Abstract
Although amyloid fibrils are generally considered to be causative or contributing agents in amyloid diseases, several amyloid fibrils are also believed to have biological functions. Among these are fibrils formed by Pmel17 within melanosomes, which act as a template for melanin deposition. We use solid-state NMR to show that the molecular structures of fibrils formed by the 130-residue pseudo-repeat domain Pmel17:RPT are polymorphic even within the biologically relevant pH range. Thus, biological function in amyloid fibrils does not necessarily imply a unique molecular structure. Solid-state NMR spectra of three Pmel17:RPT polymorphs show that in all cases, only a subset (∼30%) of the full amino acid sequence contributes to the immobilized fibril core. Although the repetitive nature of the sequence and incomplete spectral resolution prevent the determination of unique chemical shift assignments from two- and three-dimensional solid-state NMR spectra, we use a Monte Carlo assignment algorithm to identify protein segments that are present in or absent from the fibril core. The results show that the identity of the core-forming segments varies from one polymorph to another, a phenomenon known as segmental polymorphism.
Introduction
Interest in amyloid fibrils arose originally from their involvement in neurodegenerative and aging-related diseases, including Alzheimer's disease, Parkinson's disease, Huntington's disease, and type 2 diabetes. More recently, several examples of amyloid fibrils with biological functions have been identified, including extracellular curli fibrils of Escherichia coli and other bacteria, which have an adhesive function (1), and HET-s fibrils of Podospora anserina, which are required for heterokaryon incompatibility (2). Mammalian Pmel17 fibrils, which are proposed to function as templates to which melanin is deposited within melanosomes, and possibly to protect cells from adverse effects of the reactive groups that comprise this important pigment (3–7), are the subject of the experiments described below.
As our understanding of the molecular basis of amyloid formation has progressed, two surprising aspects of amyloid structures have emerged: 1) Amyloid fibrils are often polymorphic at the molecular level, meaning that a single amino acid sequence can adopt multiple distinct and self-propagating fibril structures. In many cases, different polymorphs have obviously different morphologies in transmission electron microscope (TEM) images (8,9). In all cases, different polymorphs have different solid-state NMR spectra (9–11). 2) The core structure of an amyloid fibril may not include the entire amino acid sequence. Segments that form the relatively rigid and ordered cross-β structure that characterizes all amyloid fibrils can coexist with segments that are mobile and disordered (10,12), and even with domains that retain globular structures and enzymatic functions (13,14).
Here, we report results from solid-state NMR studies of fibrils formed by Pmel17:RPT, a 130-residue domain of Pmel17 that forms amyloid fibrils within the biologically relevant acidic pH range of melanosomes and is required for in vivo fibril formation by the full-length protein (4,7,15). Our results bear on both aspects of fibril structure described above. We show that Pmel17:RPT fibrils are highly polymorphic at biologically relevant pH, even though Pmel17 is a functional amyloid. Subtle variations in growth conditions lead to fibrils that are readily distinguishable in solid-state NMR spectra. This observation is in contrast to the behavior of HET-s fibrils, which exhibit a single, highly uniform structure at biologically relevant pH (12). Moreover, we show that Pmel17:RPT polymorphs differ in the identity of the protein segments that form the fibril core. This mechanism of polymorphism was termed segmental polymorphism by Wiltzius and co-workers (16). Similarly direct experimental evidence for segmental polymorphism in amyloid fibrils, as opposed to other types of structural variations (10,17), has not been described previously.
Materials and Methods
Fibril growth
His-tagged Pmel17:RPT was expressed in E. coli and purified as previously described (4), except that cells were grown in media containing 98% 13C-glucose and 99% 15N-amonium chloride. Three polymorphs were prepared as follows: 1) P1 fibrils were grown from 120 μM Pmel17:RPT in 125 mM potassium acetate, pH 5.0, with continual rotary agitation at room temperature. 2) P2 fibrils were prepared by seeded growth, starting from parent fibrils that were grown under nominally the same conditions as P1, although solid-state NMR spectra revealed the parent fibrils to be inhomogeneous. Parent fibrils were broken into fragments (i.e., seeds) with lengths < 100 nm by probe sonication (Branson (Danbury, CT) sonifier model 250, lowest power, 1 min), and seeds were added to a monomeric 120 μM Pmel17:RPT solution. The monomeric solution was prepared initially in 10 mM TrisHCl at pH 7.5, where Pmel17:RPT has been shown to be monomeric (4), and then acidified by addition of 1 M potassium acetate, pH 5.0, to reach a final 125 mM potassium acetate concentration immediately before seeding. The absence of spontaneous aggregation and the presence of fibril seeds before and after seeding were verified by TEM. Fibril growth was monitored by both TEM and Thioflavin T fluorescence, and found to be complete in 7–10 days. The seeded-growth protocol was repeated for three successive generations. All three P2 generations exhibited identical solid-state NMR spectra. The data described below were obtained from the second P2 generation. 3) P3 fibrils were grown from 60 μM Pmel17:RPT in 100 mM sodium chloride, 25 mM potassium acetate, pH 5.5, with continual agitation in a shaking incubator at 37°C.
Fibrils with 13C labels only at backbone carbonyl sites of Leu or Met residues were grown under P1 conditions from Pmel17:RPT that was expressed with selectively labeled media, as previously described (18).
The fibril growth conditions described above led to structurally homogeneous samples. An exhaustive screening of growth conditions was not performed.
