Abstract
Genome-enabled prediction of breeding values using high-density panels (HDP) can be highly accurate, even for young sires. However, the cost of the assay may limit its use to elite animals only. Low-density panels (LDP) containing a subset of single nucleotide polymorphisms (SNP) may give reasonably accurate predictions and could be used cost-effectively with young males and females. This study evaluates strategies for selecting subsets of SNP for several traits, compares predictive ability of LDP with that of HDP, and assesses the benefits of including parent average (PA) as a predictor in models using LDP. Data consisting of progeny-test predicted transmitting ability (PTA) for net merit and 6 other traits of economic interest from 4,783 Holstein sires were evaluated using testing and training sets with regressions on their high-density genotypes and parent averages for net merit index. Additionally, SNP subsets of different sizes were selected using different strategies, including the “best” SNP based on the absolute values of their estimated effects from HDP models for either the trait itself or lifetime net merit, and evenly spaced (ES) SNP across the genome. Overall, HDP models had the best predictive ability, setting an upper bound for the predictive ability of LDP sets. Low-density panels targeting the SNP with strongest effects (for either a single trait or lifetime net merit) provided reasonably accurate predictions and generally outperformed predictions based on evenly spaced SNP. For example, evenly spaced sets would require at least 5,000 to 7,500 SNP to reach 95% of the predictive ability provided by HDP. On the other hand, this level of predictive ability can be achieved with sets of 2,000 SNP when SNP are selected based on magnitude of estimated effects for the trait. Accuracy of predictions based on LDP can be improved markedly by including parent average as a fixed effect in the model; for example, a set with the 1,000 best SNP using the parent average achieved the 95% of the accuracy of a HDP model.
Keywords: genomic selection, parent average, low-density panel, single nucleotide polymorphism
INTRODUCTION
Progeny testing combines phenotypes and pedigree information to arrive at predictions of the transmitting abilities of selection candidates. These predictions can be highly precise, especially for animals with large numbers of progeny. However, progeny testing is expensive and increases generation interval. Dense panels of molecular markers such as SNP are now available for many plant and livestock species, and these are advancing research on whole-genome selection, as well as changing breeding practices across species. Molecular markers show sharing of chromosome segments between individuals. Such information can be used to arrive at earlier prediction of transmitting ability (Meuwissen et al., 2001), and these predictions can attain high reliability (VanRaden et al., 2009; Weigel et al., 2009).
However, the cost of high-density panels (HDP) is still high: as of today, genotyping one animal with the Illumina BovineSNP50 Bead Chip (Illumina Inc., San Diego, CA) costs approximately $225, limiting the use of this technique to highly valuable animals. Hence, interest exists in developing low-density panels (LDP) that could be widely adopted in the population. Designing these LDP requires careful selection of SNP. Weigel et al. (2009) evaluated the predictive ability of LDP for net merit in US Holsteins. The early predictive ability based on LDP increased steadily as the number of markers in the model increased, and SNP subsets containing markers selected based on estimates of their effects outperformed subsets of evenly spaced (ES) markers.
In practice, selection goals typically include several traits, and an optimal subset of SNP for one trait may not be optimal for other traits. For this reason, Habier et al. (2009) proposed using ES markers to represent the entire genome and to follow haplotypes within families to impute missing SNP, as opposed to selected specific chromosomal regions. Our study addresses the challenge of selecting a unique subset of informative SNP for many traits, noting that some selection strategies may outperform ES designs in this regard. Additionally, to be a valuable contribution, the predictive ability achieved by LDP should be higher than early predictions attained with parent averages (PA).
The present study (1) evaluates predictive ability of different strategies for selecting subsets of SNP for 7 economically relevant traits in dairy cattle; (2) compares those predictions with predictions obtained from HDP and with PA; and (3) evaluates the effect of including PA as a covariate in SNP-based models for genome-assisted evaluations for net merit.
