Abstract
Alzheimer's disease is associated with the abnormal self-assembly of the amyloid-β (Aβ) peptide into toxic β-rich aggregates. Experimental studies have shown that hydrophobic nanoparticles retard Aβ fibrillation by slowing down the nucleation process; however, the effects of nanoparticles on Aβ oligomeric structures remain elusive. In this study, we investigate the conformations of Aβ(16-22) octamers in the absence and presence of a single-walled carbon nanotube (SWCNT) by performing extensive all-atom replica exchange molecular-dynamics simulations in explicit solvent. Our simulations starting from eight random chains demonstrate that the addition of SWCNT into Aβ(16-22) solution prevents β-sheet formation. Simulation starting from a prefibrillar β-sheet octamer shows that SWCNT destabilizes the β-sheet structure. A detailed analysis of the Aβ(16-22)/SWCNT/water interactions reveals that both the inhibition of β-sheet formation and the destabilization of prefibrillar β-sheets by SWCNT result from the same physical forces: hydrophobic and π-stacking interactions (with the latter playing a more important role). By analyzing the stacking patterns between the Phe aromatic rings and the SWCNT carbon rings, we find that short ring–centroid distances mostly favor parallel orientation, whereas large distances allow all other orientations to be populated. Overall, our computational study provides evidence that SWCNT is likely to inhibit Aβ(16-22) and full-length Aβ fibrillation.
Introduction
Alzheimer's disease (AD) is characterized by amyloid deposits predominantly composed of the 40- to 42-residue-long amyloid-β peptides (Aβ40/Aβ42) (1). X-ray diffraction data show that these deposits consist of fibrils that display a cross-β structure with the β-strands perpendicular and the interstrand hydrogen bonds (H-bonds) parallel to the fibril axis (2). In vitro fibrillation of Aβ is described by a nucleation-elongation process characterized by a lag phase associated with the formation of a critical nucleus, after which fibril growth proceeds rapidly (3). Before nucleation occurs, Aβ can form a variety of metastable intermediate oligomeric states with different association numbers and structures, such as small oligomers, Aβ-derived diffusible ligands (ADDLs), and protofibrils with increasing β-sheet structure (4,5).
Although the molecular mechanism of AD pathogenesis remains elusive, there is increasing experimental evidence that the key pathological species are transient β-rich oligomers, which therefore represent therapeutic targets for treatment of AD (5,6). The goal of preventing Aβ β-sheet formation by means of peptide-based inhibitors (7), small molecules (8), or proteins (9) has been pursued for many years and is still a very active field of research. However, investigators have not yet been able to use the proteins designed in these studies to construct therapeutic agents against AD, in part because of the problem of blood-brain barrier permeability (10). Recently, a new class of agents emerged from the observation that the binding of proteins to nanoparticle surfaces affects protein folding and aggregation (11,12). Experimental studies indicate that fullerene (13), carbon nanotubes (14,15), and polymeric (16) and fluorinated nanoparticles (17) inhibit/promote amyloid formation depending on their surface physicochemical properties by increasing/decreasing the lag phase time for nucleation, but they all leave the elongation phase invariant, suggesting a surface-modulated nucleation mechanism (15,16). There is strong evidence that the protein's intrinsic stability also affects the surface-mediated nucleation process. For example, Linse et al. (15) demonstrated that hydrophobic (HP) copolymeric NiPAM:BAM nanoparticles accelerate the fibrillation of the β2-microglobulin protein, which is β-sheet-rich in its native form, whereas they retard the fibrillation of Aβ40 (16), an intrinsically disordered peptide. Based on fibrillation kinetics experiments, Linse et al. suggested that binding of monomeric Aβ40 and prefibrillar oligomers to the nanoparticle surfaces prevents fibrillation. Despite these observations, however, details about the Aβ−nanoparticle interaction and the structures of the oligomeric species prenucleation remain to be determined. Such knowledge is important for elucidating the effects of HP nanoparticles on Aβ aggregation.
Single-walled carbon nanotubes (SWCNTs) are HP, tubular nanostructures with diameters of only 0.4–3.5 nm. They can easily enter into the cytoplasm and nucleus through the lipid bilayer (18,19). No short-term (20) or long-term (21) cytotoxicity of SWCNTs has been reported. These contradictory toxic effects clearly need to be clarified and understood. As a first step toward understanding the effects of HP nanoparticles on the low-molecular-weight aggregates of Aβ, we studied an SWCNT and an octamer of the 16-22 fragment of Aβ, Aβ(16-22) as a model system. Our choice of an octamer was motivated by recent all-atom molecular-dynamics (MD) studies indicating that the minimum nucleus size consists of at least eight Aβ(16-22) peptides based on the stability of preformed β-sheet assemblies (22,23). Our choice of the Aβ(16-22) peptide was based on the following factors: First, computational determination of all-atom Aβ monomeric equilibrium structures is feasible in explicit solvent (24), but one is faced with a convergence issue for all-atom dimers to pentamers (25,26). In contrast, the length of Aβ(16-22) (seven amino acids) enables the computational study of all-atom octamers at equilibrium in the presence of an SWCNT in explicit solvent. Second, Aβ(16-22) peptide, which includes the central HP core, CHC (LVFFA), is recognized as being essential for Aβ fibrillation (27,28) and also forms amyloid fibrils with antiparallel β-strands in isolation (29). Third, the KLVFF motif is a primary target in the search for aggregation inhibitors for AD therapeutics (30,31). Finally, investigators have examined the dynamics and thermodynamics of Aβ(16-22) oligomerization without nanoparticles using coarse-grained (CG, up to 8-mer) (32,33) and all-atom (up to 6-mer) simulations (34–36) starting from random conformations or preformed β-sheets, thus providing structural information for comparison.
Although recent computational studies have focused on the aggregation of amyloid peptides in the presence of nanoparticles using a CG nanoparticle model (37) or implicit/explicit lipid models (38–40), to our knowledge, this is the first computational study to investigate the structures, thermodynamics, and interactions of eight Aβ(16-22) chains with an SWCNT using all-atom replica exchange MD simulations (REMD) in explicit solvent. We performed a total of three independent REMD simulations starting from an amorphous Aβ(16-22) octamer with/without SWCNT and a prefibrillar β-sheet octamer with SWCNT.
Materials and Methods
The Aβ(16-22) peptide consists of seven residues (Ac-KLVFFAE-NH2) blocked by acetyl and amine groups as determined experimentally (29). To mimic the experimental neutral pH condition, the side chains of Lys and Glu are charged (Lys+ and Glu−). Three REMD runs were performed on three different systems: random-Aβ(16-22), random-Aβ(16-22)+SWCNT, and sheet-Aβ(16-22)+SWCNT. Each system was placed in a rectangular box of SPC water molecules (41) with a minimum distance to the water box wall of 0.9 nm. The total numbers of atoms for the three systems are 19,717, 19,663, and 18,682, respectively.