Electron microscopy
A 5 μl fibril aliquot (diluted to ∼2 μM protein) was applied to a carbon-film-coated copper grid and then blotted to a thin layer of liquid. The sample was stained with 5 μl of 3% uranium acetate solution, blotted, rinsed once with buffer, and blotted again before it was dried in air. Images were obtained at 56,000–71,000× magnification with an FEI (Hillsboro, OR) Morgagni transmission electron microscope operating at 80 kV.
NMR
Fibrils were pelleted at 50,000 × g and transferred into thin-wall 3.2 mm Varian (Palo Alto, CA) magic-angle spinning (MAS) NMR rotors (10–12 mg of fibrils in a 32 μl sample volume). NMR spectra were recorded at 14.1 T (599.08 MHz, 150.65 MHz, and 60.71 MHz for 1H, 13C and 15N, respectively) with the use of a three-channel Varian MAS probe and a Varian InfinityPlus NMR spectrometer. The sample temperatures during the NMR measurements were 5–10°C, controlled by cold nitrogen gas.
Two-dimensional (2D) 13C-13C NMR spectra were recorded with MAS at 12.5 kHz, using either 2.56 ms finite-pulse radiofrequency-driven recoupling (fpRFDR) or 20 ms rotation-assisted diffusion (RAD) exchange periods (19–21). The 13C π pulses were 25.0 μs during fpRFDR. fpRFDR was used for P2 fibrils, and RAD was used for P1 and P3 fibrils. The proton decoupling fields were 100 kHz, with two-pulse phase modulation (TPPM) (22). The total measurement time was ∼18 h, using 2.0 s recycle delays, 128 complex t1 points, and 30.6 μs t1 increments. 2D 1H-13C INEPT NMR spectra were recorded with MAS at 4 kHz. Total INEPT periods for 1H-13C polarization transfer due to one-bond scalar couplings were 4 ms, chosen to maximize 13C NMR signals in the presence of T2 relaxation. The WALTZ sequence (23) was used for proton decoupling, with 12.5 kHz decoupling fields. The total measurement time was ∼60 min, using 1.0 s recycle delays, 100 complex t1 points, and 75.0 μs t1 increments.
Three-dimensional (3D) NCOCX and NCACX spectra were recorded with MAS at 12.5 kHz, using 4 ms frequency-selective cross-polarization periods for 15N-13CO or 15N-13Cα polarization transfer after t1, and 20 ms RAD periods for 13C-13C polarization transfer after t2. Forty-eight complex t1 points were acquired with a 160.0 μs increment, and 64 t2 points were acquired with an 80.0 μs increment, using time-proportional phase incrementation (TPPI) (24). The proton TPPM decoupling fields were 90 kHz. 3D CONCA spectra were also recorded with MAS at 12.5 kHz using 4 ms frequency-selective 13CO-15N and 15N-13Cα cross-polarization periods after t1 and t2, respectively. Thirty-two complex t1 points were acquired with a 160.0 μs increment, and 96 t2 points were acquired with an 80.0 μs increment, using TPPI. The total measurement time for each 3D spectrum was ∼4 days, using 1.0 s recycle delays.
As discussed below, chemical shift assignments consistent with the solid-state NMR data were obtained with the MCASSIGN2 program (25–27). Input tables (see Table S1, Table S2, and Table S3 in the Supporting Material) were created by manual analysis of the 3D spectra, using Sparky (available at http://www.cgl.ucsf.edu/home/sparky/). 2D 13C-13C NMR spectra were used to identify sets of 13C NMR frequencies arising from the same residue and to assist with residue-type assignments. 100 MCASSIGN2 runs were performed for each Ppmel17:RPT fibril morphology. The score function in MCASSIGN2 penalizes assignments with bad connections (i.e., inconsistencies in chemical shift assignments among different 3D spectra), assignments with numerous edges (i.e., gaps between segments that have assigned signals), and assignments that do not use all available solid-state NMR signals. For P1, P2, and P3 fibrils, the number of high-scoring runs was 44, 66, and 38, respectively. Discarded runs had either bad connections or large numbers of edges. Table S4, Table S5, and Table S6 include only the unique assignments, i.e., assignments that were the same in all high-scoring runs. MCASSIGN2 is described in detail elsewhere (26) and is available upon request (e-mail to robertty@mail.nih.gov).
Measurements of 13C-13C dipole-dipole couplings in selectively labeled Pmel17:RPT fibrils were performed with the PITHIRDS-CT dipolar recoupling technique (28) at a 100.4 MHz 13C NMR frequency, using the same experimental conditions as in previous applications of this technique to amyloid fibrils (10,27).
Results
Electron microscopy of Pmel17:RPT fibrils
As described above, three Pmel17:RPT fibril samples (P1, P2, and P3) were produced from uniformly 15N,13C-labeled Pmel17:RPT under somewhat different conditions. Negatively stained TEM images in Fig. 1 show similar morphologies and widths (10–14 nm) for all three fibril samples. P2 fibrils (Fig. 1, b and e) have the greatest lengths due to the absence of agitation during growth. All three samples were found to accelerate melanin polymerization in vitro by nearly identical factors (Fig. S1).
Figure 1.
