Abstract
Recent studies suggest that Asiatic wild asses (Equus hemionus) are being increasingly poached in a commercial fashion. Part of the meat is believed to reach the meat markets in the capital Ulaanbaatar. To test this hypothesis, we collected 500 meat samples between February and May 2006. To differentiate between domestic horse (Equus caballus) and wild ass meat, we developed a restriction fragment length polymorphism (RFLP) assay based on the polymerase chain reaction (PCR). We amplified and sequenced a cytochrome b fragment (335 bp) and carried out a multialignment of the generated sequences for the domestic horse, the Asiatic wild ass, the domestic donkey (Equus asinus) and the Przewalski’s horse (Equus ferus przewalskii). We detected a species-specific restriction site (AatII) for the Asiatic wild ass, resulting in a specific restriction fragment length polymorphism (RFLP) band pattern. This RFLP assay represents a rapid and cost-effective method to detect wild ass meat. All of the 500 meat samples we collected and analysed within this pilot project proved to be domestic horsemeat as declared by the sales people. Thus, either the assumption that wild ass meat is sold as “cheap horse meat” is wrong, or we picked the wrong markets, products or season.
Keywords: Asiatic wild ass, domestic horse, illegal meat market, Mongolia, restriction fragment length polymorphism (RFLP)
Introduction
Numbers and distribution range of the Asiatic wild ass (Equus hemionus) have undergone a dramatic decline over the last 100 years. With an estimated population of 20,000 animals (Lhagvasuren, 2007), Mongolia remains the last and most important stronghold of the wild ass. Most probably no more than 5,000 individuals remain outside of Mongolia and northern China (Blank, 2007; Jowkar pers. comm., 2007; Lukarevski & Gorelov, 2007; Shah & Quershi, 2007; Yang, 2007).
In the IUCN Equid Action Plan the status of E. hemionus is qualified as “insufficiently known” and the species is listed as vulnerable (Feh et al., 2002). It is also listed in appendix I of the Convention on International Trade of Endangered Species (CITES) and in 2002 was included in appendix II of the Convention of Migratory Species (CMS or Bonn Convention). In Mongolia, it has received full protection since 1953 (Clark et al. 2006). However, due to human population growth in conjunction with severe winters in the past years, the occurrences of herder - khulan conflicts appear on the increase (Kaczensky et al., 2006).
Competition for pastures and water and poaching for meat seem to be increasingly becoming a problem in Mongolia (Kaczensky et al., 2006; Stubbe et al., 2005; Stubbe et al., 2007). For some local people, wild ass meat seems to provide a substitute or even a cheap alternative to meat from domestic animals (Kaczensky, 2007; P. Kaczensky, unpubl. data). In 2005, a national survey based on questionnaires, suggested that up to 4,500 wild asses might be poached each year throughout their distribution range in Mongolia (Wingard & Zahler, 2006).
Wildlife hunting (legal and illegal) in Mongolia seems to become more and more commercialized. Species poached for meat, like the Asiatic wild ass or the Mongolian gazelle (Procapra gutturosa) are believed to be offered on the meat markets of the urban centres, particularly in Ulaanbaatar. Many locals claimed that wild ass meat is either sold on markets as “cheap horse meat” or used to prepare meals in the restaurant gers along the Mongolian-Chinese border. Another assumption is that wild ass meat might be used by sausage factories (Wingard & Zahler, 2006). Because it is impossible to visually distinguish between wild ass and domestic horse meat, we applied molecular methods to test whether wild ass meat is indeed offered under the synonym of “cheap horse meat” on selected meat markets in Ulaanbaatar. The study was a pilot project to establish a time and cost efficient method to differentiate between the meat of Asiatic wild asses and those of other equids (E. caballus, E. ferus przewalskii, E. asinus) potentially offered on the meat markets of central Asia.
Materials and Methods
Sample collection
Between February and May 2006, we randomly collected a total of 500 samples of “cheap horse meat” four times each month from five different meat markets in Ulaanbaatar. The markets were: Bayanzurkh, Narantuul, Bombogor, Horoolol and Shonhor. All samples were labelled with the collection date and ID for the market and stored in 90% ethanol (Figure 1).