Random-Aβ(16-22)
The starting state of the Aβ(16-22) octamer with random character for each chain, shown in Fig. 1 A, is the final conformation generated in a 14-ns MD simulation at 420 K in water starting from fully extended peptides.
Figure 1.
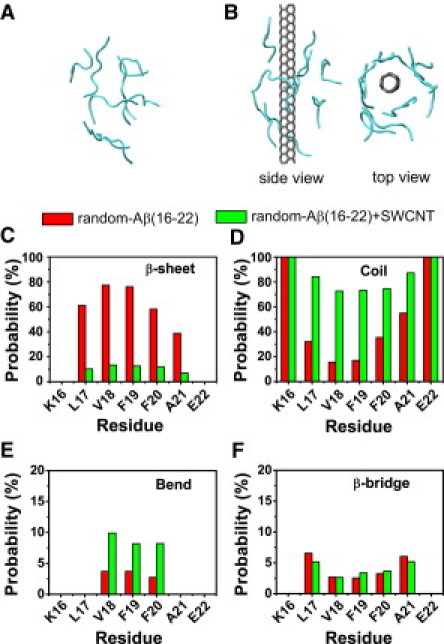
Calculated secondary structure probability of each residue in the REMD runs for random-Aβ(16-22) and random-Aβ(16-22)+SWCNT at 310 K for (C) β-sheet, (D) coil, (E) bend, and (F) β-bridge. The initial random states for the two runs are given in panels A and B. Two different views of the initial state for the random-Aβ(16-22)+SWCNT run are shown in B.
Random-Aβ(16-22)+SWCNT complex
The initial state of the Aβ(16-22) octamer in the complex is the same as in the Aβ(16-22) system. An SWCNT with a diameter of 0.542 nm is placed at the center of eight peptide chains. Its length is set to 4.25 nm to provide a sufficient surface for the Aβ(16-22) peptides to adsorb.
Sheet-Aβ(16-22)+SWCNT complex
This Aβ(16-22) β-sheet bilayer is taken from cluster 8 in Fig. 2 A and an SWCNT is placed between the two sheets (see Fig. 5 A). The SWCNT is the same size as the one in the random-Aβ(16-22)+SWCNT complex.
Figure 2.
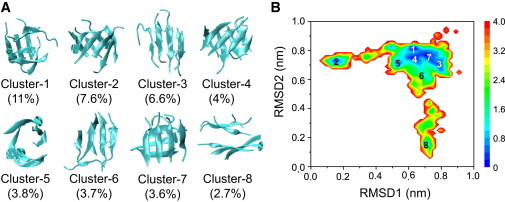
Structure analysis and FEL of Aβ(16-22) octamers in aqueous solution without SWCNT at 310 K. (A) Representative structures for the first eight most-populated clusters. (B) FEL (in kcal/mol) versus RMSD1 (Cα-RMSD relative to the β-barrel; cluster 2), and RMSD2 (Cα-RMSD relative to the parallel β-sheet bilayer; cluster 8).
Figure 5.
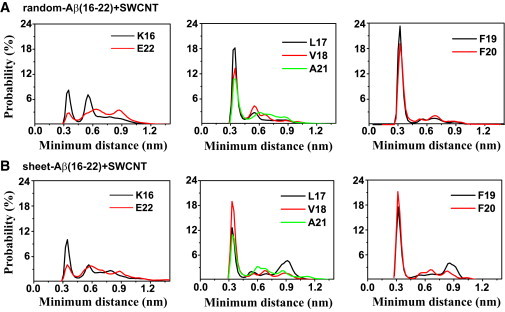
Probability distribution of the minimum distance between the side chain of each residue and SWCNT surface in the 310 K trajectory of the two REMD runs with SWCNT. The data are averaged over the last 80 ns for random-Aβ(16-22)+SWCNT and the last 40 ns for sheet-Aβ(16-22)+SWCNT.
REMD simulations
The REMD simulations are performed with the use of the GROMACS software package (42). In accordance with several computational studies of Aβ(16-22) (43–45), the GROMOS96 43A1 force field (46) is used to describe intra- and intermolecular interactions. To avoid high-pressure artifacts at elevated temperature, and following the work of Seibert et al. (47), we carry out our REMD simulations in the NPT ensemble using 40 replicas, each of 110 ns duration, at temperatures exponentially spaced between 310 K and 420 K. The swap time between two neighboring replicas is 1 ps, which is large compared with the integrated autocorrelation time of the potential energy of the system at the lowest temperature 310 K (0.51 ps) (48,49) (see the Supporting Material). The acceptance ratio is ∼17% (Fig. S1). To maintain the temperatures at the chosen values, velocity rescaling coupling method (50) is used with a coupling constant of 0.1 ps. The pressure is kept at 1 bar with a coupling constant of 1 ps via Berendsen's method (51). Constraints are applied for bond lengths using the SETTLE algorithm (52) for water, and the LINCS algorithm (53) for the peptides and SWCNT. This allows an integration time step of 2 fs. The cutoff for van der Waals interactions is 1.4 nm. A reaction-field correction (with a cutoff of 1.4 nm) with dielectric permittivity ε = 78 is used for the long-range electrostatic interactions. The carbon atoms of SWCNT are uncharged in accordance with Hummer et al. (54), and the Lennard-Jones parameters for the peptide-SWCNT and water-SWCNT interactions are obtained using the Lorentz-Berthelot rule.
Analysis
We perform the analysis using our in-house-developed codes and the GROMACS facilities. We discard the first 30 ns of each REMD to remove the bias of the initial states, except when mentioned otherwise. The structural properties of each system are therefore based on a total of 3.2 μs.
We analyze the REMD trajectories using several parameters, including the secondary structure content (via the DSSP program) (55), the free-energy landscape (FEL), the percentage of various sizes of β-sheet, the connectivity length (CL), and the probability density function (PDF) of the angle between two neighboring strands in each β-sheet. A detailed description of these parameters is given in the Supporting Material.
The Aβ(16-22) octameric structures are clustered according to the method of Daura et al. (56) with a Cα root mean-square deviation (RMSD) cutoff of 0.3 nm using residues L17-A21, with K16 and E22 excluded because of their high flexibilities. We calculate all of the Cα-RMSDs by neglecting the chain identifier of each peptide (see the Supporting Material for the procedure used to calculate the chain-independent RMSD).
