Abstract
Smad2 and Smad3 (Smad2/3) are essential signal transducers and transcription factors in the canonical transforming growth factor-β (TGF-β) signalling pathway. Active Smad2/3 signalling in the nucleus is terminated by dephosphorylation and subsequent nuclear export of Smad2/3. Here we report that protein phosphatase PPM1A regulates the nuclear export of Smad2/3 through targeting nuclear exporter RanBP3. PPM1A directly interacted with and dephosphorylated RanBP3 at Ser 58 in vitro and in vivo. Consistently, RanBP3 phosphorylation was elevated in PPM1A-null mouse embryonic fibroblasts. Dephosphorylation of RanBP3 at Ser 58 promoted its ability to export Smad2/3 and terminate TGF-β responses. Our findings indicate the critical role of PPM1A in maximizing exporter activity of RanBP3 for efficient termination of canonical TGF-β signalling.
Keywords: TGF-β signalling, Smads, PP2Cα, phosphorylation, termination
Introduction
In eukaryotes, regulation of signalling mediators/effectors in the nucleus is one of the principal mechanisms that govern duration and strength of signalling. Smads are a family of structurally related proteins with intrinsic nuclear shuttling ability that primarily serve as signalling effectors for the ligands of transforming growth factor-β (TGF-β) superfamily (Hill, 2009). On ligand binding to its receptor at the cell membrane, TGF-β signalling is activated by a short phosphorylation cascade, from receptor phosphorylation to subsequent phosphorylation of the receptor-activated Smads (R-Smads, such as Smad2/3), and ultimately regulates the transcription of TGF-β target genes (ten Dijke & Hill, 2004; Feng & Derynck, 2005; Massagué et al, 2005). Tight control of these genes underscores the importance of TGF-β signalling in regulating many biological processes from development to pathogenesis including cancer (Massagué, 2008; Heldin et al, 2009; Wu & Hill, 2009).
Although the level of nuclear R-Smads reflects the strength of TGF-β signalling at any given time, the nuclear export of R-Smads in terminating TGF-β signalling remains poorly understood. The identification of nuclear phosphatases and/or transporters for R-Smads shows two sequential steps during Smad2/3 export. In the nucleus, PPM1A/PP2Cα, a member of the PPM family of Ser/Thr protein phosphatases, specifically recognizes phosphorylated Smad2/3 and remove its C-terminal SXS phosphorylation (Lin et al, 2006). Subsequent nuclear export of the un/dephosphorylated Smad2/3 is carried out by RanBP3 (Dai et al, 2009) and/or Exportin 4 (Kurisaki et al, 2006).
Recently, Yoon et al (2008) reported that protein kinases ribosomal S6 kinases (RSKs) and AKT phosphorylate RanBP3 at Ser 58. This phosphorylation attenuates nuclear import of ribosomal protein L12 (Yoon et al, 2008). As RanBP3 mainly resides in the nucleus (Mueller et al, 1998; Hendriksen et al, 2005; Dai et al, 2009), whether and how its phosphorylation modulates the nuclear export role of RanBP3 remain unexplored. Here we report that the Ser 58 phosphorylation inhibited the ability of RanBP3 to export Smad2/3 and terminate TGF-β transcriptional responses. Moreover, we provide clear evidence that PPM1A is a bona fide RanBP3 phosphatase to regulate RanBP3 activity. These results indicate the dual roles of PPM1A in dephosphorylating both cargo Smad2/3 and exporter RanBP3 for efficient termination of Smad2/3 signalling.
