people in Korea.
Keywords: Hepatitis C Virus; Chronic Infection; Receptors, KIR; Korea
INTRODUCTION
Infection with hepatitis C virus (HCV) can result in both acute and chronic hepatitis. Acute infection rarely causes hepatic failure, but often leads to chronic infection (1). Approximately 20%-30% of chronically infected individuals develop liver cirrhosis (LC) over a 20-30 yr period, and a smaller percentage progress to hepatocellular carcinoma (HCC) (2). A recent survey showed that the worldwide mortality from HCV-related LC and HCC will double or triple within the next two decades (1). In Korea, the prevalence of HCV infection in general population is estimated to be approximately 1%, but it is reported to be as high as 80% in intravenous drug users among young adults in urban areas (3, 4).
In HCV infection, hepatic damage is caused primarily by the immune response, not by a direct cytopathic mechanism (5). Natural killer (NK) cells are a type of lymphocytes that plays an important role in the host defense against HCV infection (6, 7). They are critical effectors of early innate immunity toward virus infected cells (7). It has been reported that more than 40% of lymphoid cells in the liver are NK cells, compared with only 13% in peripheral blood cells (6, 7). NK cell function is determined by the net effect of signals to receptor families, including activating and inhibitory killer cell immunoglobulin-like receptors (KIRs) (8, 9). KIRs are a family of transmembrane glycoproteins expressed on the surface of NK cells and a certain subset of T cells, and they function as key regulators of the development, tolerance and activation of NK cells (9). NK cells play an important role in a wide range of diseases, and there have been many studies relating KIR genotypes to disease susceptibility, including viral infections such as HCV, autoimmune and inflammatory conditions, tumor immunity, preeclampsia, and recurrent spontaneous abortion (10, 11).
Combinations of KIRs form haplotypes with different inherent balances between inhibition and activation of NK cells (9). Until recently, only 16 KIR genes and pseudogenes have been identified, and group A and group B haplotypes were characterized by a dominance of genes encoding inhibitory and activating receptors respectively (11).
The ligands for KIRs are human leukocyte antigen (HLA) class I molecules (8). Several inhibitory KIR ligands are known, including the Bw4 motif common to a number of HLA-B and HLA-A alleles and a dimorphic motif that defines NK cell-mediated alloreactivity to known HLA-C alleles (12). Among KIRs, KIR2DL1, KIR2DL2, and KIR2DL3 bind their cognate HLA-C allotype C1 or C2 with different affinities (13). Among these combinations, KIR2DL1/HLA-C2 is known to provide the strongest inhibition of NK cells, and interactions of KIR2DL2/HLA-C1 and KIR2DL3/HLA-C2 follow in strength (14). Since people carry several different types of KIR genes and there are two HLA-C allotypes, various combinations of KIR and HLA-C occur, each with a different influence on either activating or inhibiting NK cell function (13, 14).
Among inhibitory combinations, KIR2DL3/HLA-C1 homozygosity provides the weakest inhibition of NK cells (13, 14). A protective association of the inhibitory receptor KIR2DL3 with HLA-C1 ligand and its effect on the course of HCV infection has been reported (15). The receptor-ligand combination of two copies of KIR2DL3 and HLA-C1 homozygosity provides weaker inhibitory signals than other inhibitory KIR and HLA-C receptor-ligand combinations and thus may render a more responsive NK cell phenotype (15). This effect was significant in the patients with low HCV exposure dose, and even more pronounced in patients who had acquired HCV infection from intravenous drug use than in patients who had received a blood transfusion and whose innate immune system might be lethargic (15).
Regarding innate immunity, HCV was shown to induce early changes in the expression of many intrahepatic genes, including genes involved in the type I interferon responses (12). However, these type I interferon responses in the liver do not correlate with the outcome of HCV infection. There were proposed mechanisms for the lack of correlation, including that specific HCV proteins might interfere with the function of NK cells (16, 17). In this context, when inhibitory KIRs and their HLA class I ligands dominate, NK cell function may be impaired and HCV infection is more likely to become chronic (15).