Finally, we probe the Aβ(16-22)−SWCNT interactions in detail by determining the probability distribution of the minimum distance between the side chain of each residue and the SWCNT surface. We also analyze the π-stacking interactions between the peptides and the nanotube surface by calculating the PDF of the distance between the centroids of the Phe aromatic ring and its closest SWCNT carbon ring, and the PDF of the angle between the two rings. We calculate the interplanar angle by determining the angle between the surface normals of the two rings. All representations of the studied systems are drawn with the use of the VMD program (57).
Results
Before characterizing the 3D structures and the energy landscapes of the Aβ(16-22) peptides with and without SWCNT at 310 K starting from an amorphous, random coil octamer, we verify the convergence of the two REMD simulations. To that end, we compare the probability distributions of end-to-end distance of all chains in a random-Aβ(16-22) (Fig. S2 A) or random-Aβ(16-22)+SWCNT (Fig. S2 B) system using the 30–70 ns and 70–110 ns data. As can be seen in the figures, the distributions overlap very well when the two independent time intervals are used. It should be noted that the shapes of the probability distributions of the end-to-end distance in the two systems look similar, with a small difference between the full widths at half-maximum (FWHMs) and heights of the peak. The FWHMs and heights of the peaks are (0.23 nm, 12%) and (0.30 nm, 19%) without and with SWCNT, respectively. These data indicate that the peptide−SWCNT interactions slightly influence the global dimension of each chain for such a short peptide as Aβ(16-22). We also calculate the β-sheet contents using the 30–70 ns and 70–110 ns data. The calculated β-sheet probabilities within the two time intervals are 42.1% and 47.0% for Aβ(16-22) without SWCNT versus 7.8% and 8.1% with SWCNT. Overall, the differences in β-sheet content and end-to-end distance distribution within the two time intervals are <5% in magnitude, indicating that our REMD simulations for the two systems have reasonably converged.
The presence of SWCNT significantly reduces β-sheet content
We calculate the secondary structure (β-sheet, coil, bend, β-bridge, and α-helix) probability of each residue at 310 K. From Fig. 1, C–F, one can see that the secondary structure content of each residue is strongly affected by SWCNT. Note that the helix content is negligible in both simulations (data not shown). The CHC region spanning L17–A21 has 39–75% probabilities to adopt β-sheet states (Fig. 1 C) in the Aβ(16-22) system, with the highest probability of 75% for V18 and F19, versus 8–15% in Aβ(16-22)+SWCNT complex. Alternatively, the coil percentage for CHC is much lower with SWCNT (18–55%) than without SWCNT (75–90%). The β-bridge percentage of the residues V18-F19-F20 is roughly the same (8%) in both systems (Fig. 1 E), and the residues V18-F19-F20 have similar bend signals (5% with SWCNT versus 9% without SWCNT (Fig. 1 F). Averaged over all conformations within 30–110 ns, the numbers of backbone H-bonds are 30 and 10 in the random-Aβ(16-22) and random-Aβ(16-22)+SWCNT systems, respectively. Taken together, these results demonstrate that the presence of SWCNT significantly prevents β-sheet formation of Aβ(16-22) peptides.
Aβ(16-22) peptides form various β-sheet-rich octamers without SWCNT, and the presence of SWCNT shifts equilibrium toward random coil aggregates
To investigate the atomic structures of the peptides in the absence/presence of SWCNT, we perform a chain-independent RMSD-based cluster analysis using a total of 40,000 conformations for each replica. Using a Cα-RMSD of 0.3 nm, we separate the octamers in random-Aβ(16-22) and random-Aβ(16-22)+SWCNT systems at 310 K (replica 1) into 166 and 136 clusters, respectively. The large number of clusters reflects the very high structural heterogeneity of the Aβ(16-22) octamers at physiological temperature. As expected, the number of clusters increases with temperature (Fig. S3) and becomes >1000 for T ≥ 391.57 K (replica index ≥ 30). The centers of the first eight most-populated clusters of Aβ(16-22) octamers, and their populations with and without SWCNT at 310 K are respectively shown in Figs. 2 A and 3 A. These clusters represent 43.0% and 39.4% of all conformations in the absence and presence of SWCNT, respectively.
Figure 3.
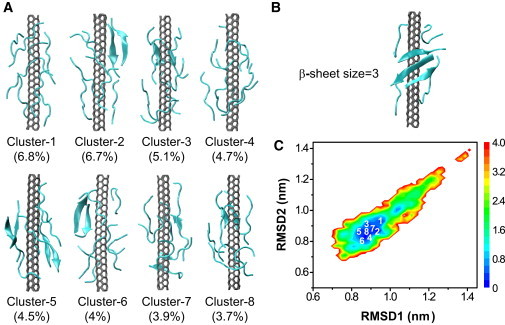
Structure analysis and FEL of Aβ(16-22) octamers with SWCNT at 310 K. (A) Representative structures for the first eight most-populated clusters. (B) Representative structures containing the largest β-sheet (i.e., a three-stranded sheet). (C) FEL (in kcal/mol) versus RMSD1 (Cα-RMSD relative to the β-barrel; cluster 2 in Fig. 2A) and RMSD2 (Cα-RMSD relative to the parallel β-sheet bilayer; cluster 8 in Fig. 2A).
Without SWCNT, the clusters in Fig. 2 A display various β-sheet-rich conformations. The first cluster consists of disordered β-sheet-rich aggregates. The second and fifth clusters contain five- to eight-stranded β-barrels with mixed parallel/antiparallel strands. The third, sixth, and eighth clusters contain parallel β-sheet bilayers consisting of 4+3 and 4+4 β-sheet sizes. Finally, the fourth and seventh clusters display two orthogonal β-sheets. The parallel and the orthogonal β-sheet bilayers have populations of 13.0% (6.6% + 3.7% + 2.7%) and 7.6% (4% + 3.6%), respectively.
In the presence of SWCNT, as shown in Fig. 3 A, the Aβ(16-22) aggregates are fully amorphous, with clusters 1 and 4 being free of any β-sheet, and clusters 2, 3, 5, and 6–8 having two-stranded β-sheets. Aggregates containing three-stranded β-sheets (Fig. 3 B) are very rare (with a population of 0.3%), and aggregates containing four-stranded β-sheets are negligible (<0.01%). Overall, in contrast to the peptides in aqueous solution without SWCNT, none of the aggregates display parallel β-sheet bilayer, orthogonal β-sheet bilayer, or β-barrel structures.