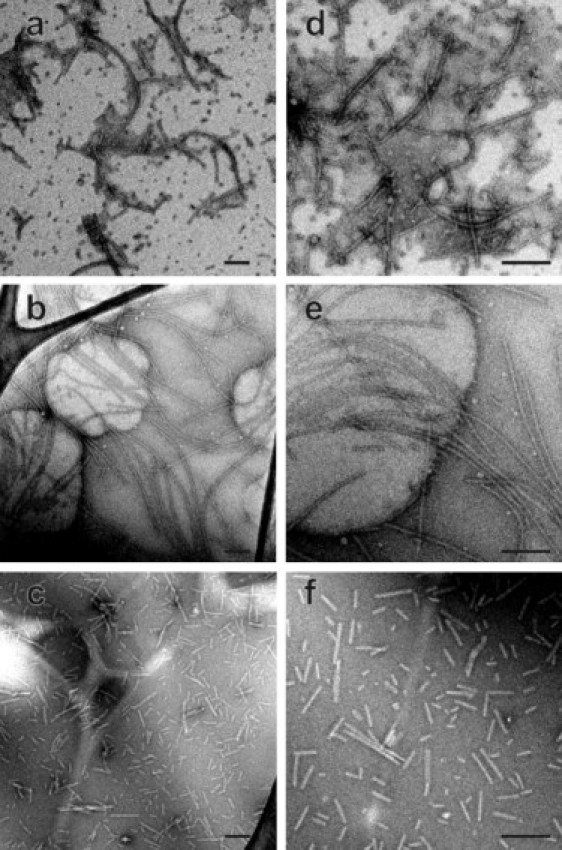
TEM images of negatively stained Pmel17:RPT fibrils. Images are shown for P1 (a and d), P2 (b and e), and P3 (c and f) samples. Scale bars: 200 nm.
Solid-state NMR reveals polymorphism at the molecular structural level
The 2D 13C-13C NMR spectra in Fig. 2 show dramatic differences in 13C NMR chemical shifts, as revealed by the crosspeak positions and intensity patterns. These differences imply that the molecular structures within P1, P2, and P3 fibrils are different, despite their similar appearances in TEM images. Some of the clearest differences occur in spectral regions that contain Cα/Cβ crosspeaks of Ala residues (region around 50 ppm/20 ppm in Fig. 2, d–f), Leu and Met residues (region around 54 ppm/42 ppm), and Ser and Thr residues (region around 70 ppm/60 ppm). Close examination of the spectra of P1, P2, and P3 fibrils shows that they represent three distinct structures rather than three different proportions of the same structures. Of importance, the density of crosspeaks in all of the 2D 13C-13C NMR spectra is low for a protein with 130 residues, implying that the immobile fibril core contains only a subset of the Pmel17:RPT sequence.
Figure 2.
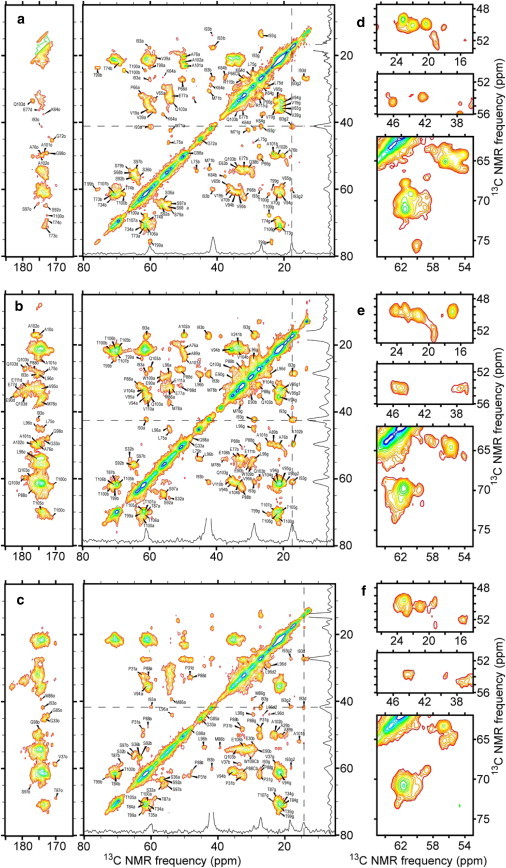
2D 13C-13C solid-state NMR spectra of Pmel17:RPT fibrils. Spectra of P1 (a and d), P2 (b and e), and P3 (c and f) fibrils are shown. Differences among these spectra reflect the polymorphism of Pmel17:RPT fibrils, with molecular structural differences resulting from different fibril growth conditions. 1D slices at chemical shifts of the dashed horizontal and vertical lines illustrate the NMR linewidths and signal/noise ratios. Subsets of the definite 13C chemical shift assignments are shown, with a, b, g, d, e, and o indicating shifts for α-, β-, γ-, δ-, ε-, and carbonyl carbons, respectively, along the horizontal frequency dimension.
The 2D 1H-13C INEPT NMR spectra recorded under conditions appropriate for solution NMR reveal signals from many mobile residues (Fig. S2). 1H and 13C chemical shifts in the INEPT spectra are close to random coil values (29), indicating the presence of dynamically disordered segments outside the fibril core.
