Abstract
Many RNAs show polarized or otherwise non-random subcellular distributions. To create a method for genome-wide genetic screens for RNAs with asymmetric subcellular distributions, we have combined methods for gene tagging and live imaging of messenger RNA (mRNA). A pilot screen in a highly polarized, differentiated cell in the Drosophila larva, the branched terminal cell of the tracheal system, demonstrates the feasibility of the method for identifying new asymmetrically localized mRNAs in vivo.
Introduction
Many cell types use messenger RNA (mRNA) localization as a way of enriching proteins at specific subcellular locations. The spatio-temporal regulation of protein synthesis in specific subcellular niches, achieved through localizing mRNA and subsequent onsite translation, has an important role in regulating distinct cellular functions (Gao, 1998; Moccia, 2003; Jansen & Kiebler, 2005; St Johnston, 2005; Mili et al, 2008). An in situ hybridization screen in the Drosophila embryo showed an unexpectedly large number (70%) of expressed transcripts with a polarized distribution (Lecuyer, 2007). The main oxygen-delivering cells in Drosophila, the terminal cells of the tracheal system, represent a highly polarized cell type with a branched morphology. They respond to the need for oxygen in the surrounding tissue by outgrowth of branches, often at sites very distant from the nucleus. Similar to neurites, their terminal branches are fine structures embedded in the tissues they provide with oxygen. Although localized mRNAs are likely to have a central role in these branched cell types for their polarization and for their response to local cues, no suitable tools are now available to identify these transcripts.
Methods for studying RNA localization such as in situ hybridization and injecting labelled RNA (Rodriguez et al, 2007) have contributed significantly in the analysis of asymmetric distribution of RNA, but they are not ideally suited for all cell types. For example, in neurons, where thin extensions might be embedded in the surrounding tissue, it can be difficult to detect the mRNA of interest because of background signal from transcripts expressed in surrounding tissue.
The MS2–MS2 coat protein (MCP) binary system (Bertrand, 1998) relies on tagging RNA with fluorescent proteins in vivo. It comprises an mRNA tagged with MS2–RNA stem loops, and the MCP, which can bind to the stem loops, fused to a fluorescent protein (Bertrand, 1998; Forrest & Gavis, 2003). As it requires making individual mRNA–MS2 transgenes constructed from known mRNAs, it does not allow efficient screening for new mRNAs. We therefore developed a method that combines transposon tagging of genes with MS2 stem loops in Drosophila with the cell-type-specific induction of gene expression together with the detection of their transcripts by green fluorescent protein (GFP).
Results And Discussion
Screening tools
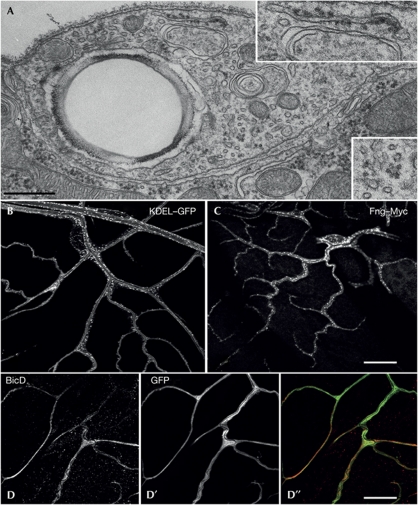
Before setting up a screen to identify mRNA in the branches of tracheal cells, we wanted to ascertain that the cellular machinery to translate mRNA was present in these sites. We find that even the finest, most distal ramifications of the branches contain the components of the translational and co-translational machinery. Specifically, we confirmed the presence of ribosomes, cytoplasmic polysomes, rough endoplasmic reticulum, Golgi bodies, PolyA-binding protein and BicaudalD (Clark et al, 2007; Coutelis & Ephrussi, 2007) in terminal branches at a great distance from the nucleus (Fig 1). Thus, the conditions to translate localized mRNA exist in terminal branches.
Figure 1.