Figs. 2 B and 3 C show the FELs of the peptides in Aβ(16-22) and Aβ(16-22)+SWCNT systems, respectively, as a function of RMSD1 and RMSD2. RMSD1 and RMSD2 are the Cα-RMSDs of all conformations with respect to the β-barrel (cluster 2) and parallel β-sheet bilayer (cluster 8) shown in Fig. 2 A, respectively, which deviate from each other by a Cα-RMSD of 0.7 nm. The locations of the first eight clusters are labeled on each free-energy surface. In the case of Aβ(16-22) in solution, these clusters are delocalized on three basins (Fig. 2 B), whereas in the presence of SWCNT, all clusters belong to one attraction basin (Fig. 3 C).
The differences in the Aβ(16-22) aggregates are further probed by the probability of the β-sheet sizes at 310 K (Fig. S4 A). We find that without SWCNT, the two- and three-stranded β-sheets have relatively high probabilities (20.6% and 13.6%), followed by four-stranded β-sheets (10.5%). With SWCNT, the population decreases rapidly with β-sheet size: two-stranded (6.7%), three-stranded (0.3%), and four-stranded (negligible, <0.01%). Differences are more pronounced for larger sizes of β-sheets. The probabilities of five-, six-, seven-, and eight-stranded β-sheets are respectively 3.2%, 2.5%, 1.1%, and 1.4% without SWCNT, whereas they vanish in the presence of SWCNT. A structural inspection shows that these five- to eight-stranded β-sheets consist of open or closed β-barrels as well as β-sheet bilayers.
Finally, to explore the orientation preference of the strands in different sizes of β-sheets, we plot the PDF of the angle between two neighboring strands in a sheet as a function of β-sheet size (Fig. S4 B). Without SWCNT, the strands are predominantly antiparallel in a two-stranded β-sheet, in good agreement with previous computational studies (34,35). For sheet sizes of 3 and 4, the ratio of antiparallel to parallel strand alignment decreases, but the antiparallel alignment is still preferred. When the sheet size increases to ≥5, the probabilities of antiparallel and parallel alignments become similar. In the presence of SWCNT, the antiparallel orientation is more populated for sheet sizes of 2 and 3.
Discussion
We performed two 110-ns all-atom REMD simulations in explicit solvent starting from Aβ(16-22) octamer with random character for each chain without and with SWCNT. Without SWCNT, our simulation shows that the Aβ(16-22) octamer has an average β-sheet content of 44.5% and mainly visits disordered β-rich states and, to a lesser extent, structures with cross-β and β-barrel characteristics. The structure of the β-barrel is not unique, and the barrel topology includes five- to eight-stranded β-barrel-like structures. We expect that the probabilities of the ordered β-rich structures might be lower using the OPLS force field, and this remains clearly to be determined on the Aβ(16-22) octamer. A previous computational study on two peptides showed that GROMOS96 overestimates the stability of the β conformation, and OPLS generates a better balance between α-helical and β-sheet structures (58). A recent REMD study on Aβ(16-22) dimer and trimer showed that GROMOS96 favors β-sheet structures, and OPLS predicts diverse structures (59).
We also observed these cross-β structures and amorphous aggregates in our recent CG MD study of the Aβ(16-22) octamer with the two termini of the peptide chain unblocked (36). The formation of a β-barrel-like structure was also revealed in previous computational studies on Aβ(16-22) (33), KFFE (60), β2m(83-89) (the 83-89 fragment of β2-microglobulin) (61) in implicit solvent and generic sequences (62), and β2m(83-89) in explicit solvent (63).
In the presence of SWCNT, our extensive REMD simulation starting from the same random Aβ(16-22) state shows that the peptides adopt mostly disordered coil states, with an average β-sheet content of 7.9%. The parallel and orthogonal β-sheet bilayers are not populated. This result demonstrates that SWCNT strongly inhibits Aβ(16-22) β-sheet formation, which consequently would significantly increase the lag phase time for nucleation. Our result is fully consistent with recent experimental studies conducted by Cabaleiro-Lago et al. (16), who reported that HP copolymeric NiPAM:BAM nanoparticles retard Aβ40 fibrillation by slowing down the nucleation process. In the same study, they showed that fibrillation of Aβ40 can be reversed by the addition of NiPAM:BAM nanoparticles up to a particular time point before mature fibrils appear. To determine whether SWCNT can destabilize prefibrillar Aβ(16-22) β-sheets, we performed a third REMD run on the sheet-Aβ(16-22)+SWCNT system.
SWCNT destabilizes ordered prefibrillar β-sheets and induces the formation of disordered coil aggregates
The results of the third REMD run at 310 K are given in Fig. 4. We first monitor the β-sheet probability of each residue at four time intervals: 0–20 ns, 20–50 ns, 50–80 ns, and 80–110 ns (Fig. 4 B). The β-sheet contents of the residues L17-V18-F19-F20 significantly decrease with simulation time, reducing to 38%, 46%, 46%, and 28% from their initial values of 67%, 86%, 84%, and 45%, respectively. The average β-sheet content within 0–20 ns is 60.3%, and it drops to 31.5% within an 80–110 ns time period.
Figure 4.
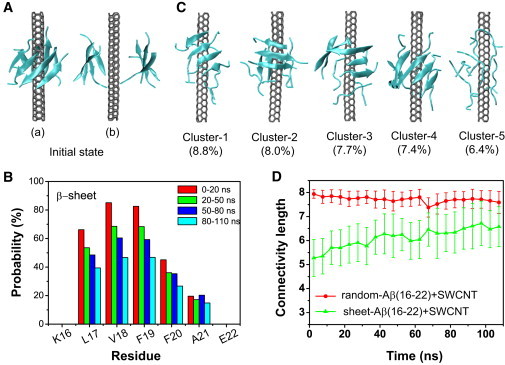
Structural analysis of Aβ(16-22) octamers generated in the 310 K REMD trajectory starting from a prefibrillar β-sheet octamer (cluster 8 in Fig. 2A) with an SWCNT placed between the two β-sheets. (A) Two different views of the initial state, where b is the perpendicular view of a. (B) β-sheet content of each residue within four time intervals: 0–20 ns, 20–50 ns, 50–80 ns, and 80–110 ns. (C) Representative structures for the first five most-populated clusters. (D) Time evolution of the CL of the Aβ(16-22) octamer in the sheet-Aβ(16-22)+SWCNT system. For comparison, the time evolution of the CL in the random-Aβ(16-22)+SWCNT system is also given. The number of CLs is averaged every 5 ns.