Figure 1.
Sample collection and labelling of meat samples from five selected markets in Ulaanbaatar between February and May 2007.
DNA preparation
For establishing the RFLP assay, tissue of three individuals of the domestic Mongolian horse and 12 individuals of the Asiatic wild ass were used. The horse samples were taken from freshly slaughtered animals, whereas the wild ass samples were taken from fresh carcasses of free-ranging specimens encountered during fieldwork throughout the Gobi 2002-2006 (Kaczensky et al., 2006; Kaczensky et al., 2007).
DNA was isolated with NucleoSpin® Tissue Kit (Macherey-Nagel), deviating from the manufacturer’s protocol only by addition of 20μl RNAse to the lysis step and by drying the columns for 10 min before elution of DNA in 50 μl pre-warmed 5mM Tris buffer (pH 8.5). All solutions were aliquoted and stored at −20°C before preparation for further analysis.
PCR
The equid-specific primers (forward (CytB 1L): 5′-CTAATTAAAATCATCAATC-3′ and reverse (CytB 2H): 5′-AAAAGTAGGATGATTCCAAT-3′) described by Orlando et al. (2003) targets a 335-bp-long DNA fragment of the cytochrome b gene from perissodactyluśs mtDNA. Amplifications were carried out in a total volume of 25 μl, containing 2 μl template DNA, 0.2 μM of each primer (CytB 2H/ CytB 1L), 0.2 mM dNTPs, 1x-PCR buffer (10x BD buffer: pH 9.4-9.5 800mM Tris-HCl, 200 mM (NH4)2SO4; Solis BioDyne Inc., Estonia), 3 mM MgCl2 (Solis BioDyne Inc., Estonia), 0.1 μg bovine serum albumin (BSA, Fermentas Inc.) and 0.5 U of Taq-Polymerase (FIREPol®, Solis BioDyne Inc., Estonia). The PCR profile on an Eppendorf PCR Mastergradient thermal cycler was as follows: 94°C for 3 min for denaturation, 35 cycles of amplification (94°C for 30 s denaturation, 50°C for 30 s annealing, 72°C for 30 s elongation) and final extension at 72°C for 10 minutes. PCR products were examined by electrophoresis through a 1.8% ethidium bromide stained agarose gel.
Sequence analysis and identification of restriction sites
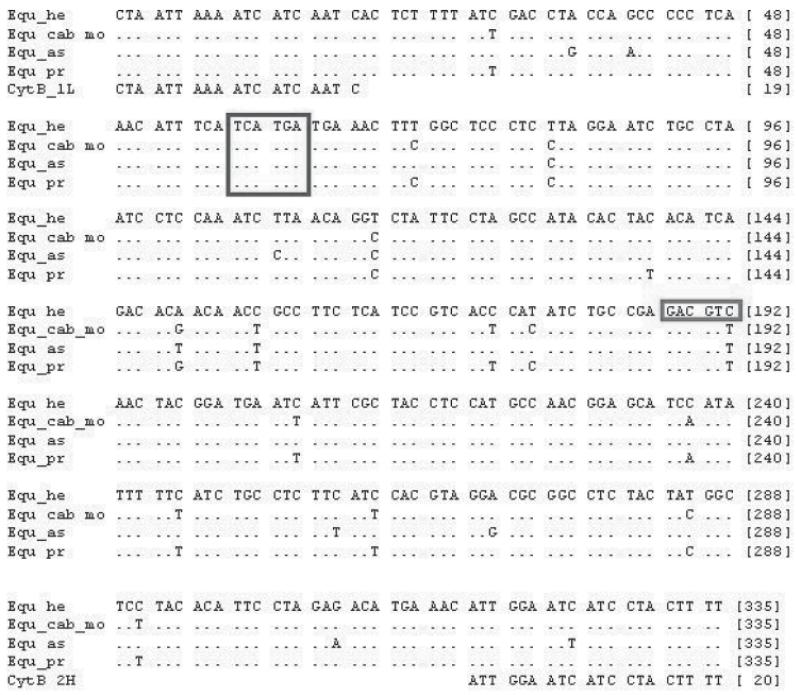
We sequenced the amplified 335 bp long PCR products from two specimens each of E. caballus and E. hemionus both forward and reverse. We separated the PCR products by electrophoresis on a native 8% PAA-gel (29:1 Bis/Acrylamide) at 150 mAh, and visualised them with ethidium bromide under UV-light (366 nm). We used the crush and soak method (Maxam and Gilbert, 1977) for purification, eluted in 15 μl 5 mM Tris (pH 