Distribution of translation and co-translation machinery in terminal branches. (A) Electron micrograph showing a cross-section of a terminal branch about 100 μm distal to the nucleus. Insets: blow-up of regions with rough endoplasmic reticulum (ER) and ribosomes. (B,C) Distribution of ‘KDEL’ peptide sequence fused to green fluorescent protein (GFP) as an ER marker and Fng–Myc as a Golgi marker. The reporter constructs were expressed using the btl–Gal driver. (D) Immunostaining against BicD. The tracheal cells are marked with cytoplasmic GFP (D′). (D′′) Overlay of D and D′. Scale bars: A, 200 nm; B,C, 50 μm and D, 10 μm.
To introduce the MS2 RNA stem loop tag randomly into the Drosophila genome, we modified a version of the P-element EP transformation vector, which can be used to induce tissue-specific expression of genes (Rørth, 1996). This vector inserts a GAL4-inducible promoter into the genome, thereby allowing the expression of genes downstream of the insertion site with the help of tissue-specific GAL4-driver lines. We modified the ‘EPg’ version of this vector by integrating six copies of the MS2-binding site downstream of the GAL4-inducible upstream activating sequence promoter (the ‘EP–MS2-vector’; Fig 2A). A significant advantage of P-element transformation vectors is the fact that they preferentially integrate into or very close to the transcription start site of genes (Spradling, 1995). Thus, most of the integrations lead to transcription of target genes with the RNA tag included as part of their 5′ untranslated regions.
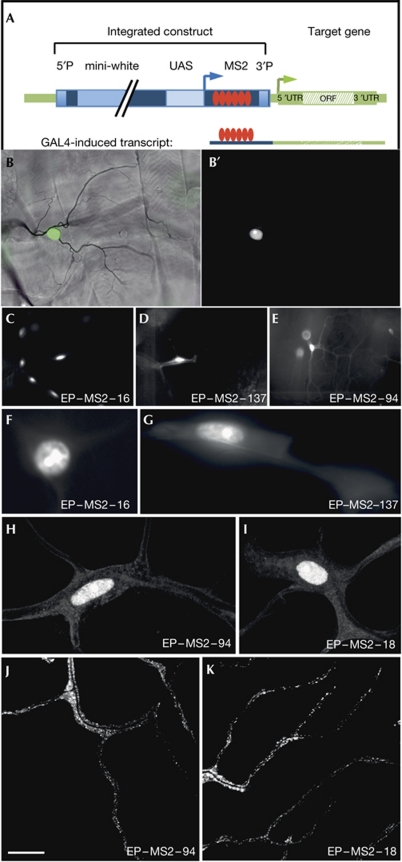
Figure 2.
MS2–green fluorescent protein (GFP) pilot screen. (A) Schematic of the modified EP transposon vector. The EP vector (blue) includes the white marker gene, the yeast upstream activating sequence (UAS) and a minimal transposase promoter from which the gene downstream of the integration site (genomic DNA shown in green) is transcribed. These sequences are flanked by the 5′ and 3′ P-elements. We have cloned six repeats of the MS2 stem loop motif (red) between the transcriptional start site and the 3′ P-element so that the motifs will be integrated into the 5′ ends of targeted genes if the P-elements insert upstream of the gene. (B,B′) MS2 coat protein (MCP)–GFP in third-instar larval terminal cells. (B) Bright-field image overlaid with a fluorescent image of a single terminal cell expressing the MCP–GFP construct. (B′) Nuclear MCP–GFP in the same cell. (C–E) Positive and negative insertions from the EP–MS2 pilot screen. Low-magnification (dissecting microscope) fluorescent image of terminal cells expressing MCP–GFP and MS2-tagged transcripts. Lines in which the GFP was seen only in the vicinity of the nucleus were scored as negative (C,D). In the positive line (E), in addition to the signal in the nucleus, MCP–GFP extends into the branches of the terminal cells. (F–K) High-resolution ( × 63 objective) images of terminal cells (nucleus and branches proximal to nucleus) expressing MCP–GFP- and MS2-tagged mRNAs from negative (F,G) and positive (H,I) lines. In the positive lines, the MCP–GFP is in a punctate pattern throughout the cells (J,K), including the terminal tips of the branches. Scale bar, 20 μm. ORF, open reading frame; UTR, untranslated region.