To identify the immobilized, core-forming segments in Pmel17:RPT fibrils, we recorded 3D NCACX, NCOCX, and CONCA spectra of the three samples (Fig. S3). These spectra connect backbone 15N chemical shifts of residue k with 13C chemical shifts of residue k, with 13C chemical shifts of residue k-1, and with 13C chemical shifts of both residue k-1 and residue k, respectively. We first attempted to make site-specific chemical shift assignments by the standard approach of tracing the sequential connectivities of 13C and 15N chemical shifts in the 3D spectra by hand. Although it was possible to find plausible connectivity pathways for many 3D crosspeaks, these pathways were not unique. Consequently, unambiguous chemical shift assignments could not be determined manually. The chief difficulties were the presence of spectral overlap, the repetitive nature of the Pmel17:RPT sequence, and the fact that only a subset of the sequence contributes to the NMR signals. As an alternative, we applied a computational approach, using the Monte Carlo/simulated annealing (MCSA) algorithm that we recently introduced for problems of this type (25–27), implemented in the program MCASSIGN2. As input to MCASSIGN2, we prepared tables of multidimensional signals from the 3D spectra (Table S1, Table S2, and Table S3), which include the chemical shifts, uncertainties in the shifts, and possible residue-type assignments. We obtained lists of possible residue-type assignments for each signal by careful examining the 3D and 2D spectra using the known 13C chemical shift ranges for various residues. The number of distinct multidimensional signals in each 3D spectrum ranges from 31 (NCOCX spectrum of P2 fibrils) to 42 (NCOCX spectrum of P3 fibrils), further confirming that the fibril cores contain only subsets of the full protein sequence. As previously described (25,26), the MCSA algorithm searches for assignments of multidimensional signals to individual residues in the protein sequence that satisfy the expected agreement among sets of chemical shifts in each pair of 3D spectra, while minimizing the number of unassigned signals and the number of gaps that separate segments of the protein sequence to which signals are assigned.
Fig. 3 summarizes the MCSA assignment results from 100 independent MCASSIGN2 runs for each sample. Only high-scoring runs that produced final assignments with no bad connections (i.e., no disagreements among chemical shift assignments in different 3D spectra) are included in Fig. 3. In these successful runs, the total number of assigned signals ranged from 100 to 106 for P1 fibrils (from a total of 112 signals in the three 3D spectra), 95 to 98 for P2 fibrils (from a total of 98), and 120 to 124 for P3 fibrils (from a total of 124). Thus, relatively few signals were unassigned, although not all runs produced the same residue-specific assignments. Residues with at least one backbone assignment (amide 15N, 13Cα, or 13CO) in every successful MCASSIGN2 run are underlined in the amino acid sequence above each panel in Fig. 3. These residues definitely contribute strong signals to the solid-state NMR data, and hence (by definition) are contained in the immobilized fibril core. Residues with no assignments for any backbone sites in any run are struck through. These residues are apparently sufficiently mobile to avoid contributing to the solid-state NMR signals, and hence are outside the fibril core. Unmarked residues have backbone assignments in some (but not all) successful MCASSIGN2 runs, meaning that we cannot determine definitely whether they are contained in the fibril core from the available data.
Figure 3.
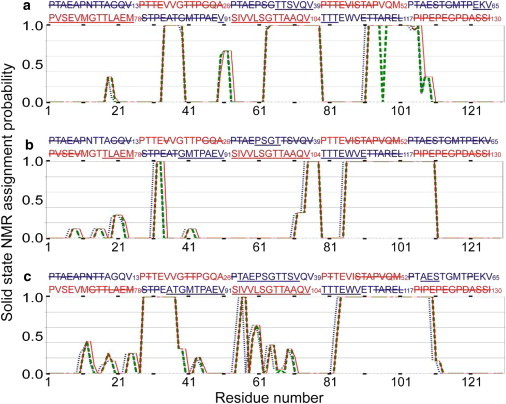
Results of Monte Carlo/simulated annealing chemical shift assignment runs for Pmel17:RPT fibrils. Plots for (a) P1, (b) P2, and (c) P3 fibrils show the fraction of high-scoring MCASSIGN2 runs that assigned signals to backbone 15N (thin lines), 13Cα (heavy dashed lines), or backbone 13CO (dotted lines) sites of each residue in the Pmel17:RPT sequence. In the sequence above each plot, residues that never have any assignments, and hence do not contribute to the solid-state NMR signals, are struck through. Residues that always have at least one assignment, and hence definitely contribute to the solid-state NMR signals, are underlined. Shaded lettering emphasizes the pseudo-repetitive nature of the Pmel17:RPT sequence.
Pmel17:RPT fibril polymorphs have distinct core-forming segments
The results in Fig. 3 imply certain common structural features in the three Pmel17:RPT fibril polymorphs. Residues 91–107 always have backbone assignments and hence are contained in the core of P1, P2 and P3 fibrils. Residues 1–6, 27, 46–49, 62, 79–81, and 113–130 never have backbone assignments and hence are outside the fibril core in the three polymorphs. It is possible that residues 1–27, 40–54, 58–62, 79–82, and 112–130 are outside the fibril core in the three polymorphs. However, the core-forming segments in the three Pmel17:RPT fibril polymorphs are clearly not identical. For example, residues 28–30 and 55–57 are in the core of P3 fibrils but outside the core of P1 and P2 fibrils; residues 36 and 37 are in the core of P1 and P3 fibrils but outside the core of P2 fibrils; residues 63–70 are in the core of P1 fibrils but outside the core of P2 fibrils; residues 74–78 are in the core of P1 and P2 fibrils but outside the core of P3 fibrils; and residues 85–90 are in the core of P2 and P3 fibrils but outside the core of P1 fibrils. These results demonstrate that Pmel17:RPT fibrils exhibit segmental polymorphism (16).