8.5) and sequenced on the automatic ABI 377. We additionally obtained sequence information of the cytochrom b gene fragment for the domestic donkey (GenBank Accession No. X97337) and the Przewalski’s horse (GenBank Accession No. DQ223534) from the National Centre of Biotechnology Information (NCBI) database. These were implicated in a multialignment using CLUSTALX (Thompson et al. 1997). Species-specific restriction sites of the equid cytochrome b sequences were identified with the program NEBcutter Version 2.0 (Vincze et al., 2003) (Figure 2)
Figure 2.
Sequence alignment of a 335bp fragment of the mitochondrial cytochrom b gene from E. hemionus (Equ_he), E. caballus (Equ_cab_mo), E. asinus (Equ_as), E. ferus przewalskii (Equ_pr) and the equid specific primer pair (CytB_1L, CytB_2H). Recognition sites of the two restriction enzymes are highlighted with black frames, i.e. PagI T ↓CATGA and AatII GACGT↓C.
The restriction enzymes AatII (target sequence: GACGT↓C) and PagI (target sequence: T↓ CATGA) were selected by the following criteria:
-
-
Both restriction enzymes produce easily distinguishable differences in RFLP banding profiles.
-
-
AatII restriction site is discriminatory for E. hemionus (+) versus the other three equids (−).
-
-
PagI cuts all tested equid sequences and thus failure of restriction, e.g. through inhibitors, can be excluded.
-
-
AatII plus PagI are compatible for double digestion.
Restriction fragment patterns were expected to produce the following RFLP pattern:
-
-
Asiatic wild ass: 59/131/145 bp
-
-
Domestic Mongolian horse: 59/276 bp
-
-
Domestic donkey: 59/276 bp
-
-
Przewalski’s horse: 59/276 bp
Restriction Fragment Length Polymorphism (RFLP) analysis
To test whether RFLP analysis is really diagnostic for species identification, we analysed three samples from E. caballus and 12 from E. hemionus from different geographical regions in Mongolia.
We performed restriction enzyme incubation with AatII plus PagI in 15μl double digestion volumes according to the manufacturer’s instruction (Fermentas Inc.). 10 μl of the PCR product was digested with 1 U PagI (Fermentas Inc.), 1 U AatII (Fermentas Inc.), 1x restriction buffer green (Fermentas Inc.) and 0.1 μg BSA for 1½ h at 37°C. The digested PCR products were separated on a 1.8% ethidium bromide stained agarose gel and visualized by ultraviolet irradiation. For size reference, a pUC19 DNA/ MspI (HpaII) marker (Fermentas Inc.) was used.