Results and Discussion
RanBP3 phosphorylation modulates TGF-β signalling
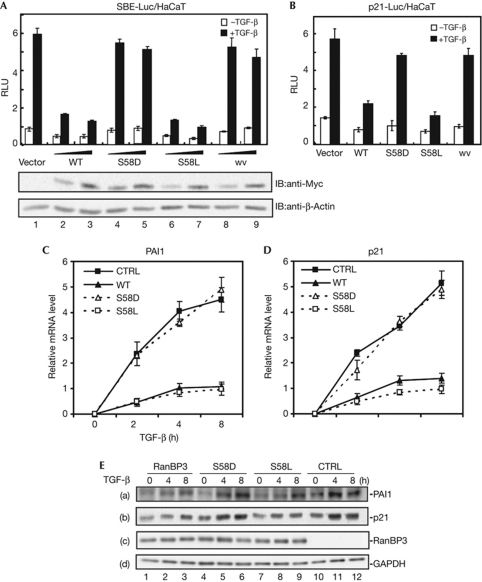
We previously reported that RanBP3 mediates nuclear export of Smad2/3 in TGF-β signalling in a Ran binding-dependent manner (Dai et al, 2009). As RanBP3 is regulated by phosphorylation at Ser 58 (Yoon et al, 2008), we sought to investigate whether RanBP3 phosphorylation modulates TGF-β signalling. To this end, we generated the phosphorylation-mimic mutant RanBP3-S58D and phosphorylation-dead mutant RanBP3-S58L. The effects of these mutants on TGF-β signalling were first examined using a synthetic TGF-β responsive reporter SBE-Luc (Zawel et al, 1998). As shown in Fig 1A, the phosphorylation-dead mutant RanBP3-S58L profoundly inhibited the SBE-Luc expression to the same extent as wild-type (WT) RanBP3. Consistently, RanBP3 and RanBP3-S58L mutant showed similar inhibitory effects on the promoter activity of the gene encoding cyclin-dependent kinase inhibitor p21WAF1/CIP1 (Fig 1B), one of the key effectors mediating TGF-β-induced growth inhibition (Datto et al, 1995). However, phosphorylation-mimic mutant RanBP3-S58D and the Ran-binding mutant RanBP3-wv could not block either SBE-luc (Fig 1A) or the p21-luc reporter response (Fig 1B).
Figure 1.
S58D substitution impairs the inhibitory function of RanBP3 in transforming growth factor-β (TGF-β) signalling. (A) Effect of RanBP3 phosphorylation mutants on SBE-Luc response. HaCaT cells were transfected with indicated plasmids and treated with 2 ng/μl TGF-β for 20 h, and cell lysates were subjected to reporter assays. Values and error bars represent average and standard deviation of three independent experiments. The expression level of RanBP3 (wild type, WT) and its mutants were examined by western blotting using Myc antibody (bottom). (B) Effect of RanBP3 phosphorylation mutants on the natural p21 promoter (p21-Luc) activity in HaCaT cells. Values and error bars represent average and standard deviation of four independent experiments. (C) Quantitative real-time reverse transcription–polymerase chain reaction (qRT–PCR) analysis of p21 messenger RNA (mRNA). HaCaT cells stably expressing Flag-tagged RanBP3 (WT, S58D, S58L) and parental HaCaT cells (CTRL) were treated with TGF-β (2 ng/μl) for up to 8 h, and were subjected to total RNA extraction. Values and error bars represent average and standard deviation of three independent experiments. (D) qRT–PCR analysis of PAI1 mRNA. Values and error bars represent average and standard deviation of three independent experiments. (E) Western blotting analysis of p21 and PAI1. HaCaT cells stably expressing Flag-tagged RanBP3 (WT, S58D, S58L) and parental HaCaT cells (CTRL) were treated with TGF-β (2 ng/μl) for 8 h and cell lysates collected at indicated times. The levels of p21, Plasminogen activator inhibitor 1 (PAI1) and RanBP3 proteins were examined by western blotting using indicated antibodies. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) blot serves as a loading control. SBE, Smad-binding element; RLU, Relative luciferase unit; wv, RanBP3 Ran-binding mutant.
To extend our analysis at the physiological level, we generated HaCaT cell lines stably expressing RanBP3 or its mutants at similar expression levels (Fig 1E, blot c). In parent HaCaT cells (CTRL), TGF-β treatment for 8 h induced rapid accumulation of p21 (Fig 1C) and plasminogen activator inhibitor 1 (PAI1) messenger RNA (Fig 1D). Overexpression of RanBP3 and RanBP3-S58L significantly attenuated accumulation of p21 and PAI1 messenger RNA, whereas gain-of-phosphorylation mutant RanBP-S58D had no effect (Fig 1C,D). Consistently, TGF-β-induced p21 and PAI1 protein expression was inhibited in the cells stably expressing RanBP3-S58L mutant (Fig 1E, lane 7–9), but not RanBP3-S58D mutant (Fig 1E, lane 4–6). These results indicate that Ser 58 phosphorylation of RanBP3 profoundly dampens its role in terminating TGF-β signalling.