Table S4, Table S5, and Table S6 list the 15N and 13C chemical shifts for residues that have unambiguous assignments (i.e., the same assignment in all successful MCASSIGN2 runs). In Fig. S4 we plot secondary 13C shifts. Most (but not all) nonglycine residues have secondary shifts consistent with β-strand conformations, i.e., negative secondary shifts for 13CO and 13Cα sites and positive secondary shifts for 13Cβ sites. Residues with non-β-strand shifts are not the same in all three polymorphs. For example, non-β-strand shifts are observed at S97 and T100 in P1 fibrils; at T99, T100, A102, and A103 in P2 fibrils; and at S97, T99, and A101 in P3 fibrils. Thus, it is likely that conformational differences also occur even within core segments that are common to all three polymorphs.
2D 1H-13C INEPT NMR spectra of P1 and P2 fibrils in Fig. S2 include signals that are assigned to mobile Thr, Val, and Ala residues that precede prolines, based on the effect of the proline on random coil 13C chemical shifts (29). From Fig. 3, these mobile Thr residues could be T22, T61, T80, and T87 in P1 fibrils, and they could be T22, T61, and T80 in P2 fibrils, as these residues are definitely outside or possibly outside the fibril cores. Mobile Val residues could only be V13 in P1 fibrils, and they could be V13, V39, and V65 in P2 fibrils. Mobile Ala residues could be A5, A26, and A48 in both P1 and P2 fibrils. In general, the 2D 1H-13C INEPT NMR spectra are consistent with results in Fig. 3, as the residue types that contribute to these spectra occur in segments identified as being outside the fibril cores in Fig. 3.
Fibril formation is accompanied by neutralization of Glu side chains
Pmel17:RPT forms fibrils only below pH ≈ 6 (4,5), corresponding to the acidic environment within melanosomes (reported to be below pH 5 (30)). Pmel17:RPT fibrils dissolve at neutral pH (4,5). Reversible, pH-dependent fibril formation is attributable to neutralization of a subset of the Glu side chains. Neutralization changes 13Cδ chemical shifts of Glu residues from above 183 ppm to below this value (31,32). Glu Cγ/Cδ crosspeaks with 13Cδ chemical shifts above 183 ppm are absent from 2D 13C-13C spectra of all three Pme17:RPT fibril polymorphs (Fig. 2). Thus, it appears that all Glu residues within immobilized segments of Pmel17 are neutralized, including side chains of E63, E69, and E77 in P1 fibrils; side chains of E77, E90, E108, and E111 in P2 fibrils; and side chains of E30, E56, E90, and E108 in P3 fibrils. The pKa values of Glu side chains in proteins are commonly in the 3.5–5.0 range. In Pmel17:RPT fibrils, a subset of the Glu side chains are neutralized at the high end of this pH range, presumably because the positive free-energy change associated with neutralization of monomers is compensated for by the negative free energy of fibrillation (5). The pH dependence of Pmel17:RPT fibrillation may be biologically significant because aggregation outside of melanosomes could lead to cytotoxicity.
Measurements of 13C-13C dipole-dipole couplings in selectively 13C-labeled Pmel17:RPT fibrils, shown in Fig. S5, suggest that the β-sheets in these fibrils have a parallel structure, as found in numerous other amyloid and prion fibrils (9,10,18,27,33–35). Uncertainties regarding the number of labeled sites that participate in the cross-β core structure prevent us from drawing definitive conclusions from these measurements. If Glu side chains within the core were not neutralized, one would expect electrostatic repulsions between neighboring Pmel17:RPT molecules to destabilize a parallel β-sheet structure.
Discussion
Polymorphism is a property shared by many amyloid and prion fibrils, including disease-associated systems such as β-amyloid (Aβ) (8–10,17), α-synuclein (11), and β2-microglobulin (36) fibrils. Polymorphism is generally believed to be the basis for self-propagating strains of prions, both for mammalian PrP (37,38) and for yeast prions (39–42). Polymorphism in 40-residue Aβ fibrils has been shown to result from variations in overall symmetry and the conformations in non-β-strand segments (10), but not in the identity of the residues in the fibril core. Polymorphism in fibrils formed by the D23N mutant of Aβ has been shown to include variations in the β-sheet structure, which has always been found to be parallel in full-length, wild-type Aβ fibrils (9,10,33) but can be either parallel or antiparallel in D23N-Aβ fibrils (17). Fibrils formed by short Aβ fragments can have structures that are pH-dependent (43), but variations in the core-forming residues for a given amino acid sequence have not been reported. For mammalian PrP, fibrils with distinct morphologies formed in vitro from recombinant protein appear to have similar core-forming segments (27,35). Proteolysis experiments on infectious PrP prions suggest that distinct strains may have different cores (44,45), but it is not yet known whether infectious PrP has an amyloid structure. Distinct strains of the yeast prion Sup35NM have been shown to exhibit distinct infrared absorption spectra (suggesting differences in β-sheet content), differences in electron paramagnetic resonance (EPR) spectra (suggesting localized variations in spin-labeled side-chain mobilities), differences in hydrogen/deuterium (H/D) exchange rates in certain segments, and different sensitivities to site-specific mutations (39,40,42).