The SWCNT-induced destabilization of β-sheet bilayer is illustrated by the time evolution of the CL at 310 K (Fig. 4 D). For comparison, the time evolution of CL in the random-Aβ(16-22)+SWCNT system at 310 K is also given. The CL of the Aβ(16-22) octamer in the sheet-Aβ(16-22)+SWCNT system increases from its initial value of 5.2 to 6.7 in the last 30 ns of the simulation. Although the CL and the β-sheet content in sheet-Aβ(16-22)+SWCNT system do not reach the same values of 7.7 and 7.9% as in the random-Aβ(16-22)+SWCNT system within the 110-ns REMD run, their time evolutions reflect a dissociation of prefibrillar β-sheet structure by SWCNT and a propensity for random coil aggregates. We also reveal these features by performing a cluster analysis for 20,000 conformations generated within 70–110 ns using a Cα-RMSD of 0.3 nm. The centers of the first five most-populated clusters, representing 38.3% of all conformations, are shown in Fig. 4 C. It should be noted that the initial β-sheet bilayer structure, which becomes the 17th cluster, represents only 2.0% of the 20,000 conformations. The orthogonal β-sheet bilayer is not populated. Although the first four clusters are β-rich octamers, two to four of the eight chains are in random state. Moreover, all of the chains in cluster 5 are in a coil state. Of interest, a five-stranded open β-barrel wrapping around SWCNT is observed in cluster 4. The total population of five-, six-, seven-, and eight-stranded barrels is 2.1%, implying that barrel topologies are likely metastable states of Aβ(16-22) oligomers in the presence of SWCNT. Of note, we also observed an SWCNT-induced β-barrel formation starting from a preformed β-sheet bilayer in our recent all-atom MD study of Aβ(25-35) peptide (64).
Overall, although our simulation starting from an Aβ(16-22) β-sheet bilayer is not fully converged, it demonstrates that the presence of SWCNT destabilizes β-sheet structures and induces the formation of disordered coil aggregates as well as metastable β-barrel-like structures. This result indicates that the fibrillation of Aβ16-22 can be reversed at its early stage by the addition of SWCNT. Given that REMD simulations accelerate the sampling of MD simulations by an order of magnitude (47), our results indicate that full destabilization of the Aβ(16-22) β-sheet octamer by SWCNT is a slow process, taking >1.1 μs.
SWCNT inhibits Aβ(16-22) β-sheet formation through both HP and π-stacking interactions
To understand the physical driving forces underlying the β-sheet inhibition and destabilization by SWCNT, we plot in Fig. 5 the probability distribution of the minimum distance between the side chain of each residue and SWCNT surface in the two REMD runs at 310 K (Fig. 5, A and B). Two probability peaks are seen for each residue, with a dominant peak centered at 0.35 nm for the HP residues L17-V18-F19-F20-A21. The probabilities of these HP residues at 0.35 nm are all greater than those of K16 and E22. In particular, the aromatic residues F19 and F20 have the highest probability at 0.35 nm. These data indicate that the HP residues L17–A21 have stronger interactions with SWCNT than the hydrophilic residues K16 and E22, and, remarkably, the interaction between the two aromatic residues (F19 and F20) and SWCNT is the strongest in both systems. Our results are consistent with electron microscope images showing that peptides containing aromatic rings have strong affinity with carbon nanotubes (65), and recent atomic force microscopy and optical absorption spectroscopy experiments indicating that π-stacking interactions play an important role in peptide-SWCNT interactions (66).
To investigate the detailed interactions between the aromatic rings of Phe and SWCNT, we plot in Fig. 6 A the PDF of the centroid distance (d) between the Phe ring and its closest SWCNT carbon ring. There are three peaks centered at 0.37, 0.5, and 0.75 nm, with a dominant peak at 0.37 nm. To probe the favorable orientation between the two rings around the three different distances, we calculate the PDF of the interplanar ring angle at a centroid distance of 0.3 ≤ d ≤ 0.45 nm (Fig. 6 B), 0.45 < d ≤ 0.65 nm (Fig. 6 C), and d ≥ 0.65 nm (Fig. 6 D). A peak at 15° is observed in Fig. 6 B, indicating that the two rings have a strong preference to be parallel-aligned when d < 0.45 nm. For 0.45 < d ≤ 0.65 nm, the angle is mainly distributed in the range of 60–90°, with almost equal population for each angle in this range, indicative of both perpendicular (T-shaped) and staggered ring organizations. When d > 0.65 nm, the PDF curve is almost flat for the angle in the range of 30–90°, with a rather broad peak around 50°, implying that the two rings can align in any orientation, including parallel and T-shaped ones, with a slight preference for a 50° orientation. These results suggest that there are many possible stacking patterns between Phe aromatic ring and SWCNT carbon ring, and the exact stacking orientation depends on their centroid distance. In particular, a parallel-aligned π-stacking geometry is likely to be the most favorable one at a short distance. This is supported by the PDF curve of the angle for the ring centroid distance in the full distance range (Fig. S5), where a peak appears at 15°. Of interest, the analysis of π-stacking in a group of nonhomologous proteins with known x-ray crystal structures suggests a parallel-displaced π-stacking to be the major organization of π-π interactions in proteins (67).
Figure 6.
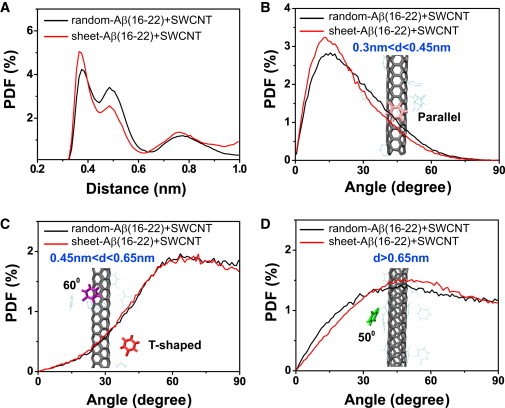
Analysis of π-stacking interactions between the Phe aromatic ring and the SWCNT carbon ring in random-Aβ(16-22)+SWCNT and sheet-Aβ(16-22)+SWCNT systems at 310 K. (A) The PDF of the centroid distance (d) between the Phe ring (each Phe residue is considered) and its closest SWCNT carbon ring. (B–D) The PDF of the angle between the two rings with a centroid distance of (B) 0.3 ≤ d ≤ 0.45 nm, (C) 0.45 < d ≤ 0.65 nm, and (D) d ≥ 0.65 nm. The selection of the three distances is based on the PDF peak in panel A.