The second component is a fusion protein of the MCP with GFP under the control of the upstream activating sequence promoter. The MCP–GFP fusion protein includes a nuclear localization sequence to ensure that the protein is concentrated in the nucleus unless bound to the mRNA. Transgenic flies were generated with the construct cloned into the UAST transformation vector (Brand & Perrimon, 1993). For the analysis in tracheal cells, we created a chromosome carrying a trachea-specific GAL4 construct (btl-GAL4; Vincent et al, 1998), together with the MCP–GFP marker construct. The subcellular localization of MCP–GFP in a tracheal cell is shown in Fig 2B. In the absence of a coexpressed tagged mRNA, MCP–GFP is concentrated in the nucleus.
Pilot screen
We evaluated the tools we produced by performing a pilot screen according to the procedure shown in supplementary Fig S2 online. The initial generation of transgenic insertion lines yielded seven different EP–MS2 insertions in the X chromosome, which could be used as the starting point (‘ammunition’ for mobilization) for transposition to new locations within the genome. The most efficiently transposable insertion of these, EP–MS266 (supplementary Fig S1 online), was used for the generation of 250 new insertion lines. Among them, 27 lines could not be propagated. The remaining 223 lines were tested in the pilot screen by coexpressing the tagged genes with MCP in tracheal cells and screening third-instar larvae for the presence of fluorescence in terminal cell branches.
A total of 212 lines showed a distribution of GFP that was not significantly different from that of control crosses without the EP–MS2 construct, with signal only in or around the nuclei (‘negative lines’, Fig 2C,D,F,G; supplementary Fig S3 online). In 11 lines, a clear GFP signal was detectable in the branches of terminal cells (examples in Fig 2E,H,I). These 11 lines were selected for more thorough analysis, along with four control lines that showed no signal in the branches.
High-resolution microscopy (Zeiss, Apotome, × 63 lens) revealed that the fluorescent signal in the positive lines was distributed throughout the branches of terminal cells in a punctate pattern (Fig 2J,K), with no significant differences among the 11 lines. Negative lines occasionally showed some GFP in the cytoplasm near the nuclei, but never at distant sites. In these cases, the signal showed a diffuse distribution rather than the puncta seen in the positive lines (Fig 2F–I). The insertion positions of the P elements in the 11 positive lines were determined by inverse polymerase chain reaction (PCR; Bellen, 2004; supplementary Table S1 online). One insertion could not be mapped, another was inserted in a sequence that is present in several copies in the genome, and the remaining nine were inserted in or near seven different genes. Three of the identified genes have previously been shown to produce mRNAs that localize to specific subcellular domains. Btsz mRNA localizes apically in epithelial cells in the ovary, the blastoderm and the eye imaginal discs (Serano & Rubin, 2003); mRNAs for CG9924 and lola were also found to localize apically in embryonic epithelia in a genome-wide in situ hybridization screen (Lecuyer, 2007). Two of the seven genes, lola and ATPalpha, were known to be expressed in the tracheal system (Giniger et al, 1994; Goeke, 2003; Paul et al, 2007).
Analysis of negative results
Although we assume that the 212 negative lines scored negative in our assay because the mRNA of the targeted lines did not localize to tracheal branches, there might be other explanations. The simplest are that the transposon might not have inserted sufficiently close to a gene, or it might have inserted in the wrong orientation, or it might have inserted near a gene in the correct orientation, but the transcribed mRNA might have been unstable. To distinguish between these possibilities and judge the efficiency of the screen, we mapped 11 of the insertion lines that did not score in our assay. Seven were inserted in a position close to or within a known gene and in the correct orientation for this gene to be transcribed (supplementary Table S1 online). In four cases, the nearest predicted or known gene downstream of the inserted transposon was more than 4 kb away. Thus, up to four of the 11 cases might have been unproductive insertions, although this might be an overestimate as in some of these cases the transposon might have inserted upstream of hitherto unannotated genes, or within as yet unidentified upstream promoters of known genes. Therefore, at the most 40% of the negatives might be explained by non-productive insertions.