Wiltzius and co-workers (16) suggested that segmental polymorphism may occur in full-length IAPP fibrils, based on results from crystallography of IAPP fragments as well as predictions of amyloid-formation propensities. Definitive evidence for segmental polymorphism in amyloid fibrils from solid-state NMR has been lacking. The experiments described above provide this evidence in the case of Pmel17:RPT fibrils. It is particularly interesting that segmental polymorphism occurs in the fibril-forming domain of a functional amyloid, where one might assume that an evolved biological function would require a single molecular structure. Solid-state NMR experiments by Wasmer and co-workers (12) showed that the functional HET-s amyloid of P. anserina has a unique structure at biologically relevant pH. In contrast, our results show that Pmel17:RPT fibrils are polymorphic in the biologically relevant pH range (4,5,30). Thus, we hypothesize that the function of Pmel17 fibrils as templates to which melanin is deposited in melanosomes does not require a specific structure, although this function may require a structural motif shared by the various Pmel17 polymorphs. Although it is possible that full-length Pmel17 fibrils within melanosomes are more structurally homogeneous than our Pmel17:RPT fibrils (direct experiments on Pmel17 fibrils from melanosomes are required to address this issue), the absence of strict structural requirements for Pmel17 fibril function is consistent with in vitro experiments that demonstrated similar effects on melanin synthesis by other, unrelated amyloids, including α-synuclein, Aβ, HET-s218-289, and Sup35NM fibrils (3,4). Moreover, the homogeneity of HET-s prion domain fibrils formed in vitro correlates with the absence of distinct full-length HET-s prion strains in vivo, and the polymorphic nature of yeast and mammalian prion protein fibrils in vitro correlates with the presence of distinct strains in vivo.
Several groups have developed computational methods for predicting the amyloid-forming segments of proteins. Fig. 4 shows predictions for Pmel17:RPT obtained with the WALTZ (46), TANGO (47), PASTA (48), and Zyggregator (49) algorithms. All four algorithms predict that residues 90–100 participate in the fibril core, in good agreement with the results shown in Fig. 3 for all polymorphs. WALTZ, PASTA, and Zyggregator predict that this core segment extends further toward the C-terminus, also in agreement with experiments. Only partial agreement between predictions and experiments is observed elsewhere in the Pmel17:RPT sequence, indicating the difficulty of making successful predictions for segments that contribute to the fibril core in some (but not all) polymorphs.
Figure 4.
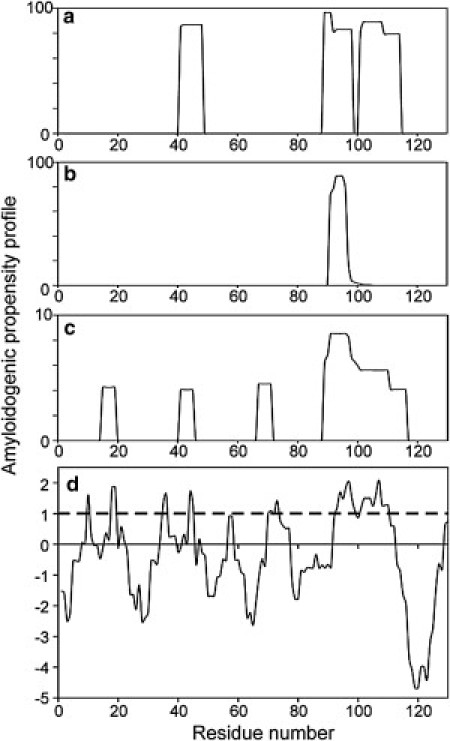
Predicted amyloidogenic propensities of the Pmel17:RPT sequence. Results from the WALTZ, TANGO, PASTA, and Zyggregator algorithms (a–d, respectively) are shown. Vertical scales are quantities produced by the prediction programs, except that the PASTA output is shown as its absolute value. For Zyggregator, values above the dashed horizontal line) indicate a strong aggregation propensity.
Solid-state NMR spectra of the three Pmel17:RPT fibril polymorphs show signals arising from between 35 and 42 residues (see Table S1, Table S2, and Table S3), or 27–32% of the amino acid sequence. Although the entire Pmel17:RPT sequence is pseudo-repetitive (consisting of 10 homologous 13-residue segments), the cross-β core structure seems not to reflect this pseudo-repetitive nature in the straightforward manner found in HET-s218-289 fibrils (12) and proposed for curli fibrils (1), because the core-forming segments in Pmel17:RPT fibrils do not correspond simply to repeat segments (Fig. 3).
For comparison, 47 out of 72 residues contribute strong signals to solid-state NMR spectra of HET-s218-289 fibrils (12,25), ∼32 residues contribute to solid-state NMR spectra of hydrated Aβ1-40 fibrils (10), <40 residues contribute to solid-state NMR spectra of PrP23-231 fibrils (27), and 30 residues contribute to solid-state NMR spectra of PrP23-144 fibrils (50). Signals from ∼40 out of 140 residues were assigned by Heise et al. (51) in solid-state NMR spectra of fibrils formed by the A53T mutant of α-synuclein, and 43 out of 99 residues were assigned by Andronesi et al. (52) in solid-state NMR spectra of fibrils formed by the K19 construct of τ. These results suggest that immobilization of 30–50 residues in the cross-β core may be a general feature of amyloid structures, regardless of the length of the polypeptide chain. On the other hand, EPR and H/D exchange studies of α-synuclein fibrils indicate somewhat longer core-forming segments (53,54). Long, straight β2-microglobulin fibrils yield solid-state NMR spectra that contain signals from at least 58 immobilized residues (36). Thus, in at least some cases, fibril cores may contain >50 residues. In the absence of detailed molecular structural models for most amyloid fibrils, the underlying reasons for the limited sizes of core-forming segments within amyloid fibrils cannot be known definitively.