In this study, the frequency of KIR genes, HLA-C allotypes, and combinations of KIRs with their HLA-C ligands were evaluated in two different groups: controls and patients with chronic HCV infection. This is the first reported study to investigate the incidence of KIR genes and combinations of KIRs with HLA-C ligands in Korean HCV carrier cohort.
MATERIALS AND METHODS
Study population
The study population consisted of 147 Korean patients with chronic HCV infection who had no history of blood transfusions. The study includes 71 males and 76 females between 20 and 78 yr of age. Patients and controls were native Koreans and informed consent to participate in this study was obtained from all patients. The controls were 159 unrelated individuals who attended the Catholic Hemopoietic Stem Cell Information Bank. The control population was matched with the HCV-infected population by each risk factor of contracting HCV. The risk factors of HCV transmission were described as acupuncture, tattooing, hemodialysis, and intravenous drug use (4). Diagnosis of chronic HCV infection was based on seropositivity for anti-HCV antibody using third-generation enzyme immunoassays (Abbott Laboratories, North Chicago, IL, USA) and the confirmation of HCV RNA using the real-time polymerase chain reaction (Biosewoom Inc. Seoul, Korea). Additionally, HCV genotype was determined by VERSANT HCV genotype assay (LiPA 2.9, Innogenetics, Ghent, Belgium). The diagnosis of chronic liver disease due to HCV infection was based on clinical or histological analyses, including standard serological assays, biochemical liver function tests, radiological imaging, and/or liver biopsy.
HLA-C genotyping
The genotyping for HLA-C was performed by the amplification refractory mutation system-polymerase chain reaction (PCR) method. Each reaction contained a primer mixture consisting of allele- or group-specific primer pairs as well as internal control primers matching nonallelic sequences. Specific amplification of the HLA-C gene was performed using 33 primers for HLA-C. PCR was carried out in a reaction (13 µL) containing 100 200 ng genomic DNA, 0.8 × buffer (40 mM KCl, 1.2 mM MgCl2, 8.0 mM Tris-HCl, pH 8.8, 0.08% Triton X-100), 5% dimethyl sulfoxide (DMSO), 200 µM of each dNTP, 0.25 U Taq DNA polymerase (Boehringer Mannheim, Germany), 1 µM of each sequence-specific primer and 0.2 µM of internal control primers. The amplifications were performed in a My Cycler thermocycler (BioRad, Hercules, CA, USA). In total, 30 cycles were used for the amplification with the following steps: heating to 96℃ for 1 min to denature the DNA, denaturation at 96℃ for 25 sec, annealing at 70℃ for 45 sec, and extension at 72℃ for 30 sec (for the first 5 cycles); 96℃ for 25 sec, 65℃ for 45 sec, 72℃ for 30 sec (for the next 21 cycles); 96℃ for 25 sec, 55℃ for 60 sec, 72℃ for 120 sec (for the last 4 cycles); and a final 1-min extension at 72℃. The presence or absence of PCR products was determined after separation on a 1.5% agarose gel containing 0.5 µg/mL ethidium bromide. HLA-C alleles were assigned to HLA-C1 or HLA-C2 groups, as defined by KIR specificity. An individual who typed solely as C2 was referred to as homozygous for C2 alleles (genotype C2/C2), whereas an individual who typed as C1 alone was referred to homozygous for C1 alleles (genotype C1/C1). The remainders were described as C1/C2 heterozygotes.