We next investigated the behaviour of transcripts that could in theory have been expressed from the inserted transposon but did not score as positive in the screen. To test whether the GAL4-driven expression of these target genes resulted in detectable levels of mRNA, we performed reverse transcription (RT)–PCR experiments on a small subset of four positive lines (btsz, lola, hr39 and CG30403) and three negative lines (CG4455, CG13985 and bif). Using a forward primer that anneals in the 3′ inverted repeat of the transgene and a reverse primer specific for the gene of interest (supplementary Fig S4A online), we found that all showed detectable transcripts (supplementary Fig S4B online), although the positive lines appeared to show higher expression levels. Thus, all of the tested genes can be transcribed; nevertheless, not all transcripts resulted in a fluorescent signal in the tracheal branches. The difference between the positive and negative lines is therefore not caused by the negative lines not producing transcripts, but more probably by a difference in the distribution or the tissue-specific stability of the mRNA. Further support for this comes from the different behaviour of the lines in different tissues (see below).
Further validation
To determine whether the distribution of the endogenous mRNAs confirmed the distribution shown by the artificially expressed MS2-tagged RNA, we carried out fluorescent in situ hybridizations (FISHs) for sample candidate genes.
We first established conditions for FISH in the larval tracheal system using two genes known to be expressed and required in terminal cells. We used probes for the genes encoding serum response factor (SRF), a transcription factor (Affolter, 1994; Guillemin, 1996) and the cytoplasmic fibroblast growth factor-signalling protein Dof (Vincent et al, 1998). We reasoned that the srf mRNA should be localized near the nucleus, whereas the mRNA for dof might be found at a distance from the nucleus. Our in situ hybridization experiments confirmed this. In addition, the mRNA for CG9924 was found in tracheal branches, validating the results from the MS2-tagged mRNA (Fig 3B–D; controls in supplementary Fig S5 online). We were not successful in obtaining in situ hybridizations for btsz, hr39 and CG30403, but confirmed by RT–PCR from isolated tracheal tissue that btsz, hr39 and CG30403 are expressed in these cells (Fig 3A). Combining our own and published results indicates that in all of the seven cases (btsz, lola, Hsp70Aa, CG9924, ATPalpha, hr39 and CG30403) the endogenous RNA of the tagged gene is expressed in tracheal cells. This indicates that, unexpectedly, the screen preferentially identified genes for which the endogenous RNA is also present in tracheal cells, although the expression of the tagged transgenes was artificially induced by GAL4. Antibodies are available for Dof and ATPalpha. They show that the proteins are also found in the terminal branches (supplementary Fig S5E,F online).
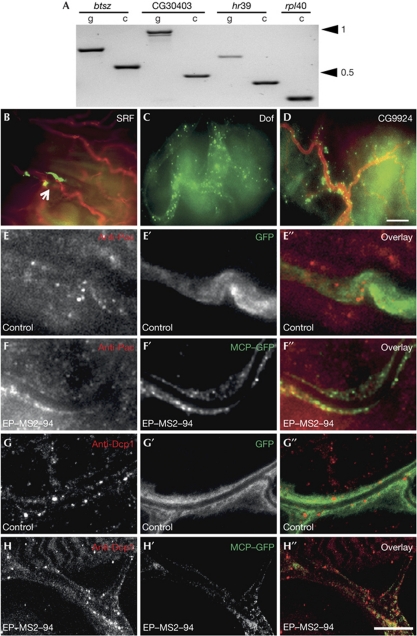
Figure 3.