The work described above demonstrates an approach for identifying core-forming segments in amyloid fibrils that is an alternative to H/D exchange (39,54–56), proteolysis (57,58), or scanning mutagenesis (42,59). Our approach rests on the assumptions that immobilized protein segments will produce strong signals under standard solid-state NMR measurement conditions, and mobile segments will not—assumptions that are supported by numerous previous studies (12,25,27,50,52,60). Although immobilized segments may also exist outside the cross-β core (for example, if a globular protein domain is tightly tethered to the core (13)), the 13C NMR chemical shifts allow us to confirm that the detected signals arise from the characteristic β-sheet structure of an amyloid core. In contrast, the association between H/D exchange and core structure is less definitive, particularly if the H/D exchange rates are not analyzed quantitatively, because even the β-sheets in stable proteins can exhibit H/D exchange on the timescale of minutes to hours, depending on the solvent conditions (61). When the cross-β core is comprised of β-strands that are separated by flexible loops, proteolysis experiments may underestimate the core-forming segments (for example, if digestion of exposed loop segments leads to unraveling and further digestion of the core structure). The effects of mutagenesis on fibril formation may also be difficult to interpret, particularly because the introduction of an amino acid substitution within a core-forming segment of one polymorph may simply drive the fibril structure toward a different polymorph.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Supporting Material
References
- 1.Barnhart M.M., Chapman M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maddelein M.L., Dos Reis S., Saupe S.J. Amyloid aggregates of the HET-s prion protein are infectious. Proc. Natl. Acad. Sci. USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler D.M., Koulov A.V., Kelly J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGlinchey R.P., Shewmaker F., Wickner R.B. The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc. Natl. Acad. Sci. USA. 2009;106:13731–13736. doi: 10.1073/pnas.0906509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfefferkorn C.M., McGlinchey R.P., Lee J.C. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc. Natl. Acad. Sci. USA. 2010;107:21447–21452. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watt B., van Niel G., Marks M.S. N-terminal domains elicit formation of functional Pmel17 amyloid fibrils. J. Biol. Chem. 2009;284:35543–35555. doi: 10.1074/jbc.M109.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoashi T., Muller J., Hearing V.J. The repeat domain of the melanosomal matrix protein PMEL17/GP100 is required for the formation of organellar fibers. J. Biol. Chem. 2006;281:21198–21208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- 8.Goldsbury C.S., Wirtz S., Frey P. Studies on the in vitro assembly of Aβ1-40: implications for the search for Aβ fibril formation inhibitors. J. Struct. Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 9.Petkova A.T., Leapman R.D., Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 10.Paravastu A.K., Leapman R.D., Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heise H., Hoyer W., Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasmer C., Lange A., Meier B.H. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 13.Loquet A., Bousset L., Böckmann A. Prion fibrils of Ure2p assembled under physiological conditions contain highly ordered, natively folded modules. J. Mol. Biol. 2009;394:108–118. doi: 10.1016/j.jmb.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Baxa U., Speransky V., Wickner R.B. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc. Natl. Acad. Sci. USA. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGlinchey R.P., Shewmaker F., Wickner R.B. Repeat domains of melanosome matrix protein Pmel17 orthologs form amyloid fibrils at the acidic melanosomal pH. J. Biol. Chem. 2011;286:8385–8393. doi: 10.1074/jbc.M110.197152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiltzius J.J.W., Landau M., Eisenberg D. Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tycko R., Sciarretta K.L., Meredith S.C. Evidence for novel β-sheet structures in Iowa mutant β-amyloid fibrils. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shewmaker F., Kryndushkin D., Wickner R.B. Two prion variants of Sup35p have in-register parallel β-sheet structures, independent of hydration. Biochemistry. 2009;48:5074–5082. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J. Chem. Phys. 2001;114:8473–8483. [Google Scholar]
- 20.Bennett A.E., Rienstra C.M., Griffin R.G. Homonuclear radio frequency-driven recoupling in rotating solids. J. Chem. Phys. 1998;108:9463–9479. [Google Scholar]
- 21.Morcombe C.R., Gaponenko V., Zilm K.W. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J. Am. Chem. Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 22.Bennett A.E., Rienstra C.M., Griffin R.G. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 23.Shaka A.J., Keeler J., Freeman R. An improved sequence for broad-band decoupling: WALTZ-16. J. Magn. Reson. 1983;52:335–338. [Google Scholar]
- 24.Drobny G., Weitekamp D.P., Pines A. Multiple quantum spectrum of oriented hexane-d6. Chem. Phys. 1986;108:179–185. [Google Scholar]
