Abstract
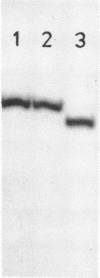
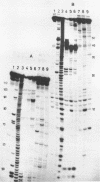
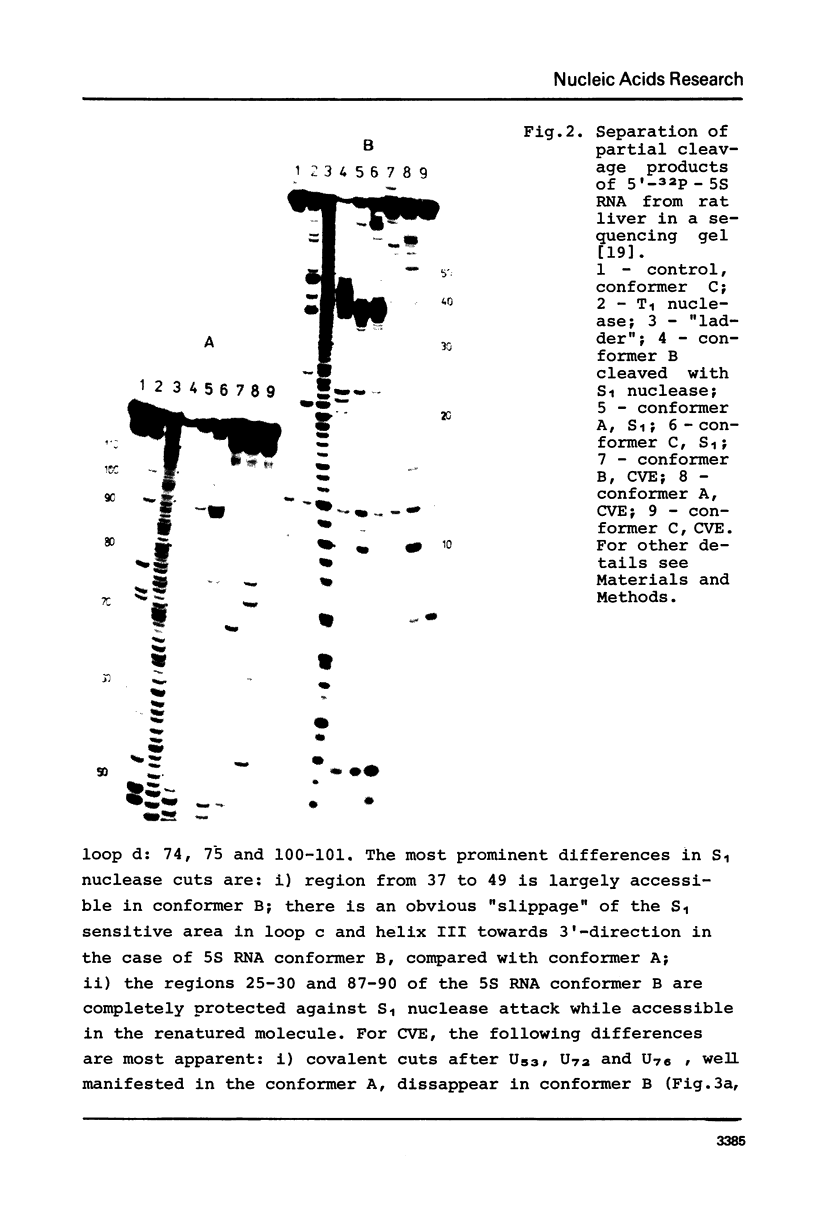
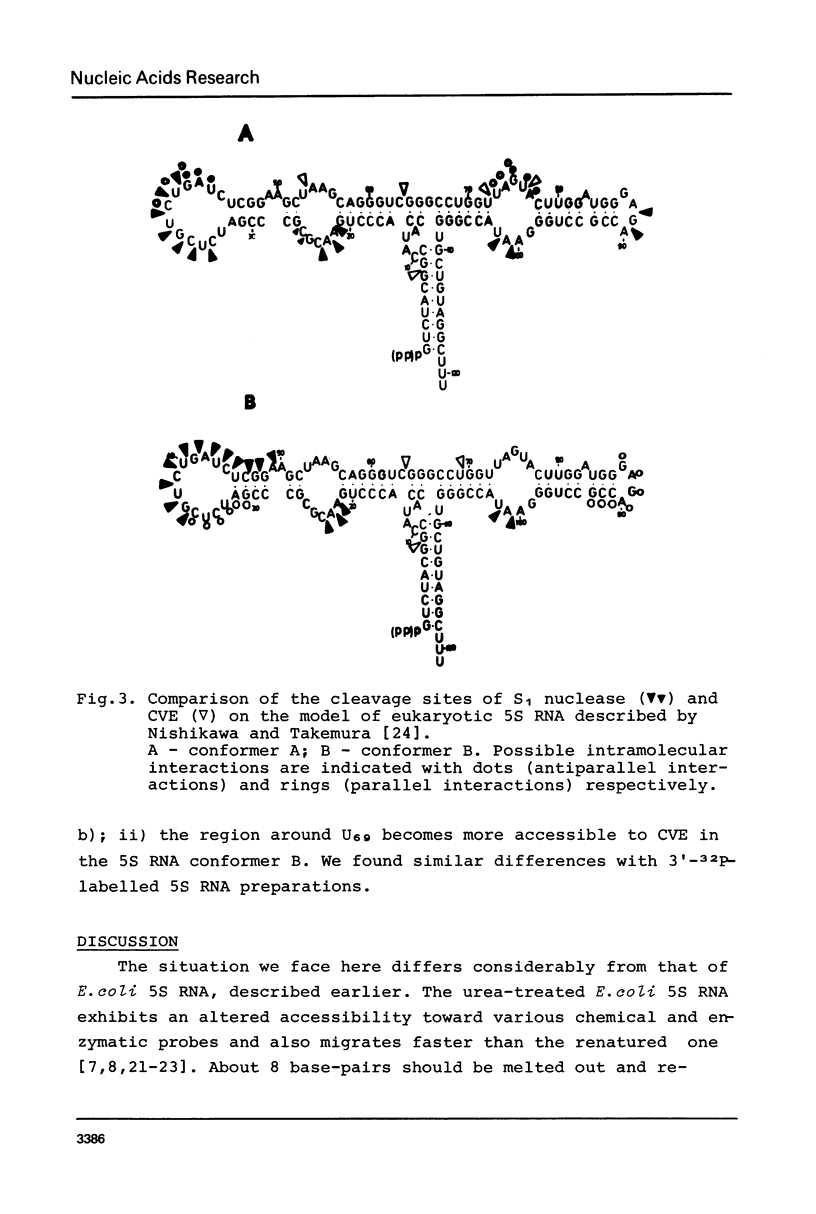
Three different conformers of rat liver 5S ribosomal RNA were investigated by partial nuclease cleavage technique using S1 nuclease and cobra venom endoribonuclease (CVE) as conformational probes. Urea-treated and renatured 5S RNA co-migrate on non-denaturing gels, but exhibit distinct differences in their nuclease cleavage patterns. The most prominent differences in S1 nuclease and CVE accessibility of these conformers are located in region 30-50 and around nucleotides 70 and 90. The third form of 5S RNA with higher electrophoretic mobility was generated by EDTA treatment. The cleavage patterns of this 5S RNA conformer are similar to that characteristic for the renatured 5S RNA. The results demonstrate the difference in secondary structure and possibly different tertiary base-pairing interactions of 5S RNA conformers.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert M., Bellemare G., Monier R. Selective reaction of glyoxal with guanine residues in native and denatured Escherichia coli 5S RNA. Biochimie. 1973;55(2):135–142. doi: 10.1016/s0300-9084(73)80385-2. [DOI] [PubMed] [Google Scholar]
- Aubert M., Scott J. F., Reynier M., Monier R. Rearrangement of the conformation of Escherichia coli 5S RNA. Proc Natl Acad Sci U S A. 1968 Sep;61(1):292–299. doi: 10.1073/pnas.61.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt A., Ebel J. P. The secondary structure of the protein L1 binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the L11 mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981 Jan 24;9(2):293–307. doi: 10.1093/nar/9.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm S., Fabian H., Venyaminov SYu, Matveev S. V., Lucius H., Welfle H., Filimonov V. V. Structural analysis of the A and B conformers of Escherichia coli 5 S ribosomal RNA by infrared spectroscopy. FEBS Lett. 1981 Sep 28;132(2):357–361. doi: 10.1016/0014-5793(81)81197-0. [DOI] [PubMed] [Google Scholar]