Finally, it is instructive to look at the role of water in all of the REMD simulations. To that end, we calculated the number of water molecules within 0.36 nm of every residue at five different stages of the simulations (Fig. S6). Previous experimental (68) and computational studies (44,64) showed that the expulsion of interfacial water molecules is a key event in the aggregation of amyloid peptides such as Aβ(16-22) (44,68). Consistent with those studies, fast dehydration occurs in the early stage of Aβ(16-22) oligomerization in solution without SWCNT (Fig. S6 A). The number of contacting water molecules with the five HP residues decreases within 2 ns and remains almost constant after 5 ns. Remarkably, a rapid dehydration process is also observed both in random-Aβ(16-22)+SWCNT and sheet-Aβ(16-22)+SWCNT systems (Fig. S6, B and C), indicating that the expulsion of water is an obligatory step in the interactions of Aβ(16-22) peptide with SWCNT. As shown in Fig. S6 D, within the 70–110 ns time period, the hydration status of the Aβ(16-22) peptide with and without SWCNT is quite similar, which implies that the water-peptide interaction is similar in magnitude among the three different systems, and the significant decrease in β-sheet content of Aβ(16-22) aggregates in the presence of SWCNT results from the SWCNT-peptide interactions.
Conclusions
In summary, based on three 110-ns all-atom REMD simulations on Aβ(16-22) octamers starting from two different conformations in the absence and presence of SWCNT, we propose the following inhibition mechanisms of SWCNT on Aβ(16-22) nucleation (Fig. 7): Without SWCNT, random Aβ(16-22) chains form various β-sheet-rich oligomers. The interaction of SWCNT with amorphous aggregates prevents β-sheet formation and leads to only disordered coil aggregates at equilibrium. The interaction of SWCNT with a prefibrillar β-sheet bilayer destabilizes the β-sheet structure and induces the formation of disordered coil aggregates.
Figure 7.
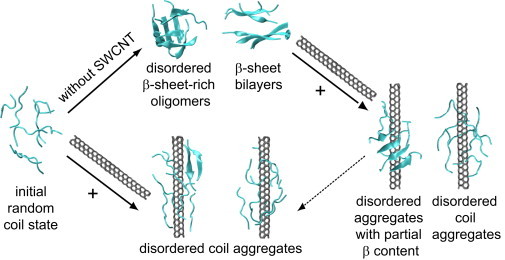
Proposed inhibition mechanism of Aβ(16-22) nucleation by SWCNT, based on simulation results from three different REMD runs. All of the conformations are from the 110-ns REMD simulations.
An analysis of the probability distribution of the minimum distance between the side chain of each residue and the SWCNT surface provides evidence for strong HP interactions between the CHC residues L17–A21 with SWCNT, among which residues F19–F20 show the strongest interactions with SWCNT through π-stacking interactions. The interpeptide HP interactions and notably the aromatic stacking interactions have been demonstrated to be important for amyloid formation of several peptides, including Aβ(16-22) (34,69,70). However, certain experimental observations show that mutations that replace the aromatic side chains of F19 and F20 with leucines or isoleucines promote aggregation of the Aβ42 peptide, indicating that aromatic π-stacking is not critical, at least for the full-length Aβ peptide (71). The strong HP and π-stacking interactions between SWCNT and Aβ(16-22) peptide would interfere with the peptide-peptide interactions that are responsible for Aβ(16-22) aggregation, thus slowing down the nucleation process. The SWCNT used in this study has a diameter of 0.542 nm. It is expected that the increase in the SWCNT diameter would change the Aβ(16-22)−SWCNT interaction, thus affecting the inhibition extents of SWCNT on Aβ(16-22) aggregation. Further studies are needed to understand the effect of SWNT diameter on Aβ(16-22) aggregation.
The full-length Aβ42 peptide contains one hydrophilic patch spanning residues 1–15, and two HP regions spanning residues 17–21 and 30–42 separated by a hydrophilic linker, residues 22–29. Obviously, due to the hydrophilic character of the N-terminal region and of the linker, these two regions are not to be the most probable sites for Aβ42-SWCNT interactions. Region 30–42 with the amino acids A-I-I-G-L-M-V-G-G-V-V-I-A is certainly another probable binding site; however, this fragment is free of any aromatic residues. On the basis of our simulations, we hypothesize that the HP L17–A21 region is the most probable site for SWCNT interactions, and the resulting propensity of residues 17–21 to adopt random coil conformation upon binding to SWCNT is likely to reduce the β-sheet formation of the same residues in the Aβ40/42 oligomers, thereby prolonging the lag time for Aβ nucleation. Our hypothesis is supported by a previous experimental study in which results from a Thioflavin T fluorescence assay showed that fullerene strongly inhibits Aβ40 aggregation at the early stage by specifically binding to the KLVFF motif (13). Overall, our findings provide significant insight into the inhibition mechanism of HP nanoparticles (notably SWCNT) against the aggregation of Aβ(16-22) and full-length Aβ, and provide what to our knowledge are new clues for the development of drug candidates against AD.
Acknowledgments
This work was supported by the Program for New Century Excellent Talent in University (NCET-08-0125), the National Science Foundation of China (11074047), and the Research Fund for the Doctoral Program of Higher Education of China. P.D. received support from the Centre National de la Recherche Scientifique, the University of Paris Diderot, and the Institut Universitaire de France. Simulations were performed at the Réseau Québécois de Calcul de Haute Performance, Montreal, Quebec, Canada (with the support of Prof. Normand Mousseau) and the National High-Performance Computing Center of Fudan University.