RanBP3 phosphorylation inhibits Smad2/3 export
We next determined how phosphorylation impairs the ability of RanBP3 to inhibit TGF-β signalling. We found that Ser 58 mutations did not affect RanBP3–Smad2/3 interaction (supplementary Fig S1 online), nor did it change the Smad-binding specificity of RanBP3 (supplementary Fig S2 online). These data implies that RanBP3 phosphorylation does not inhibit Smad2/3 export through disruption of the Smad2/3–RanBP3 association. Because the RanBP3 mutants resided exclusively in the nucleus, we examined the effects of these mutants on nuclear accumulation of Smad2/3 in HaCaT cells. Quantitative analysis showed that nuclear Smad3 decreased in 66.7% (n=24) and 62.5% (n=24) of cells transiently expressing RanBP3 or RanBP-S58L, respectively, but in only 8.3% (n=24) of RanBP3-S58D-positive cells (Fig 2A). In agreement with this notion that RanBP3-S58D could not inhibit nuclear accumulation of Smad2/3, epidermal growth factor (EGF) or insulin stimulation, which induces RanBP3 phosphorylation (Yoon et al, 2008), induced Smad2 nuclear accumulation in HaCaT cells (supplementary Fig S3 online). Together, these results indicate that phosphorylation or its mimicry disables RanBP3 to inhibit TGF-β signalling.
Figure 2.
RanBP3-S58D fails to promote nuclear export of Smad2/3. (A) Microscopic analysis of Smad2/3 nuclear accumulation. HaCaT cells were transfected with Myc-tagged RanBP3 or mutants. After 24 h transfection, cells were treated with transforming growth factor-β (TGF-β, 2 ng/μl) for 2 h and fixed. Endogenous Smad2/3 (red) was immunostained by using anti-Smad2 (left) or Smad3 (right) antibody, respectively. The arrow indicates the cells with ectopically expressed Myc-RanBP3 or its mutants detected by anti-Myc immunostaining (green). 4,6-Diamidino-2-phenylindole (DAPI) stains the nucleus (blue). Intensity of nuclear Smad3 among these cells was quantified using National Institutes of Health Image software. Far right panel shows the percentage of cells with decreased nuclear Smad3 level (red bar) within 24 randomly counted cells. (B) Western blot analysis of Smad2/3 export. Cells were collected at the indicated times (depicted on the right) and the cytoplasmic/nuclear fractions were extracted. Successful fractionation was confirmed by western blot analysis of the cytoplasmic marker glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and nuclear marker Lamin A/C. Smad2/3 in the cytoplasmic or nuclear fractions were examined by anti-Smad2/3 western blotting. SB431542 is a TGF-β type I receptor inhibitor. (C) Quantitative analysis of Smad2 export. Details are described in Methods. Values and error bars represent average and standard deviation of three independent experiments. The expression level of RanBP3 and mutants were examined by western blot (bottom). The β-actin blot serves as a loading control. (D) In vitro import assay of Smad2 import in permeabilized cells. Details are described in Methods. Nuclear imported Smad2 was examined by western blot using anti-Smad2 antibody. The Lamin A/C blot serves as a nuclear marker and loading control. WT, wild type.
To specifically evaluate Smad2/3 export, we compared the redistribution of nuclear Smad2/3 into the cytoplasm in HaCaT cells in the presence of overexpressed RanBP3 mutants. TGF-β-induced nuclear accumulation of Smad2/3 was reflected by the decreased level of Smad2/3 in the cytoplasm and the increased level of Smad2/3 in the nucleus. After removal of TGF-β and additional treatment with the TβRI inhibitor SB431542, which blocks phosphorylation and nuclear import of new Smad2/3, the level of cytoplasmic Smad2/3 gradually increased in 30 min mostly due to Smad2/3 nuclear export (Fig 2B, lane 1–4). Stable expression of WT RanBP3 or RanBP3-S58L in HaCaT cells promoted the nuclear Smad2/3 export (supplementary Fig S4 online, lanes 5–12 versus lanes 1–4). When comparing the two phosphorylation mutants of RanBP3, we observed a more rapid accumulation of the cytoplasmic Smad2/3 in the S58L-expressing stable cells than that in S58D-expressing stable cells (Fig 2B, lanes 5–12). We further compared RanBP3 and RanBP3 mutants in a highly sensitive quantitative export assay (Dai et al, 2009). In agreement with the results above, RanBP3-S58D was ineffective in exporting Smad2, whereas unphosphorylatable RanBP3-S58L showed the same activity as WT RanBP3 in promoting Smad2 export (Fig 2C). In conclusion, S58 phosphorylation disrupts the ability of RanBP3 to mediate nuclear export of Smad2/3.