MATERIALS AND METHODS
Data
Data were provided by the USDA-ARS Animal Improvement Programs Laboratory (Beltsville, MD) and consisted of 4,703 sires genotyped with the Illumina BovineSNP50 Bead Chip and their progeny-test PTA for milk, fat, and protein production, lifetime net merit, productive life, SCS, and daughter pregnancy rate (DPR). Net merit is a measure of the expected lifetime net profit and is a linear combination of PTA for protein yield (16%), fat yield (19%), productive life (22%), SCS (10%), udder conformation (7%), foot and leg conformation (4%), body size (6%), DPR (6%), and calving ability (5%) (Cole et al., 2010). Many of those traits are genetically correlated. The production traits (milk, fat, and protein) have positive and strong genetic correlations between them (between 0.45 and 0.81) and have moderate positive correlations with both productive life (around 0.09) and SCS (approximately 0.18). Daughter pregnancy rate, on the other hand, correlates negatively with all of the other traits (approximately −0.32 for production traits) except productive life, where the correlation is positive (0.51). Somatic cell score correlates negatively with all of the other traits here evaluated (approximately −0.30 with DPR and productive life, and −0.10 with the other traits) (Cole et al., 2010). The reliability of PTA ranged from 49 to 99%, where 16% of sires had reliability less than 80%, 68% were between 80 and 90%, and 16% were greater than 90%. Models were trained with data from sires born before 1999 (n = 3,305) using PTA computed in 2003. The predictive ability of models was assessed using 2008 PTA of bulls born between 1999 and 2003 (n = 1,398). Single nucleotide polymorphisms with minor allele frequencies below 5% or with more than 10% missing values were removed; after editing 32,518 markers were available. Missing SNP were imputed using samples from the empirical marginal distribution of marker genotypes. Specifically, let pj be the estimated allele frequency of the jth marker, and z1ij and z2ij be 2 samples drawn from a Bernoulli distribution with success probability equal to pj for the ith sire. Then, we imputed xij using xij = z1ij + z2ij, (xij = 0, 1, 2) as codes for the SNP genotypes. In addition to the aforementioned information, standardized 2003 PA for net merit was obtained for 3,715 sires, 2,821 from the training set (85%) and 893 from the testing set (64%).
Models
Methods for genome-enabled predictions must be able to estimate an enormous number of marker effects from a much smaller number of phenotypic observations (p ≫ n). In an ordinary least squares approach, not all marker effects can be estimated (Lande and Thompson, 1990), and marker-by-marker tests lead to biased estimates and multiple testing problems. Consideration of marker effects as random addresses this issue, and it has been shown that marker effect estimates may be better when the variance of the effects is assumed to be heterogeneous (Meuwissen et al., 2001). Different methods have been proposed to deal with these problems. In the Bayesian Least Absolute Selection and Shrinkage Operator (LASSO), the prior distribution assigned to the marker effects is double exponential. Compared with the normal distribution, it has a larger peak at zero and heavier tails (Tibshirani, 1996), and it produces marker-specific shrinkage. This prior can be represented as an infinite mixture of scaled normal distributions (Park and Casella, 2008). In this study, Bayesian LASSO regression was used in all models that were fitted.
The PTA for each of the traits y = (y1,…,y4,703)′ were standardized to unit sampling variance. If yi is PTA of the ith subject and
and
are the (sample) mean and standard deviation, respectively, then the standardized PTA was defined as . Later, the standardized PTA were regressed on SNP genotypes, xi = (xi1,…,xip)′, i∈ (1,…,4,703) using a Bayesian LASSO model (Park and Casella, 2008) that was extended to accommodate fixed effects. For net merit, some models also included standardized PA (p̄i) as a fixed effect (computed as the average of the parental PTA for 2003). In the Bayesian LASSO, the likelihood function is
where β0 is an effect common to all subjects; β1 =(β1,1,…, β1, p)′ is a vector of marker effects; β2 is a regression coefficient for p̄i (included in only some models for net merit); and is a residual variance. The prior distribution of model unknowns is as follows:
where is a normal density assigned to the jth marker effect, is a scaled-inverted chi-squared density with degrees of freedom dfε and prior scale Sε, assigned to the residual variance; is an exponential prior for the scale parameters , and p (λ|α1, α2, K) ∝ Beta (λK−1|α1, α2) is a nonconjugate prior assigned to the regularization parameter λ, with support on [0,K]; parameters α1 and α2 control the shape of this distribution, with α1 = α2 = 1 giving a uniform distribution on [0,K]; de los Campos et al. (2009) give a discussion of possible priors in Bayesian LASSO models. Samples from the posterior distribution were obtained using the Gibbs sampler described in de los Campos et al. (2009) and were implemented in the R environment (R Development Core Team, 2009). In our application, dfε = 1, Sε = 0.5, α1 = α2 = 1.4, and K = 500. Inferences were based on 55,000 samples obtained after discarding 10,000 as burn-in. Convergence was evaluated by visual inspection of trace plots.