Validation of candidates. (A) Expression of candidate genes in tracheal cells: polymerase chain reaction products for btsz, CG30403, hr39 and rpl40 as positive control using genomic DNA (g) or complementary DNA prepared from larval tracheal RNA (c) as template; 1- and 0.5-kb size bands are indicated. (B–D) Fluorescent in situ hybridization with RNA probes against endogenous messenger RNAs (mRNAs) of srf, dof and CG9924 on larval tracheal cells. The arrowhead in B marks the position of the nucleus of the tracheal cell. (E–H) Parts of terminal branches expressing cytoplasmic green fluorescent protein (GFP; E′,G′) or MS2 coat protein (MCP)–GFP (F′,H′) together with the MS2 mRNA from line EP–MS2–51. Larval fillets were stained with antibodies against Pacman (E,F) or Dcp1 (G,H). Pacman- and Dcp1-positive puncta are seen in the tracheal cell, as well as in the underlying muscle tissue (E–H). E″, F″, G″ and H″—overlays. Scale bars: B–D, 30 μm and E–H, 10 μm; SRF, serum response factor.
RNA transported over long distances in cells, for example in neurons, is often seen in particles, and the punctate staining we observed was therefore not entirely unexpected. We nevertheless tested whether the puncta might be stress granules, perhaps caused by overexpression of two transgenes, by immunostaining for Pacman (Drosophila Ribonuclease 1), a marker for stress granules and P bodies (Till, 1998; Anderson & Kedersha, 2006), and the decapping enzyme Dcp1 (Lin et al, 2006). In both wild-type larvae and in larvae expressing the tagged mRNA of candidate EP–MS2–94, tracheal branches contain Pacman-positive granules. However, the tagged mRNA puncta are distinct from these Pacman-positive granules (Fig 3E–F″). They might represent RNA transport or storage particles. Dcp1 was detectable in some of the particles, which might indicate that some of them could be stress granules (Fig 3G–H″).
Functional relevance of identified genes
To test whether the seven genes identified in the screen were required for tracheal development or physiology, we knocked down their expression in the tracheal system using in vivo RNA interference. In six cases, this resulted in lethality (Hsp70Aa, lola, CG9924, CG30403, ATPalpha and bitesize), suggesting that these genes do indeed have a role in the tracheal system. No effect was seen for Hr39.
General suitability for tissue-specific screens
To test the general suitability of this system to screen for asymmetric RNA distribution in vivo, we analysed some of the lines in oocytes and neurons. We tested all 12 positive lines and 63 negative lines in oocytes. In this experiment, the MCP–GFP construct had been made in the UASp vector to enable efficient expression in the germ line, and the signal was enhanced by using anti-GFP antibodies to detect GFP in fixed ovaries. In line with EP–MS2–152, the signal was concentrated at the anterior pole of the late oocyte (Fig 4F′), whereas no signal was seen in the oocyte cytoplasm in any of the other lines (Fig 4E′). This line had scored negative in tracheal cells, and conversely none of the lines that had been scored as positive in tracheal cells showed a localized signal in the oocyte. The frequency of lines showing polarized mRNA distribution in the oocyte is unexpectedly low, as many different mRNAs are deposited in the oocyte. This low frequency could either reflect the limited sensitivity of our method or it could suggest that only relatively few mRNAs show a tight subcellular localization in the oocyte.
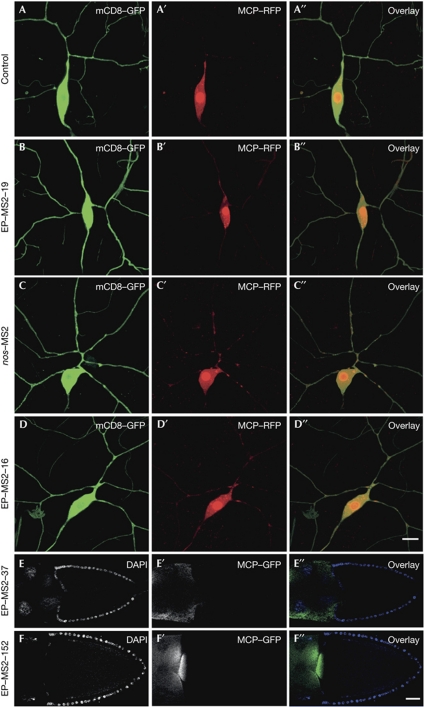
Figure 4.