Killer cell immunoglobulin-like receptor genotyping
The KIR alleles were determined by PCR using sequence-specific primers (PCR-SSP) methodology. Inhibitory KIR were typed for KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5, KIR3DL1, KIR3DL2 and KIR3DL3 alleles, and activating KIRs for KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5 and KIR3DS1 alleles. Specific amplifications of the KIR genes were performed using 50 forward and reverse primers. PCR was carried out with primers in a reaction (10 µL) containing 1 × buffer, 0.1-0.5 µM of each nucleotide primer, 2.5 mM dNTPs, 100 ng genomic DNA and 0.5 U Taq DNA polymerase (Boehringer Mannheim). The amplifications were carried out in a My Cycler thermocycler (Bio-Rad). In total, 35 cycles of PCR were completed using the following steps: 25 sec at 91℃, 45 sec at 65℃, 30 sec at 72℃ (first 4 cycles); 25 sec at 91℃, 45 sec at 60℃, 30 sec at 72℃ (next 26 cycles); 25 sec at 91℃, 60 sec at 55℃, 120 sec at 72℃ (last 5 cycles), and finally a 10 min extension at 72℃. The presence or absence of PCR products was determined after separation on a 2% agarose gel containing 0.5 µg/mL ethidium bromide.
Statistical analysis
All numerical data were described as the mean and standard error or as the median and range. Chi-square tests were used to compare the clinical parameters. In comparing the numbers of KIRs, independent t-test and chi-square tests were used. Statistical analyses were performed using the SPSS software (SPSS 15.0; SPSS Inc., Chicago, IL, USA). Two-tailed analysis was used, and results with a P value under 0.05 were considered statistically significant.
Ethics statement
This study was approved by the institutional review board of Seoul St. Mary's Hospital (IRB No. KC11TISI0415). All subjects provided signed informed consent for research, and the use of the leftover specimens for this study is covered under an institutional review board protocol.
RESULTS
Demographic and clinical characteristics of the study population
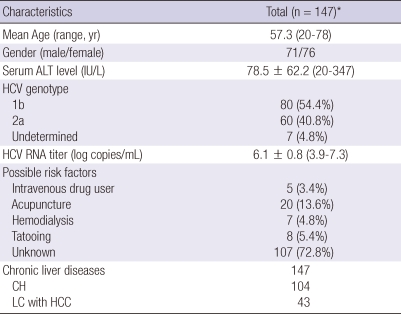
Demographic and clinical characteristics of patients with chronic HCV infection enrolled in the study are shown in Table 1. To identify risk factors for HCV infection in the Korean population, a detailed evaluation of patient histories was performed. Patients with a history of blood transfusions were excluded. Most chronically HCV-infected patients (72.8%) reported no history of risk factors of HCV infection. Reported risk factors for HCV transmission were acupuncture (13.6%), tattooing (5.4%), hemodialysis (4.8%), and intravenous drug use (3.4%). Patients did not undergo antiviral therapy.
Table 1.
Demographic and clinical characteristics of the study population
*This population did not undergo antiviral therapy. HCV, hepatitis C virus; ALT, alanine aminotransferase; CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
Frequency of KIR genes, HLA-C allotypes, and combinations of KIRs with their HLA-C ligands
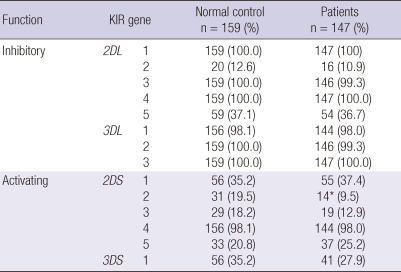
Regarding the frequency of KIR genes, the results of our study showed higher frequencies of A haplotypes and lower frequencies of B haplotypes in both the control group and patients with chronic HCV infection (Table 2). However, difference in KIR genotypes was detected between the control group and chronic HCV carriers (Table 2). Among inhibitory KIRs, there were no significant differences between the control group and patients with chronic HCV infection, whereas the frequency of KIR2DS2 of activating group in patients with chronic HCV infection was significantly lower than the control group (14/147 [9.5%] vs 31/159 [19.5%], P < 0.01). There were no statistically significant differences in the frequency of other activating KIR genes.