Supporting Material
References
- 1.Pitschke M., Prior R., Riesner D. Detection of single amyloid β-protein aggregates in the cerebrospinal fluid of Alzheimer's patients by fluorescence correlation spectroscopy. Nat. Med. 1998;4:832–834. doi: 10.1038/nm0798-832. [DOI] [PubMed] [Google Scholar]
- 2.Serpell L.C., Sunde M., Fraser P.E. The protofilament substructure of amyloid fibrils. J. Mol. Biol. 2000;300:1033–1039. doi: 10.1006/jmbi.2000.3908. [DOI] [PubMed] [Google Scholar]
- 3.Lomakin A., Teplow D.B., Benedek G.B. Kinetic theory of fibrillogenesis of amyloid β-protein. Proc. Natl. Acad. Sci. USA. 1997;94:7942–7947. doi: 10.1073/pnas.94.15.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh D.M., Hartley D.M., Teplow D.B. Amyloid β-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J. Biol. Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 5.Klein W.L. Aβ toxicity in Alzheimer's disease: globular oligomers (ADDLs) as new vaccine and drug targets. Neurochem. Int. 2002;41:345–352. doi: 10.1016/s0197-0186(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 6.Cleary J.P., Walsh D.M., Ashe K.H. Natural oligomers of the amyloid-β protein specifically disrupt cognitive function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 7.Soto C., Sigurdsson E.M., Frangione B. β-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: implications for Alzheimer's therapy. Nat. Med. 1998;4:822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]
- 8.Scherzer-Attali R., Pellarin R., Segal D. Complete phenotypic recovery of an Alzheimer's disease model by a quinone-tryptophan hybrid aggregation inhibitor. PLoS ONE. 2010;5:e11101. doi: 10.1371/journal.pone.0011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi T., Mihara H. Peptide and protein mimetics inhibiting amyloid β-peptide aggregation. Acc. Chem. Res. 2008;41:1309–1318. doi: 10.1021/ar8000475. [DOI] [PubMed] [Google Scholar]
- 10.Poduslo J.F., Curran G.L., Berg C.T. Macromolecular permeability across the blood-nerve and blood-brain barriers. Proc. Natl. Acad. Sci. USA. 1994;91:5705–5709. doi: 10.1073/pnas.91.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colvin V.L., Kulinowski K.M. Nanoparticles as catalysts for protein fibrillation. Proc. Natl. Acad. Sci. USA. 2007;104:8679–8680. doi: 10.1073/pnas.0703194104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fei L., Perrett S. Effect of nanoparticles on protein folding and fibrillogenesis. Int. J. Mol. Sci. 2009;10:646–655. doi: 10.3390/ijms10020646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J.E., Lee M. Fullerene inhibits β-amyloid peptide aggregation. Biochem. Biophys. Res. Commun. 2003;303:576–579. doi: 10.1016/s0006-291x(03)00393-0. [DOI] [PubMed] [Google Scholar]
- 14.Ghule A.V., Kathir K.M., Ling Y.-C. Carbon nanotubes prevent 2,2,2 trifluoroethanol induced aggregation of protein. Carbon. 2007;45:1586–1589. [Google Scholar]
- 15.Linse S., Cabaleiro-Lago C., Dawson K.A. Nucleation of protein fibrillation by nanoparticles. Proc. Natl. Acad. Sci. USA. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabaleiro-Lago C., Quinlan-Pluck F., Linse S. Inhibition of amyloid β protein fibrillation by polymeric nanoparticles. J. Am. Chem. Soc. 2008;130:15437–15443. doi: 10.1021/ja8041806. [DOI] [PubMed] [Google Scholar]
- 17.Saraiva A.M., Cardoso I., Brezesinski G. Controlling amyloid-β peptide(1-42) oligomerization and toxicity by fluorinated nanoparticles. ChemBioChem. 2010;11:1905–1913. doi: 10.1002/cbic.201000237. [DOI] [PubMed] [Google Scholar]
- 18.Shi Kam N.W., Jessop T.C., Dai H. Nanotube molecular transporters: internalization of carbon nanotube-protein conjugates into mammalian cells. J. Am. Chem. Soc. 2004;126:6850–6851. doi: 10.1021/ja0486059. [DOI] [PubMed] [Google Scholar]
- 19.Porter A.E., Gass M., Welland M. Direct imaging of single-walled carbon nanotubes in cells. Nat. Nanotechnol. 2007;2:713–717. doi: 10.1038/nnano.2007.347. [DOI] [PubMed] [Google Scholar]
- 20.Ren H.-X., Chen X., Huang X.-J. Toxicity of single-walled carbon nanotube: how we were wrong. Mater. Today. 2010;13:6–8. [Google Scholar]
- 21.Kaiser J.P., Roesslein M., Wick P. Carbon nanotubes—curse or blessing. Curr. Med. Chem. 2011;18:2115–2128. doi: 10.2174/092986711795656171. [DOI] [PubMed] [Google Scholar]
- 22.Röhrig U.F., Laio A., Petronzio R. Stability and structure of oligomers of the Alzheimer peptide Aβ16-22: from the dimer to the 32-mer. Biophys. J. 2006;91:3217–3229. doi: 10.1529/biophysj.106.088542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma B., Nussinov R. Stabilities and conformations of Alzheimer's β-amyloid peptide oligomers (Aβ 16-22, Aβ 16-35, and Aβ 10-35): sequence effects. Proc. Natl. Acad. Sci. USA. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sgourakis N.G., Merced-Serrano M., Garcia A.E. Atomic-level characterization of the ensemble of the Aβ(1-42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms. J. Mol. Biol. 2011;405:570–583. doi: 10.1016/j.jmb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huet A., Derreumaux P. Impact of the mutation A21G (Flemish variant) on Alzheimer's β-amyloid dimers by molecular dynamics simulations. Biophys. J. 2006;91:3829–3840. doi: 10.1529/biophysj.106.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masman M.F., Eisel U.L., Luiten P.G. In silico study of full-length amyloid β 1-42 tri- and penta-oligomers in solution. J. Phys. Chem. B. 2009;113:11710–11719. doi: 10.1021/jp901057w. [DOI] [PubMed] [Google Scholar]
- 27.Tjernberg L.O., Näslund J., Nordstedt C. Arrest of β-amyloid fibril formation by a pentapeptide ligand. J. Biol. Chem. 1996;271:8545–8548. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 28.Nilsberth C., Westlind-Danielsson A., Lannfelt L. The ‘Arctic’ APP mutation (E693G) causes Alzheimer's disease by enhanced Aβ protofibril formation. Nat. Neurosci. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 29.Balbach J.J., Ishii Y., Tycko R. Amyloid fibril formation by A β 16-22, a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 30.Lowe T.L., Strzelec A., Murphy R.M. Structure-function relationships for inhibitors of β-amyloid toxicity containing the recognition sequence KLVFF. Biochemistry. 2001;40:7882–7889. doi: 10.1021/bi002734u. [DOI] [PubMed] [Google Scholar]
- 31.Lashuel H.A., Hartley D.M., Callaway D.J. New class of inhibitors of amyloid-β fibril formation. Implications for the mechanism of pathogenesis in Alzheimer's disease. J. Biol. Chem. 2002;277:42881–42890. doi: 10.1074/jbc.M206593200. [DOI] [PubMed] [Google Scholar]
- 32.Wei G., Mousseau N., Derreumaux P. Computational simulations of the early steps of protein aggregation. Prion. 2007;1:3–8. doi: 10.4161/pri.1.1.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y., Derreumaux P., Wei G. Thermodynamics and dynamics of amyloid peptide oligomerization are sequence dependent. Proteins. 2009;75:954–963. doi: 10.1002/prot.22305. [DOI] [PubMed] [Google Scholar]