- Cameron V., Uhlenbeck O. C. 3'-Phosphatase activity in T4 polynucleotide kinase. Biochemistry. 1977 Nov 15;16(23):5120–5126. doi: 10.1021/bi00642a027. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Farber N. M., Cantor C. R. A slow tritium exchange study of the solution structure of Escherichia coli 5 S ribosomal RNA. J Mol Biol. 1981 Feb 25;146(2):223–239. doi: 10.1016/0022-2836(81)90433-2. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Geroch M. E., Richards E. G., Davies G. A. 5 S RNA. 1. Preparation and characterisation of highly purified 5 S RNA from Escherichia coli. Eur J Biochem. 1968 Nov;6(3):325–330. doi: 10.1111/j.1432-1033.1968.tb00452.x. [DOI] [PubMed] [Google Scholar]
- Jordan B. R. Studies on 5 s RNA conformation by partial ribonuclease hydrolysis. J Mol Biol. 1971 Feb 14;55(3):423–439. doi: 10.1016/0022-2836(71)90327-5. [DOI] [PubMed] [Google Scholar]
- Kao T. H., Crothers D. M. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel've M. B., Metspalu A. Kh, Lind A. Ia, Saarma M. Iu, Villems R. L. Konformatsionnye izomery 5S RNK ribosom pecheni krysy. Mol Biol (Mosk) 1978 May-Jun;12(3):695–699. [PubMed] [Google Scholar]
- Luoma G. A., Burns P. D., Bruce R. E., Marshall A. G. Melting of Saccharomyces cerevisiae 5S ribonucleic acid: ultraviolet absorption, circular dichroism, and 360-MHz proton nuclear magnetic resonance spectroscopy. Biochemistry. 1980 Nov 11;19(23):5456–5462. doi: 10.1021/bi00564a047. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu A., Saarma M., Villems R., Ustav M., Lind A. Interaction of 5-S RNA, 5.8-S RNA and tRNA with rat-liver ribosomal proteins. Eur J Biochem. 1978 Nov 2;91(1):73–81. doi: 10.1111/j.1432-1033.1978.tb20938.x. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Nucleotide sequence of 5 S RNA from Torulopsis utilis. FEBS Lett. 1974 Mar 15;40(1):106–109. doi: 10.1016/0014-5793(74)80904-x. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Garrett R. A. Structure of 5 S ribosomal RNA from Escherichia coli: identification of kethoxal-reactive sites in the A and B conformations. J Mol Biol. 1979 Aug 25;132(4):621–636. doi: 10.1016/0022-2836(79)90378-4. [DOI] [PubMed] [Google Scholar]