Table 2.
Frequency of killer cell immunoglobulin-like receptor (KIR) genes in controls and patients with chronic hepatitis C virus infection
*P < 0.01.
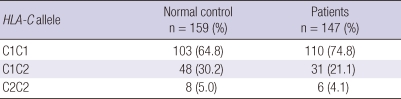
When evaluating the HLA-C allotype, this study found no statistically significant result (Table 3). HLA-C1/C1 had a higher incidence in chronic HCV carriers than in controls, but this was not statistically significant.
Table 3.
Frequency of human leukocyte antigen (HLA)-C allotypes in controls and patients with chronic hepatitis C virus infection
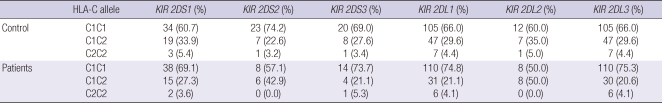
Also, no statistically significant difference was found in this study when looking at the frequency of combinations of KIRs with their HLA-C ligands between the control group and chronic HCV carriers (Table 4).
Table 4.
Frequency of combinations of killer cell immunoglobulin-like receptor (KIR) genes with their human leukocyte antigen (HLA)-C ligands in controls and patients with chronic hepatitis C virus infection
DISCUSSION
As previously mentioned, the number of KIR genes varies among individuals, resulting in a heterogeneous array of possible KIR genes. The KIR gene diversities have already been studied in many different geographic populations (18-20). According to a previous report, the frequencies of KIR sequences observed in a Korean population, compared with a Caucasian population, showed predominance of A haplotype (KIR2DL1, 2DL3, and 2DS4), while KIR genes on B haplotypes (KIR2DL5, 2DS1, 2DS2, 2DS3, 2DS5, and 3DS1) with lower frequencies (20). Likewise, our data also showed higher frequencies of A haplotypes and lower frequencies of B haplotypes in both the control group and patients with chronic HCV infection.
This study showed a significantly lower frequency of KIR2DS2 among chronic HCV carriers compared with controls in a Korean population (P < 0.01). This result can be a clue to the further investigation on HCV immunology with host genetics. KIRs are involved in innate immunity through their effects on NK cells and also in acquired immunity, because certain subsets of T cells express both the KIRs and T cell receptors (12). Strikingly, KIR2DS2 is expressed by both the innate and acquired arms of the immune system and is frequently included in the KIR repertoire of T cells, whereas other KIRs such as KIR2DL1 are expressed by fewer cells of acquired immunity arm (21, 22). In one previous study, the absence of KIR2DS2 was associated with failure of pegylated interferon and ribavirin therapy in liver transplant patients with recurrent HCV infection in Americans (23). Likewise, for future prospective study, following assumption can be suggested that KIR2DS2 might aid in HCV clearance by enhancing both the innate and acquired immune responses of people, at least in a Korean population. According to a previous study, the frequency of KIR2DS2 genotype expression was lower in patients who failed early eradication of HCV and progressed to chronic liver disease, and it was proposed that KIR2DS2 might enhance the more effective cellular response, as well as the innate immune response against HCV (23).
Regarding the HLA-C allotype, this study found no statistically significant result, whereas previous studies reported different results that HLA-C1 homozygosity may function for HCV clearance. The study by Khakoo et al. (15) suggested that HLA-C1 homozygosity might have a protective effect on HCV infected hosts, because of the capacity of these molecules to present antigens that have stronger affinities for cytotoxic T cells (15). Another recent study found that the frequency of HLA-C1 homozygosity was greater in a group of patients who achieved sustained virological response (SVR) to combined pegylated interferon and ribavirin therapy than in the non-response group, although the results were not statistically significant (24).