Results
Diagnostic value of the RFLP analysis for species differentiation
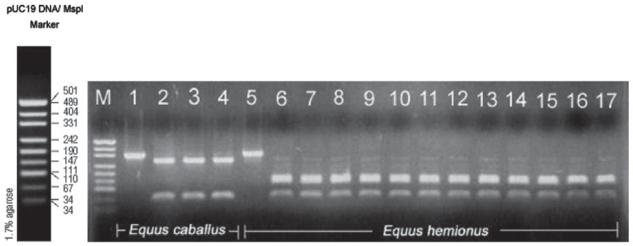
Multiple sequence alignment of the cytochrome b sequences from four equids species E. caballus, E. hemionus, E. asinus and E. ferus przewalskii revealed interspecies polymorphisms. The multialignment displayed 22 point mutations within the 335 bp sequence analysed. Applicable for discrimination of E. hemionus by RFLP analysis is the transition at the position 192 bp (C↔T) using AatII restriction enzyme. The double digestion treatment with the six-cutter restriction enzymes Pag I and Aat II resulted in different, easily distinguishable banding patterns for the equids E. caballus, E. asinus and E. ferus przewalskii (59/276 bp) and the Asiatic wild ass (59/131/145 bp) (Fig 3). The intraspecific banding patterns for the twelve reference samples of E. hemionus were consistent throughout the geographical range sampled (see Figure 3, Lane 6-17).
Figure. 3.
RFLP banding patterns of an amplified 335 bp fragment of the cytochrome b gene obtained from two different Mongolian equid species after double digestion with restriction enzymes AatII and PagI. Lane 1: PCR product E. caballus, undigested (335 bp); Lanes 2-4: E. caballus, digested (59/276 bp); Lane 5: PCR product E. hemionus, undigested (335 bp); Lanes 6-17: E. hemionus from different geographical regions in Mongolia, digested (59/131/145 bp); M: pUC19 MspI size marker.
After the initial establishment, the analysis per 10 samples will need on average 1.5 hours for lysis and DNA preparation by using the BIO&SELL nexttec™ Geneomic DNA-Isolation-Kit (fast DNA-isolation kit) and about 3 hours for the RFLP analysis (PCR, restriction and electrophoresis), with total consumable cost of approximately 40 US$. For the laboratory equipment, pipettes, a centrifuge, electrophoresis supply and a PCR machine are needed.
Presence of Asiatic wild ass meat on selected meat markets of Ulaanbaatar
None of the 500 so-called “cheap horse meat” samples from the Ulaanbaatar meat market showed E. hemionus banding pattern. All showed the banding pattern of E. caballus. Thus, all the meat sold on the five meat markets, during the timeframe surveyed, was correctly labelled.
Discussion
The RFLP assay proved to be a reliable, rapid and cost-efficient method to distinguish between the meats of different equid species. As it is impossible to visually distinguish between domestic horse and wild ass meat on a market stand, the RFLP assay provides a simple law enforcement tool for detecting poached wild ass meat. The analysis techniques could easily be established in a Mongolian lab and requires only minimal training.
Although we failed to find any wild ass meat sold on the five markets surveyed, this does not necessarily mean wild ass meat is not marketed in Ulaanbaatar. It is possible that we targeted the wrong markets or picked a season where no or little wild ass meat is offered. Furthermore it is possible that wild ass meat is not sold in its raw form, but rather enters the food market in a processed form, e.g. as an admix to sausages or as a filling for “khuushuur” (Mongolian style samosas or dough pockets filled with ground meat). Many locals in the Gobi suspect that wild ass meat is used for the latter purpose at restaurant gers along the Mongolia-Chinese border (P. Kaczensky, unpubl. data).
We suggest that further efforts are made to sample more meat markets over a longer time period in Ulaanbaatar as well as in the aimag centres within the distribution range of the wild ass (e.g. Sainshand and Dalanzadgad). Furthermore it would be good to test and validate the RFLP assay method for processed meat. If this is successful, subsequent analysis of random samples of processed meat products from Ulaanbaatar and the main Gobi markets appears warranted.
Acknowledgements
Funding for the meat sample analysis was provided by the Austrian Science Foundation (FWF, grant P18624). We would like to thank N. Enkhsaikhan for his help with shipping and export logistics. Dried skin samples from wild ass carcasses were exported from Mongolia with CITES MN0133419 and imported to the EU with CITES AT06-E-1888 and veterinary permit BMGF-74130/0270-IV/5/2006. The domestic horse meat samples were imported with veterinary permit BMGF-74130/0183-IV/5/2006.