Suitability of the EP–MS2 technique in other Drosophila tissues. (A–D″) Distribution of MS2 coat protein (MCP)–RFP in third-instar da2 neurons coexpressing the indicated tagged mRNAs and mCD8–green fluorescent protein (GFP) to visualize neurons. All constructs are expressed under the control of GAL4477. (A′) Control expressing MCP–RFP without MS2-tagged RNA. (B′) MCP–RFP signal from a negative line. (C′) nos-MS2 distribution in the dendrites of the da2 neurons. (D′) A similar punctate pattern of MCP–RFP is seen in the branches of a neuron expressing the tagged RNA of line EP–MS2–16. (E,F) Egg chambers expressing MCP–GFP and MS2-tagged RNA from a negative (E′) and a positive (F′) line. Nuclei are stained with DAPI (4,6-diamidino-2-phenylindole, blue). E″ and F″—overlays. Scale bars: A–D″, 20 μm and E–F″, 25 μm.
Two positive lines from the tracheal screen (EP–MS2–67 and EP–MS2–18) and three negative lines (EP–MS2–12, EP–MS2–16 and EP–MS2–19) were tested in a specific subset of larval peripheral neurons, the dendritic arborization (da) neurons, using the GAL4477 driver line (Grueber et al, 2003). EP–MS2–16 showed a punctate red fluorescent protein (RFP) signal outside the nucleus (Fig 4D′), which differed from the signal seen in the absence of a tagged mRNA or in lines scored as negative in neurons (Fig 4A,B). The distribution resembled that of similarly tagged nanos mRNA (Fig 4C′), which we used as control as it had previously been shown to be transported into the branches of the dendritic arbour (Brechbiel & Gavis, 2008). Moreover, in this situation, the RNA is found throughout the branches in a punctate pattern thought to represent transport or storage particles, supporting the notion that the granules we observe in tracheal cells might have a similar function. These results show that the method is suited to reveal mRNA localization in different tissues. Again, the two lines that were positive in tracheal cells were negative in neurons, and EP–MS2–16 had been scored negative both in the tracheal system and in the oocyte.
Our results show that the tagged mRNAs behave differently in different tissues. None of the 12 lines with mRNA localization in terminal branches showed a polarized distribution in oocytes or neurons, and conversely the transcripts that are localized in the oocyte or neurons show no distinct localization in tracheal cells. It is possible that in some cases these apparent differences in localization are in fact due to differences in expression levels caused by sequences near the insertion site of the EP–MS2 transgene. Alternatively, the different localization patterns might indicate that polarized mRNA localization is restricted to those cells in which the transcript is normally expressed. If the latter is true, RNA localization signal recognition and/or the association of RNAs with cytoskeletal transport machinery might involve not only general localization factors but also factors that function in a cell-type-specific manner. The acquisition of larger data sets will make it possible to distinguish between these possibilities.
Conclusion
In summary, the method described here is suitable for identifying mRNAs with asymmetric distributions in various cell types. Although numerous genes might be missed because of low stability of the targeted mRNA, bias of P-element insertion or other factors discussed above, the new method has many advantages. As the mRNA to be tested is exclusively expressed in the cell type of interest, its visualization is not hampered by gene expression in surrounding tissues. This is particularly important for rare cells, or for cells that are embedded in other tissues, such as neurons or tracheal cells, and which might share gene expression with surrounding tissues. Moreover, our screen is independent of the abundance of the natural mRNA, as it uses a strong promoter for the expression of the tagged mRNA. A further advantage is that the P-element insertion provides an immediate genetic handle on the targeted gene.
Methods
Fly maintenance, tissue fixation, antibody staining and molecular techniques were performed by following standard protocols. Lists of stocks and reagents can be found in the supplementary information online.