In addition, there was no statistically significant difference regarding the frequency of combinations of KIRs with their HLA-C ligands. This result is also different from the data of previous studies (15, 25). According to the report mentioned earlier, individuals inoculated with a low HCV viral load, who had the receptor-ligand combination of two copies of KIR2DL3 and HLA-C1 homozygosity, were more likely to recover from HCV infection than individuals with any other genotype (15). Another recent study showed that patients who exhibited SVR after combined therapy had an increased frequency of KIR2DL3/HLA-C1 homozygosity compared with non-responders (25). In particular, although the protective effects of HLA-C1/KIR2DL3 were previously reported to be increased in patients with no history of blood transfusion (15), as our Korean study group, we did not observe any significant associations.
We acknowledge that our study has several limitations. First, the small patient population, particularly in the genetic study, limits the generalization of the conclusion. Also, the patient population in the genetic study is likely to be biased. Next, for the control group, patients who spontaneously cleared themselves of HCV would be a proper population for comparison with the chronically infected patients. The control group of our study consisted of 159 unrelated individuals, matched with the infected population by each risk factor of contracting HCV. But this group cannot represent the cleared group of subjects. To overcome these limitations of our study, further prospective study with a larger scale is required. Also, functional studies to correlate anti-HCV activity of NK cells with KIR will be necessary to further understand the role of KIR in HCV clearance.
In conclusion, this study shows a lower frequency of KIR2DS2 among patients with chronic HCV infection than controls in a Korean population. This may be attributed to the impairment of both the innate and the acquired immune responses.
Footnotes
This study was supported, in part by a grant of the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A092258), and also by Bio R&D program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0018167).
AUTHOR SUMMARY
Frequency of Killer Cell Immunoglobulin-like Receptors (KIRs) in Korean Patients with Chronic HCV Infection
Pil Soo Sung, Hee Baeg Choi, Su-Yeon Kim, Sung Woo Hong, Chung-Hwa Park, Myeong Jun Song, Sung Won Lee, Chan Ran Yoo, Sang Wook Choi, Nam Ik Han, Tai-Gyu Kim and Seung Kew Yoon
Killer cell immunoglobulin-like receptors (KIRs) are the receptors of natural killer (NK) cells that mediate innate immunity. A protective association of the inhibitory receptor KIR2DL3 with HLA-C1 ligand and its effect on the course of HCV infection was reported previously. Here we investigate the frequency of KIR genes, HLA-C allotypes and combinations of KIRs with HLA-C ligands in Korean HCV carrier cohort. The data showed that the frequency of KIR2DS2 in patients with chronic HCV infection was significantly lower than controls. Our study suggests that KIR2DS2 might aid in HCV clearance by enhancing immune responses.
References
- 1.Bostan N, Mahmood T. An overview about hepatitis C: a devastating virus. Crit Rev Microbiol. 2010;36:91–133. doi: 10.3109/10408410903357455. [DOI] [PubMed] [Google Scholar]
- 2.Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29:908–914. doi: 10.1002/hep.510290311. [DOI] [PubMed] [Google Scholar]
- 3.Lim YS. Current status of liver disease in Korea: hepatitis C. Korean J Hepatol. 2009;15:S25–S28. doi: 10.3350/kjhep.2009.15.S6.S25. [DOI] [PubMed] [Google Scholar]
- 4.Shin HR. Epidemiology of hepatitis C virus in Korea. Intervirology. 2006;49:18–22. doi: 10.1159/000087258. [DOI] [PubMed] [Google Scholar]