References
- Blank DA. Asiatic wild ass in Israel. Exploration into the Biological Resources of Mongolia. 2007;10:261–266. [Google Scholar]
- Clark EL, Munkhbat J, Dulamtseren S, Baillie JEM, Batsaikhan N, Samiya R, Stubbe M, editors. Mongolian Red List of Mammals. Zoological Society of London; London: 2006. (Regional Red List Series Vol. 1). (compilers & editors) (In English and Mongolian) [Google Scholar]
- Feh C, Shah N, Rowen M, Reading RP, Goyal SP. Status and action plan for the Asiatic wild ass (Equus hemionus) In: Moehlman PD, editor. Equides: Zebras, Asses and Horses. IUCN Publication Services Unit; Cambridge, United Kingdom: 2002. pp. 62–71. Available from: http://iucn.org. [Google Scholar]
- Kaczensky P. Wildlife value orientations of rural Mongolians. Human Dimensions in Wildlife. 2007;12:317–329. [Google Scholar]
- Kaczensky P, Sheehy DP, Johnson DE, Walzer C, Lkhagvasuren D, Sheehy CM. Mongolia Discussion Papers, East Asia and Pacific Environment and Social Development Departure. World Bank; Washington, D.C.: 2006. Room to roam? The threat to khulan (Wild Ass) from human intrusion. Available from: http://web.worldbank.org/WBSITE/EXTERNAL/COUNTRIES/EASTASIAPACIFICEXT/EXTEAPREGTOPENVIRONMENT/contentMDK:21074556~pagePK:34004173~piPK:34003707~theSitePK:502886,00.html. [Google Scholar]
- Kaczensky P, Enkhsaikhan N, Ganbaatar O, Walzer C. Identification of herder-wild equid conflicts in the Great Gobi B Strictly Protected Area in SW Mongolia. Exploration into the Biological Resources of Mongolia. 2007;10:99–116. [Google Scholar]
- Lhagvasuren B. Population assessment of khulan (Equus hemionus) in Mongolia. Exploration into the Biological Resources of Mongolia. 2007;10:45–48. [Google Scholar]
- Lukarevski VS, Gorelov YK. Khulan (Equus hemionus Pallas 1775) in Turkmenistan. Exploration into the Biological Resources of Mongolia. 2007;10:231–240. [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proceedings of the National Academy of Science USA. 1977;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando L, Eisenmann V, Reynier F, Sondaar P, Hänni C. Morphological Convergence in Hippidion and Equus (Amerhippus) South American Equids Elucidated by Ancient DNA Analysis. Journal of Molecular Evolution. 2003;57:29–40. doi: 10.1007/s00239-003-0005-4. [DOI] [PubMed] [Google Scholar]
- Sha N, Qureshi Q. Social organization and determinants of spatial distribution of Khur (Equus hemionus khur) Exploration into the Biological Resources of Mongolia. 2007;10:189–200. [Google Scholar]
- Stubbe A, Stubbe M, Batsajchan N, Samjaa R, Dorzderem S. First results of Wild Ass research in the South Gobi Aymag / Mongolia 2003 and 2004. Exploration into the Biological Resources of Mongolia. 2005;9:107–120. [Google Scholar]
- Stubbe M, Stubbe A, Batsajchan N. Morphology, reproduction and mortality of Equus hemionus hemionus in Mongolia. Exploration into the Biological Resources of Mongolia. 2007;10:117–132. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Research. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingard JR, Zahler P. Mongolia Discussion Papers, East Asia and Environment and Social Development Department. World Bank; Washington, D.C.: 2006. Silent Steppe: The illegal Wildlife Trade Crisis. Available at: http://web.worldbank.org/WBSITE/EXTERNAL/COUNTRIES/EASTASIAPACIFICEXT/MONGOLIAEXTN/contentMDK:21021328~pagePK:141137~piPK:141127~theSitePK:327708,00.html. [Google Scholar]
- Yang W. An overview on the state of Equus hemionus in whole China. Exploration into the Biological Resources of Mongolia. 2007;10:155–158. [Google Scholar]