Microscopes, imaging and data analysis software. Leica TCS SP2, Zeiss Axioplan 2-imaging, Zeiss Apotome and Leica M2 16FA were used for microscopy, and Quantix (Photometrix) and Axiocam HRm (Zeiss) cameras for imaging. Images were acquired with Leica Confocal Software LCS, Axiovision Rel 4.6 (Zeiss) and Axiovision 1 (Zeiss) and edited in Adobe Photoshop Adobe Systems and ImageJ.
EP–MS2 and NLS–MCP–eGFP transgenes. The EP–MS2 constructs were generated in the EPg vector (gift from Pernille Rørth; Mata et al, 2000), which is similar to the standard EP vector (Rørth, 1996), but contains the transposase promoter instead of the Hsp70 basal promoter. A 773-bp, T4-polymerase-blunted BamH1/Nsi1 fragment from pSL–MS2 (Bertrand, 1998), which contains six repeats of the MS2 stem loop fragment (6 × MS2–BS), was inserted in the blunt-ended PstI site of EPg between the transposase promoter and the 3′ P-element.
The enhanced GFP (eGFP) coding sequence was PCR amplified from the pBI eGFP plasmid (Clontech) using the primers eGFP1 and eGFP2 (see supplementary Table S2 online). The amplicon was cloned using the Not1 and Xba1 sites into pUAST (Brand & Perrimon, 1993) and pUASp (Rørth, 1998) to generate pUAST–eGFP and pUASp–eGFP. A BamH1/Not1 fragment containing the NLS–HA–MCP sequence from pGFP–MS2/LEU (Bertrand, 1998) was cloned into the Bgl2/Not1 sites of pUAST–eGFP and BamH1/Not1 sites of pUASP–eGFP to generate the vectors pNLS–MCP–eGFP-T and pNLS–MCP–eGFP-P, respectively.
EP–MS2, pNLS–MCP–eGFP-T and pNLS–MCP–eGFP-P plasmids were injected into w[1118] embryos to create transgenic flies by standard methods.
Larval preparation for screen. For low-resolution screening under a fluorescent dissecting microscope, larvae were immobilized by drowning in normal tap water for 30 min and mounted on slides. For high-resolution microscopy, larval fillets were fixed for 20 min in 4% paraformaldehyde (PFA) and mounted in Vectashield.
Immunostainings. Third-instar larvae were filleted and fixed with 4% PFA for 20 min. Fixed fillets were washed with 0.1% PBST (PBS + 0.1% Triton X-100) three times for 10 min, followed by 1 h incubation in blocking reagent (PBST, 1% BSA). The fillets were incubated overnight in primary antibody at 4°C, washed four times (15 min each) in 0.1% PBST, incubated in fluorochrome-conjugated secondary antibodies for 2 h at room temperature (RT), washed for 2 h at RT in 0.1% PBST and mounted in Vectashield.
Ovaries were fixed for 10 min at RT in 8% methanol-free formaldehyde (Polyscience) diluted in PBS. After two short washes in 0.1% PBST, they were blocked for 1 h in 1% PBST, 0.5% BSA, incubated overnight at RT with an anti-GFP antibody in 0.3% PBST, 0.5% BSA. After two short washes with 0.1% PBST, and a 1 h wash with 0.1% PBST and 10% normal goat serum, ovaries were incubated for 2 h with a secondary antibody coupled to Alexa488 (Invitrogen).