Despite its role as a cofactor in CRM1-mediated nuclear protein export (Englmeier et al, 2001; Lindsay et al, 2001; Nemergut et al, 2002), RanBP3 can function independently of CRM1 to mediate nuclear export of Smad2/3 (this study) and β-catenin (Hendriksen et al, 2005). We observed a similar effect of S58 phosphorylation in Wnt signalling. RanBP3 and RanBP3-S58L, but not RanBP3-S58D, inhibited β-catenin-dependent reporter expression (TOPFlash) (supplementary Fig S5 online). Therefore, Ser 58 phosphorylation seems to modulate the export function of RanBP3 in a cargo-independent manner. Notably, Ser 58 resides in the N-terminal domain of RanBP3, which contains FXFG nucleoporin-binding motifs that are characteristic of nucleoporins (Mueller et al, 1998). It remains to be investigated whether and how Ser 58 phosphorylation of RanBP3 impacts cargo transport through the nuclear pore.
The negative role of RanBP3 phosphorylation in nuclear export (Fig 2) is apparently opposite to its positive function in nuclear import (Yoon et al, 2008). To examine whether RanBP3 phosphorylation also regulates Smad2 import, we examined the Smad2 nuclear import in the same stable cells using in vitro import assays (Fig 2D). Smad2 added in digitonin-permeabilized HaCaT parental cells was imported into the nucleus as monitored by the increasing amount of nuclear Smad2 (Fig 2D, lanes 4–6). Compared with HaCaT parental cells, neither RanBP3 nor phosphorylation mutants affected the nuclear accumulation of Smad2, suggesting that RanBP3 or its phosphorylation does not regulate Smad2 import.
PPM1A is a RanBP3 phosphatase
Protein kinases Akt and RSK are reported to phosphorylate RanBP3 at Ser 58 (Yoon et al, 2008). Because of the important role of phosphorylation in regulating the transport activity of RanBP3, an equally important question has been: what is the identity of the RanBP3 phosphatase? By screening the human serine/threonine phosphatome, we looked for phosphatase(s) that potently reduced the P-RanBP3 level induced by a constitutively active AKT-DDD mutant (data not shown). We found that PPM1A, which is a Smad2/3 nuclear phosphatase (Lin et al, 2006), was the strongest candidate for RanBP3 dephosphorylation.
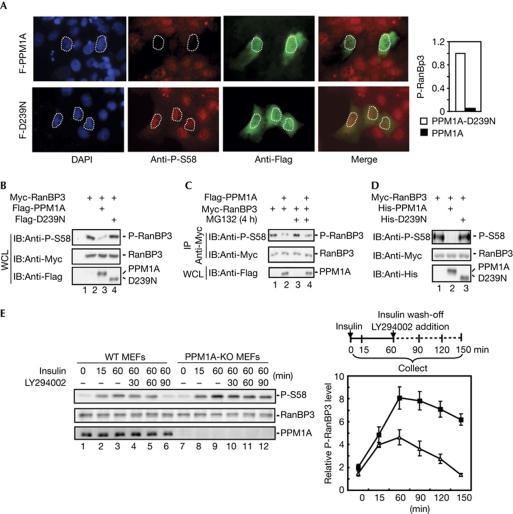
As shown in Fig 3A, expression of Flag-PPM1A (green) clearly resulted in a marked decrease in the EGF-induced nuclear P-RanBP3 level (red) in comparison with the neighbouring non-transfected cells. In sharp contrast, PPM1A-D239N, a catalytically inactive mutant of PPM1A (Lin et al, 2006), could not decrease the nuclear P-RanBP3 level, indicating that the phosphatase activity of PPM1A is required for reducing P-RanBP3 (Fig 3A). Other members in the PPM family such as PPM1F and PPM1G could not block EGF-induced nuclear P-RanBP3 accumulation, confirming that the effect of PPM1A on RanBP3 is specific (supplementary Fig S6 online). Western blotting analysis further showed that WT PPM1A, but not the mutant D239N, reduced the level of P-RanBP3 (Fig 3B). This PPM1A-induced reduction of P-RanBP3 was not due to the proteasome-dependent degradation because the proteasome inhibitor MG132 did not reverse the effect of PPM1A on the P-RanBP3 level (Fig 3C). To rule out the possibility that PPM1A activates another phosphatase in cells that could be the direct RanBP3 phosphatase, we performed an in vitro phosphatase assay using recombinant PPM1A. PPM1A, but not the D239N mutant, evidently dephosphorylated RanBP3 (Fig 3D) and Smad3 (supplementary Fig S7 online). These data strongly support PPM1A as a bona fide RanBP3 phosphatase.