- 34.Gnanakaran S., Nussinov R., García A.E. Atomic-level description of amyloid β-dimer formation. J. Am. Chem. Soc. 2006;128:2158–2159. doi: 10.1021/ja0548337. [DOI] [PubMed] [Google Scholar]
- 35.Klimov D.K., Thirumalai D. Dissecting the assembly of Aβ16-22 amyloid peptides into antiparallel β sheets. Structure. 2003;11:295–307. doi: 10.1016/s0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 36.Irbäck A., Mitternacht S. Spontaneous β-barrel formation: an all-atom Monte Carlo study of Aβ16-22 oligomerization. Proteins. 2008;71:207–214. doi: 10.1002/prot.21682. [DOI] [PubMed] [Google Scholar]
- 37.Auer S., Trovato A., Vendruscolo M. A condensation-ordering mechanism in nanoparticle-catalyzed peptide aggregation. PLOS Comput. Biol. 2009;5:e1000458. doi: 10.1371/journal.pcbi.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman R., Pellarin R., Caflisch A. Amyloid aggregation on lipid bilayers and its impact on membrane permeability. J. Mol. Biol. 2009;387:407–415. doi: 10.1016/j.jmb.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 39.Jiang P., Xu W., Mu Y. Amyloidogenesis abolished by proline substitutions but enhanced by lipid binding. PLOS Comput. Biol. 2009;5:e1000357. doi: 10.1371/journal.pcbi.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strodel B., Lee J.W.L., Wales D.J. Transmembrane structures for Alzheimer's Aβ(1-42) oligomers. J. Am. Chem. Soc. 2010;132:13300–13312. doi: 10.1021/ja103725c. [DOI] [PubMed] [Google Scholar]
- 41.Berendsen H.J.C., Postma J.P.M., Hermans J. D. Reidel Publishing; Dordrecht: 1981. Intermolecular Forces, Interaction Models for Water in Relation to Protein Hydration. 331–342. [Google Scholar]
- 42.Lindahl E., Hess B., van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 43.Soto P., Griffin M.A., Shea J.E. New insights into the mechanism of Alzheimer amyloid-β fibrillogenesis inhibition by N-methylated peptides. Biophys. J. 2007;93:3015–3025. doi: 10.1529/biophysj.107.112086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krone M.G., Hua L., Shea J.E. Role of water in mediating the assembly of Alzheimer amyloid-β Aβ16-22 protofilaments. J. Am. Chem. Soc. 2008;130:11066–11072. doi: 10.1021/ja8017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen P.H., Li M.S., Thirumalai D. Monomer adds to preformed structured oligomers of Aβ-peptides by a two-stage dock-lock mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:111–116. doi: 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Billeter S.R., Eising A.A., Tironi I.G. Vdf Hochschulverland, ETH; Zurich, Switzerland: 1996. Biomolecular Simulation: the GROMOS96 Manual and User Guide. [Google Scholar]
- 47.Seibert M.M., Patriksson A., van der Spoel D. Reproducible polypeptide folding and structure prediction using molecular dynamics simulations. J. Mol. Biol. 2005;354:173–183. doi: 10.1016/j.jmb.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 48.Periole X., Mark A.E. Convergence and sampling efficiency in replica exchange simulations of peptide folding in explicit solvent. J. Chem. Phys. 2007;126:014903. doi: 10.1063/1.2404954. [DOI] [PubMed] [Google Scholar]
- 49.Abraham M.J., Gready J.E. Ensuring mixing efficiency of replica-exchange molecular dynamics simulations. J. Chem. Theory Comput. 2008;4:1119–1128. doi: 10.1021/ct800016r. [DOI] [PubMed] [Google Scholar]
- 50.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 51.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 52.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 53.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 54.Hummer G., Rasaiah J.C., Noworyta J.P. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 55.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 56.Daura X., Gademann K., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. Engl. 1999;38:236–240. [Google Scholar]
- 57.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 58.Yoda T., Sugita Y., Okamoto Y. Comparisons of force fields for proteins by generalized-ensemble simulations. Chem. Phys. Lett. 2004;386:460–467. [Google Scholar]
- 59.Nguyen P.H., Li M.S., Derreumaux P. Effects of all-atom force fields on amyloid oligomerization: replica exchange molecular dynamics simulations of the Aβ(16-22) dimer and trimer. Phys. Chem. Chem. Phys. 2011;13:9778–9788. doi: 10.1039/c1cp20323a. [DOI] [PubMed] [Google Scholar]
- 60.Wei G., Mousseau N., Derreumaux P. Sampling the self-assembly pathways of KFFE hexamers. Biophys. J. 2004;87:3648–3656. doi: 10.1529/biophysj.104.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song W., Wei G., Derreumaux P. Self-assembly of the β2-microglobulin NHVTLSQ peptide using a coarse-grained protein model reveals a β-barrel species. J. Phys. Chem. B. 2008;112:4410–4418. doi: 10.1021/jp710592v. [DOI] [PubMed] [Google Scholar]
- 62.Bellesia G., Shea J.E. Effect of β-sheet propensity on peptide aggregation. J. Chem. Phys. 2009;130:145103. doi: 10.1063/1.3108461. [DOI] [PubMed] [Google Scholar]
- 63.De Simone A., Derreumaux P. Low molecular weight oligomers of amyloid peptides display β-barrel conformations: a replica exchange molecular dynamics study in explicit solvent. J. Chem. Phys. 2010;132:165103. doi: 10.1063/1.3385470. [DOI] [PubMed] [Google Scholar]
- 64.Fu Z., Luo Y., Wei G. Induced β-barrel formation of the Alzheimer's Aβ25-35 oligomers on carbon nanotube surfaces: implication for amyloid fibril inhibition. Biophys. J. 2009;97:1795–1803. doi: 10.1016/j.bpj.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S., Humphreys E.S., Jagota A. Peptides with selective affinity for carbon nanotubes. Nat. Mater. 2003;2:196–200. doi: 10.1038/nmat833. [DOI] [PubMed] [Google Scholar]
- 66.Zorbas V., Smith A.L., Musselman I.H. Importance of aromatic content for peptide/single-walled carbon nanotube interactions. J. Am. Chem. Soc. 2005;127:12323–12328. doi: 10.1021/ja050747v. [DOI] [PubMed] [Google Scholar]
- 67.McGaughey G.B., Gagné M., Rappé A.K. π-Stacking interactions. Alive and well in proteins. J. Biol. Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 68.Mukherjee S., Chowdhury P., Gai F. Effect of dehydration on the aggregation kinetics of two amyloid peptides. J. Phys. Chem. B. 2009;113:531–535. doi: 10.1021/jp809817s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gazit E. A possible role for π-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 70.Makin O.S., Atkins E., Serpell L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. USA. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Armstrong A.H., Chen J., Hecht M.H. Mutations that replace aromatic side chains promote aggregation of the Alzheimer's Aβ peptide. Biochemistry. 2011;50:4058–4067. doi: 10.1021/bi200268w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