Fluorescent in situ hybridization. The dorsal branches of the larval tracheal systems were not suited for in situ hybridizations as the tips of the branches are too deeply embedded in the surrounding tissues and are either damaged during preparation or not reachable for the probes. We therefore analysed tracheal branches on the midgut or on imaginal discs. Tissues from third-instar larvae were dissected in ice-cold PBS, fixed immediately for 20 min in 4% PFA (EM grade) and washed several times in 0.2% PBST. FISH was carried out as described in the study by Lecuyer et al (2008) using Alexa488-TSA (Invitrogen).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank P. Filardo for the original idea of the screen and the pSL–MS2–BS plasmid, P. Rørth for the EPg vector, S.F. Newbury, Tze-Bin Chou, DSHB, M. Freeman and VDRC for flies and reagents, I. Willaschek for assistance in the oocyte screen, and Sara Sigurbjörnsdóttir for assistance in the RT–PCR experiments. This work was supported by grants from the German Research Council to M.L. and V.R. (SFB 572/3, grants A9 and B9), National Institutes of Health grant R01 GM067758 to E.R.G., a fellowship from the Cologne International Graduate School in Genetics and Functional Genomics to J.N.N., and funding from European Molecular Biology Organization for the lab of M.L.
Footnotes
Maria Leptin is Director of EMBO. EMBO financially contributes to Maria Leptin’s research programme.
References
- Affolter M et al. (1994) The Drosophila SRF homolog is expressed in a subset of tracheal cells and maps within a genomic region required for tracheal development. Development 120: 743–753 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N (2006) RNA granules. J Cell Biol 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ et al. (2004) The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E et al. (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Brechbiel JL, Gavis ER (2008) Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr Biol 18: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A, Meignin C, Davis I (2007) A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134: 1955–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutelis JB, Ephrussi A (2007) Rab6 mediates membrane organization and determinant localization during Drosophila oogenesis. Development 134: 1419–1430 [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER (2003) Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 13: 1159–1168 [DOI] [PubMed] [Google Scholar]
- Gao FB (1998) Messenger RNAs in dendrites: localization, stability, and implications for neuronal function. Bioessays 20: 70–78 [DOI] [PubMed] [Google Scholar]
- Giniger E, Tietje K, Jan LY, Jan YN (1994) lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 120: 1385–1398 [DOI] [PubMed] [Google Scholar]
- Goeke S et al. (2003) Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat Neurosci 6: 917–924 [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN (2003) Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112: 805–818 [DOI] [PubMed] [Google Scholar]
- Guillemin K et al. (1996) The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development 122: 1353–1362 [DOI] [PubMed] [Google Scholar]
- Jansen RP, Kiebler M (2005) Intracellular RNA sorting, transport and localization. Nat Struct Mol Biol 12: 826–829 [DOI] [PubMed] [Google Scholar]
- Lecuyer E et al. (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Necakov AS, Caceres L, Krause HM (2008) High-resolution fluorescent in situ hybridization of Drosophila embryos and tissues. Cold Spring Harbor Protocols 2008: pdb.prot5019. [DOI] [PubMed] [Google Scholar]
- Lin MD, Fan SJ, Hsu WS, Chou TB (2006) Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev Cell 10: 601–613 [DOI] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rørth P (2000) Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101: 511–522 [DOI] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG (2008) Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia R et al. (2003) An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci 23: 9409–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SM, Palladino MJ, Beitel GJ (2007) A pump-independent function of the Na,K-ATPase is required for epithelial junction function and tracheal tube-size control. Development 134: 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Condeelis J, Singer RH, Dictenberg JB (2007) Imaging mRNA movement from transcription sites to translation sites. Semin Cell Dev Biol 18: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P (1996) A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA 93: 12418–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P (1998) Gal4 in the Drosophila female germline. Mech Dev 78: 113–118 [DOI] [PubMed] [Google Scholar]
- Serano J, Rubin GM (2003) The Drosophila synaptotagmin-like protein bitesize is required for growth and has mRNA localization sequences within its open reading frame. Proc Natl Acad Sci USA 100: 13368–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC et al. (1995) Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci USA 92: 10824–10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D (2005) Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol 6: 363–375 [DOI] [PubMed] [Google Scholar]
- Till DD et al. (1998) Identification and developmental expression of a 5′-3′ exoribonuclease from Drosophila melanogaster. Mech Dev 79: 51–55 [DOI] [PubMed] [Google Scholar]
- Vincent S, Wilson R, Coelho C, Affolter M, Leptin M (1998) The Drosophila protein Dof is specifically required for FGF signaling. Mol Cell 2: 515–525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.