Figure 3.
PPM1A dephosphorylates RanBP3 at Ser 58. (A) Microscopic analysis of PPM1A phosphatase activity on RanBP3 Ser 58 dephosphorylation. HaCaT cells were transfected with Flag-tagged PPM1A or phosphatase-dead mutant PPM1A(D239N). After 24 h transfection, cells were treated with epidermal growth factor (50 ng/ml) for 1 h, fixed and immunostained with anti-P-S58 antibody (red) or anti-Flag antibody (green). Quantification of nuclear P-RanBP3 (immunostaining intensity) in PPM1A/D239N-transfected cells (n=10) and non-transfected cells (n=30) is shown on the right. (B) PPM1A dephosphorylated RanBP3 in 293T cells. Cells (293T) transfected with indicated expression plasmids and whole-cell lysates (WLCs) were subjected to western blotting. (C) Proteasomal inhibitor MG132 had no effect on RanBP3 dephosphorylation. Cells (293T) were transfected with indicated expression plasmids, treated with MG132 (20 μM) for 4 h and then collected. Myc-RanBP3 was immunoprecipitated from WCL and then subjected to western blotting. (D) PPM1A dephosphorylated RanBP3 in vitro. Cells (293T) were transfected with Myc-RanBP3, and anti-Myc immunoprecipitation was carried out. In vitro phosphatase assay was used to analyse the phosphatase activity of recombinant His-PPM1A and His-PPM1A-D239N on P-RanBP3. (E) Insulin-induced phosphorylation of RanBP3 increased in PPM1A-null cells. PPM1A knockout (KO) and wild-type (WT) mouse embryonic fibroblasts (MEFs) were treated with insulin (100 nM) for 60 min, then washed three times to remove insulin and treated with PI3K inhibitor LY294002 (50 μM) for 90 min. Cells were collected at indicated times and subjected to western blot analysis using antibodies as indicated (left panel). Relative level of P-RanBP3 (P-RanBP3/RanBP3) was quantified using National Institutes of Health Image software (right panel). Values and error bars represent average and standard deviation of three independent experiments. IB, immunoblot; IP, immunoprecipitation.
To further elucidate the physiological role of PPM1A as a RanBP3 phosphatase, we took a loss-of-function approach using mouse embryonic fibroblasts (MEFs) derived from PPM1A−/− mutant mice (PPM1A-knock out (KO)) and WT control mice (WT). As expected, insulin stimulation (1 h) induced a gradual increase of P-RanBP3 in WT MEFs (Fig 3E, lanes 1–3). Notably, insulin could induce a higher level of P-RanBP3 in PPM1A-KO MEFs (Fig 3E, lanes 7–9). After being stopped by addition of LY294002 (1.5 h), insulin-induced RanBP3 phosphorylation gradually decreased in WT MEFs (Fig 3E, lanes 4–6), and yet was sustainable for longer in PPM1A-KO MEFs (Fig 3E, lanes 10–12). These results collectively showed a specific inverse correlation between PPM1A activity and nuclear RanBP3 phosphorylation level.