- 5.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 6.Doherty DG, Norris S, Madrigal-Estebas L, McEntee G, Traynor O, Hegarty JE, O'Farrelly C. The human liver contains multiple populations of NK cells, T cells, and CD3+ CD56+ natural T cells with distinct cytotoxic activities and Th1, Th2, and Th0 cytokine secretion patterns. J Immunol. 1999;163:2314–2321. [PubMed] [Google Scholar]
- 7.Liu ZX, Govindarajan S, Okamoto S, Dennert G. NK cells cause liver injury and facilitate the induction of T cell-mediated immunity to a viral liver infection. J Immunol. 2000;164:6480–6486. doi: 10.4049/jimmunol.164.12.6480. [DOI] [PubMed] [Google Scholar]
- 8.Moretta A, Biassoni R, Bottino C, Pende D, Vitale M, Poggi A, Mingari MC, Moretta L. Major histocompatibility complex class I-specific receptors on human natural killer and T lymphocytes. Immunol Rev. 1997;155:105–117. doi: 10.1111/j.1600-065x.1997.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 9.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 10.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 11.Williams AP, Bateman AR, Khakoo SI. Hanging in the balance. KIR and their role in disease. Mol Interv. 2005;5:226–240. doi: 10.1124/mi.5.4.6. [DOI] [PubMed] [Google Scholar]
- 12.Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat Rev Immunol. 2004;4:190–198. doi: 10.1038/nri1306. [DOI] [PubMed] [Google Scholar]
- 13.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 14.Boyton RJ, Altmann DM. Natural killer cells, killer immunoglobulin-like receptors and human leucocyte antigen class I in disease. Clin Exp Immunol. 2007;149:1–8. doi: 10.1111/j.1365-2249.2007.03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 16.Norman PJ, Carrington CV, Byng M, Maxwell LD, Curran MD, Stephens HA, Chandanayingyong D, Verity DH, Hameed K, Ramdath DD, Vaughan RW. Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun. 2002;3:86–95. doi: 10.1038/sj.gene.6363836. [DOI] [PubMed] [Google Scholar]
- 17.Djulejic E, Petlichkovski A, Trajkov D, Hristomanova S, Middleton D, Spiroski M. Distribution of killer cell immunoglobulin-like receptors in the Macedonian population. Hum Immunol. 2010;71:281–288. doi: 10.1016/j.humimm.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, Purcell RH, Chisari FV. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci U S A. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whang DH, Park H, Yoon JA, Park MH. Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol. 2005;66:146–154. doi: 10.1016/j.humimm.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Wang SH, Huang CX, Ye L, Wang X, Song L, Wang YJ, Liang H, Huang XY, Ho WZ. Natural killer cells suppress full cycle HCV infection of human hepatocytes. J Viral Hepat. 2008;15:855–864. doi: 10.1111/j.1365-2893.2008.01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Bergen J, Thompson A, van der Slik A, Ottenhoff TH, Gussekloo J, Koning F. Phenotypic and functional characterization of CD4 T cells expressing killer Ig-like receptors. J Immunol. 2004;173:6719–6726. doi: 10.4049/jimmunol.173.11.6719. [DOI] [PubMed] [Google Scholar]
- 23.Askar M, Avery R, Corey R, Lopez R, Thomas D, Pidwell D, Eghtesad B, Miller C, Fung J, Zein NN. Lack of killer immunoglobulin-like receptor 2DS2 (KIR2DS2) and KIR2DL2 is associated with poor responses to therapy of recurrent hepatitis C virus in liver transplant recipients. Liver Transpl. 2009;15:1557–1563. doi: 10.1002/lt.21878. [DOI] [PubMed] [Google Scholar]
- 24.Vidal-Castiñeira JR, López-Vázquez A, Díaz-Peña R, Alonso-Arias R, Martínez-Borra J, Pérez R, Fernández-Suárez J, Melón S, Prieto J, Rodrigo L, López-Larrea C. Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol. 2010;84:475–481. doi: 10.1128/JVI.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knapp S, Warshow U, Hegazy D, Brackenbury L, Guha IN, Fowell A, Little AM, Alexander GJ, Rosenberg WM, Cramp ME, Khakoo SI. Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology. 2010;51:1168–1175. doi: 10.1002/hep.23477. [DOI] [PMC free article] [PubMed] [Google Scholar]