- Richards E. G., Lecanidou R., Geroch M. E. The kinetics of renaturation of 5-S RNA from Escherichia coli in the presence of Mg 2+ ions. Eur J Biochem. 1973 Apr;34(2):262–267. doi: 10.1111/j.1432-1033.1973.tb02755.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Monier R., Aubert M., Reynier M. Some optical properties of 5S-RNA from E. coli. Biochem Biophys Res Commun. 1968 Dec 9;33(5):794–800. doi: 10.1016/0006-291x(68)90230-1. [DOI] [PubMed] [Google Scholar]
- Shinagawa M., Padmanabhan R. Inhibition of a nuclease contaminant in the commercial preparations of Escherichia coli alkaline phosphatase. Anal Biochem. 1979 Jun;95(2):458–464. doi: 10.1016/0003-2697(79)90756-5. [DOI] [PubMed] [Google Scholar]
- Speek M., Lind A. Structural analyses of E. coli 5S RNA fragments, their associates and complexes with proteins L18 and L25. Nucleic Acids Res. 1982 Feb 11;10(3):947–965. doi: 10.1093/nar/10.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots I., Metspalu A., Villems R., Saarma M. Location of single-stranded and double-stranded regions in rat liver ribosomal 5S RNA and 5.8S RNA. Nucleic Acids Res. 1981 Oct 24;9(20):5331–5343. doi: 10.1093/nar/9.20.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne R., Jordan B. R., Monier R. A common conformational feature in several prokaryotic and eukaryotic 5 S RNAs: a highly exposed, single-stranded loop around position 40. J Mol Biol. 1973 May 15;76(2):303–311. doi: 10.1016/0022-2836(73)90393-8. [DOI] [PubMed] [Google Scholar]
- Weidner H., Yuan R., Crothers D. M. Does 5S RNA function by a switch between two secondary structures? Nature. 1977 Mar 10;266(5598):193–194. doi: 10.1038/266193a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Pribula C. D., Fox G. E., Zablen L. B. The nucleotide sequence of the 5S ribosomal RNA from a photobacterium. J Mol Evol. 1975 Jun 9;5(1):35–46. doi: 10.1007/BF01732012. [DOI] [PubMed] [Google Scholar]