PPM1A directly interacts with RanBP3
To investigate how PPM1A dephosphorylates RanBP3, we examined whether PPM1A bound to RanBP3 in protein interaction assays. Their direct interaction was assessed using in vitro glutathione S-transferase (GST) binding assays. Results indicated that recombinant PPM1A bound to recombinant GST–RanBP3 fusion protein, but not GST alone (Fig 4A, lane 5 versus lane 1). We also mapped the regions of RanBP3 that could bind to PPM1A, and found RanBP3-R (residues 290–499) mainly mediated the RanBP3–PPM1A interaction (Fig 4A, lane 4). In comparison, pull-down using GST–RanBP3-N (residues 1–185) could retrieve a marginal level of PPM1A (Fig 4A, lane 2), and RanBP3-F (middle region, residues 182–292) did not interact with PPM1A (Fig 4A, lane 3).
Figure 4.
PPM1A physically interacts with RanBP3. (A) PPM1A directly interacted with RanBP3. Recombinant His-PPM1A protein was incubated with glutathione S-transferase (GST)-fused proteins of RanBP3 full-length or fragment (containing N, F and R domains) on glutathione-Sepharose beads. Retrieved proteins were separated by SDS–PAGE and examined by anti-His western blotting. GST-only protein used in lane 5 serves as negative control. (B) PPM1A preferentially interacted with RanBP3-S58D. In vitro GST pulldown assay was done as described in A. (C) RanBP3 phosphorylation at Ser 58 enhanced PPM1A–RanBP3 interaction. PPM1A-bound RanBP3 were immunoprecipitated with anti-Flag antibody and detected by anti-Myc western blotting. (D) Endogenous PPM1A interacted with RanBP3 under physiological conditions. HaCaT cells at 90% confluence were treated with insulin (100 nM) or transforming growth factor-β (TGF-β, 2 ng/μl) for 1 h. PPM1A-bound RanBP3 was immunoprecipitated with an anti-PPM1A antibody and detected by anti-RanBP3 western blotting. The anti-mouse immunoglobulin-G (IgG) in lane 4 serves as a negative control. HA, haemagglutinin; IB, immunoblot; IP, immunoprecipitation; WCL, whole-cell lysate.
We then determined whether phosphorylation influenced the interaction. We found that PPM1A bound to the phosphorylation-mimic mutant RanBP3-S58D more strongly than the phosphorylation-dead RanBP3-S58L (Fig 4B, lane 4 versus lane 5), implying that PPM1A binding to RanBP3 might depend on Ser 58 phosphorylation. To further evaluate this, we examined the RanBP3–PPM1A interaction in the presence of AKT-DDD, the constitutively active form of AKT. Overexpression of AKT-DDD induced RanBP3 phosphorylation, and increased the PPM1A interaction with RanBP3 (Fig 4C, lane 3 versus lane 2), but not with RanBP3-S58L (Fig 4C, lane 5 versus lane 4). Consistent with these results, insulin treatment enhanced RanBP3 phosphorylation in HaCaT cells (Fig 4D, lane 2 versus lane 1) and consequently the endogenous PPM1A–RanBP3 interaction (Fig 4D, right panel, lane 6 versus lane 5). These data strongly indicate that PPM1A preferentially recognizes RanBP3 in its phosphorylated form. However, neither insulin nor TGF-β stimulation affected PPM1A activity toward P-RanBP3 (supplementary Fig S8 online). Therefore, PPM1A action on RanBP3 in these signalling contexts seems to be regulated through the substrate recognition by the phosphatase in the nucleus.
In summary, we discovered that PPM1A has dual functions in terminating Smad2/3 signalling. In addition to its established role in dephosphorylating Smad2/3 (Lin et al, 2006), PPM1A also dephosphorylates RanBP3 at the phosphorylation site Ser 58. This duality of PPM1A, which has not been reported before, has significant implications. As a net result, PPM1A promotes efficient nuclear export of Smad2/3 by coupling and coordinating the two sequential steps for Smad signal termination; that is, preparing the cargo for export (in other words, Smad2/3 dephosphorylation) and increasing the nuclear exporter activity (RanBP3 dephosphorylation; Fig 5).
Figure 5.
A working model for Smad2/3 nuclear export involving RanBP3 and PPM1A. In addition to its previously demonstrated role that prepares nuclear Smad2/3 readiness for export by RanBP3, PPM1A directly binds to RanBP3 and regulates its export activity through Ser 58 dephosphorylation. TGF-β, transforming growth factor-β.
Methods
Details on plasmids, antibodies used/made in the study and other methods are provided in the supplementary information online.