- 25.Tycko R., Hu K.-N. A Monte Carlo/simulated annealing algorithm for sequential resonance assignment in solid state NMR of uniformly labeled proteins with magic-angle spinning. J. Magn. Reson. 2010;205:304–314. doi: 10.1016/j.jmr.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu K.-N., Qiang W., Tycko R. A general Monte Carlo/simulated annealing algorithm for resonance assignment in NMR of uniformly labeled biopolymers. J. Biomol. NMR. 2011;50:267–276. doi: 10.1007/s10858-011-9517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tycko R., Savtchenko R., Baskakov I.V. The α-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel β-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry. 2010;49:9488–9497. doi: 10.1021/bi1013134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J. Chem. Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 29.Wishart D.S., Bigam C.G., Sykes B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 30.Bhatnagar V., Anjaiah S., Ramaiah A. pH of melanosomes of B 16 murine melanoma is acidic: its physiological importance in the regulation of melanin biosynthesis. Arch. Biochem. Biophys. 1993;307:183–192. doi: 10.1006/abbi.1993.1577. [DOI] [PubMed] [Google Scholar]
- 31.Qin J., Clore G.M., Gronenborn A.M. Ionization equilibria for side-chain carboxyl groups in oxidized and reduced human thioredoxin and in the complex with its target peptide from the transcription factor NF κB. Biochemistry. 1996;35:7–13. doi: 10.1021/bi952299h. [DOI] [PubMed] [Google Scholar]
- 32.Tollinger M., Forman-Kay J.D., Kay L.E. Measurement of side-chain carboxyl pK(a) values of glutamate and aspartate residues in an unfolded protein by multinuclear NMR spectroscopy. J. Am. Chem. Soc. 2002;124:5714–5717. doi: 10.1021/ja020066p. [DOI] [PubMed] [Google Scholar]
- 33.Benzinger T.L.S., Gregory D.M., Meredith S.C. Propagating structure of Alzheimer's β-amyloid(10-35) is parallel β-sheet with residues in exact register. Proc. Natl. Acad. Sci. USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ladner C.L., Chen M., Langen R. Stacked sets of parallel, in-register β-strands of β2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J. Biol. Chem. 2010;285:17137–17147. doi: 10.1074/jbc.M110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cobb N.J., Sönnichsen F.D., Surewicz W.K. Molecular architecture of human prion protein amyloid: a parallel, in-register β-structure. Proc. Natl. Acad. Sci. USA. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debelouchina G.T., Platt G.W., Griffin R.G. Magic angle spinning NMR analysis of β2-microglobulin amyloid fibrils in two distinct morphologies. J. Am. Chem. Soc. 2010;132:10414–10423. doi: 10.1021/ja102775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safar J., Wille H., Prusiner S.B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 38.Jones E.M., Surewicz W.K. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 39.Toyama B.H., Kelly M.J.S., Weissman J.S. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka M., Chien P., Weissman J.S. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Diaz-Avalos R., King C.Y., Caspar D.L. Strain-specific morphologies of yeast prion amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang H.-Y., Lin J.-Y., King C.Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc. Natl. Acad. Sci. USA. 2008;105:13345–13350. doi: 10.1073/pnas.0802215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petkova A.T., Buntkowsky G., Tycko R. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J. Mol. Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 44.Bessen R.A., Marsh R.F. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Telling G.C., Parchi P., Prusiner S.B. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 46.Maurer-Stroh S., Debulpaep M., Rousseau F. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-Escamilla A.M., Rousseau F., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 48.Trovato A., Chiti F., Seno F. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PLOS Comput. Biol. 2006;2:e170. doi: 10.1371/journal.pcbi.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tartaglia G.G., Pawar A.P., Vendruscolo M. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 50.Helmus J.J., Surewicz K., Jaroniec C.P. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heise H., Celej M.S., Baldus M. Solid-state NMR reveals structural differences between fibrils of wild-type and disease-related A53T mutant α-synuclein. J. Mol. Biol. 2008;380:444–450. doi: 10.1016/j.jmb.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 52.Andronesi O.C., von Bergen M., Baldus M. Characterization of Alzheimer's-like paired helical filaments from the core domain of τ protein using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2008;130:5922–5928. doi: 10.1021/ja7100517. [DOI] [PubMed] [Google Scholar]
- 53.Chen M., Margittai M., Langen R. Investigation of α-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 54.Del Mar C., Greenbaum E.A., Woods V.L., Jr. Structure and properties of α-synuclein and other amyloids determined at the amino acid level. Proc. Natl. Acad. Sci. USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoshino M., Katou H., Goto Y. Mapping the core of the β(2)-microglobulin amyloid fibril by H/D exchange. Nat. Struct. Biol. 2002;9:332–336. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 56.Kheterpal I., Zhou S., Wetzel R. Aβ amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc. Natl. Acad. Sci. USA. 2000;97:13597–13601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kheterpal I., Williams A., Wetzel R. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 58.Sajnani G., Pastrana M.A., Requena J.R. Scrapie prion protein structural constraints obtained by limited proteolysis and mass spectrometry. J. Mol. Biol. 2008;382:88–98. doi: 10.1016/j.jmb.2008.06.070. [DOI] [PubMed] [Google Scholar]
- 59.Williams A.D., Shivaprasad S., Wetzel R. Alanine scanning mutagenesis of Aβ (1-40) amyloid fibril stability. J. Mol. Biol. 2006;357:1283–1294. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 60.Helmus J.J., Surewicz K., Jaroniec C.P. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid-state nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orban J., Alexander P., Bryan P. Hydrogen-deuterium exchange in the free and immunoglobulin G-bound protein G B-domain. Biochemistry. 1994;33:5702–5710. doi: 10.1021/bi00185a006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