In vitro phosphatase assay. The phosphatase reaction was performed at 30°C for 30 min in a buffer (150 mM Tris–HCl (pH 7.5), 30 mM MgCl2, 5 mM dithiothreitol and 1 mg/ml BSA) with recombinant PPM1A and RanBP3, and the dephosphorylation of Ser 58 was analysed by western blotting with anti-p-Ser 58 antibody.
Cell fractionation. HaCaT parental or its stable cells were collected with fractionation buffer (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.5% NP40 and protease inhibitors) for 20 min on ice, and then fractionated by centrifugation (1,000 r.p.m., 5 min) at 4°C to pellet nuclei. The supernatant were collected as the cytoplasmic fraction. The fractions were analysed by SDS–PAGE and western blotting analysis.
Quantitative Smad2 export assay. This was essentially done as described (Lin et al, 2006). Briefly, HEK293T cells were transfected with plasmids for MS2-Smad2, CAT reporter pDM128/8xMS2 and β-galactosidase plasmid (for normalization) and additional factors under examination. After 45 h transfection, cell lysates were analysed for chloramphenicol acetyltransferase activity using ELISA-based assay (Roche). Relative export activity is calculated by normalizing the CAT activity with β-galactosidase activity in each sample.
See supplementary information online for additional Methods.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Maarten Fornerod (pcDNA-RanBP3), Bryan Cullen (MS2 nuclear export assay system), Joan Massagué (MS2-Smad2) and Bert Vogelstein (WWP1-luc/p21-luc and SBE-luc). Thanks also go to members of Feng and Lin labs for helpful discussion. This research was supported by National Institutes of Health grants (X.-H.F., X.L.), National Natural Science Foundation of China grant 31090360 and NSFZ grant Z2110591 (X.-H.F.), and MOST grant 2011CBA01100 (Z.L.).
Author contributions: F.D., X.L., Z.L. and X.-H.F. designed the experiments and analysed the data. F.D. and T.S. performed the experiments. F.D., X.L., Z.L. and X.F. wrote the manuscript. All authors discussed the results and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Dai F, Lin X, Chang C, Feng XH (2009) Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-beta signaling. Dev Cell 16: 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF (1995) Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA 92: 5545–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englmeier L, Fornerod M, Bischoff FR, Petosa C, Mattaj IW, Kutay U (2001) RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep 2: 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Derynck R (2005) Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693 [DOI] [PubMed] [Google Scholar]
- Heldin CH, Landstrom M, Moustakas A (2009) Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol 21: 166–176 [DOI] [PubMed] [Google Scholar]
- Hendriksen J, Fagotto F, van der Velde H, van Schie M, Noordermeer J, Fornerod M (2005) RanBP3 enhances nuclear export of active β-catenin independently of CRM1. J Cell Biol 171: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS (2009) Nucleocytoplasmic shuttling of Smad proteins. Cell Res 19: 36–46 [DOI] [PubMed] [Google Scholar]
- Kurisaki A, Kurisaki K, Kowanetz M, Sugino H, Yoneda Y, Heldin CH, Moustakas A (2006) The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol Cell Biol 26: 1318–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X et al. (2006) PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell 125: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG (2001) Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol 153: 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J (2008) TGF-β in cancer. Cell 134: 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D (2005) Smad transcription factors. Genes Dev 19: 2783–2810 [DOI] [PubMed] [Google Scholar]
- Mueller L, Cordes VC, Bischoff FR, Ponstingl H (1998) Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett 427: 330–336 [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Lindsay ME, Brownawell AM, Macara IG (2002) Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J Biol Chem 277: 17385–17388 [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS (2004) New insights into TGF-beta-Smad signalling. Trends Biochem Sci 29: 265–273 [DOI] [PubMed] [Google Scholar]
- Wu MY, Hill CS (2009) TGF-β superfamily signaling in embryonic development and homeostasis. Dev Cell 16: 329–343 [DOI] [PubMed] [Google Scholar]
- Yoon SO, Shin S, Liu Y, Ballif BA, Woo MS, Gygi SP, Blenis J (2008) Ran-binding protein 3 phosphorylation links the Ras and PI3-kinase pathways to nucleocytoplasmic transport. Mol Cell 29: 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE (1998) Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1: 611–617 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.