A sequence of models using subsets of markers of various sizes and selected using different strategies were fitted to each of the 7 traits. Table 1 gives a summary of these models and the abbreviations used to denote them. We first fitted models using the HDP (p = 32,518 SNP) in the training set comprising 2003 PTA for milk, fat, protein, productive life, SCS, and DPR (n = 3,305 sires in the training set), as well as net merit (n = 2,821). These models are denoted as HDP (Table 1) and (in the training sets) they set an upper bound for the predictive ability expected from LDP. Subsequently, we fitted regressions to LDP of various sizes (p = 30, 50, 100, 150, 200, 250, 300, 500, 750, 1,000, 1,250, 1,500, 2,000, 2,500, 3,000, 3,500, 4,000, 4,500, 5,000, 7,500, 10,000). The SNP for the LDP were selected based on 4 strategies: (1) the top SNP (with p equal to the size of the LDP) selected based on the largest absolute value of their estimated effects obtained from the HDP model for each trait (Top.Trait); (2) a combination of the top SNP selected based on the largest absolute value of their estimated effects obtained from the HDP model for all the traits but net merit (Top.All), where each trait had approximately, as the traits (co)vary, p/6 top SNP in the platform; (3) the top SNP chosen based on the largest absolute value of their estimated effects on the HDP model for the lifetime net merit index (Top. Merit); and (4) SNP that were evenly spaced (ES) along the genome. The Top.All strategy does not assign the number of SNP based on economic weights for each trait, because the weights would be modified by the estimated SNP effects. A random set of SNP shows a lower bound for the strategies; rather than using SNP completely at random, we used ES that may have advantages for imputation, as Weigel et al. (2010) shows in a related study. We also evaluated sets of selected SNP based on the absolute value of the ratio of the estimated marker effect relative to its posterior standard deviation. However, results from this selection strategy were very similar to those obtained without standardization, and are therefore not included in this article. The Top.All, Top.Merit, and ES selection strategies yield the same chip for all traits, whereas the Top. Trait chips are trait-specific. Finally, models for net merit were fitted with and without PA included as a fixed effect. Parent average is an estimator of transmitting ability for young animals. For animals without progeny or phenotypes, the PA is equivalent to prediction using full pedigree data. Using PA in combination with the genotypic information that allows tracking of Mendelian segregation may improve genetic estimates. The PA for net merit was also used alone to predict the PTA; this model sets a lower bound for predictive ability. Predictive ability was assessed in the testing set (n = 893 and n = 1,398 sires in the testing sets for net merit and for all other traits, respectively) via the correlation between genome-based predictions and the PTA obtained with progeny testing in 2008.
Table 1.
Description of regression models: SNP selection criteria, number of SNP used (p), use of parent average (PA) as covariate, and notation for models
| SNP selection criteria (number of SNP) | PA included | Notation |
|---|---|---|
| All (high-density panel, HDP) (p = 32,518) | Yes | HDP.SNP.PA |
| No | HDP.SNP | |
| Evenly spaced (ES) (p = variable) | Yes | ES.PA |
| No | ES | |
| Preselected (Top) by largest effects on the HDP.SNP model for the specific trait (Trait) (p = variable) | Yes | Top.Trait.PA |
| No | Top.Trait | |
| Preselected (Top) by largest effects on the HDP.SNP models for all the traits (All): milk, fat, protein, productive life, SCS, and daughter pregnancy rate. Each trait contributed 1/6 of the SNP in the platform. (p = variable) | Yes | Top.All.PA |
| No | Top.All | |
| Preselected (Top) by largest effects on the HDP.SNP model for net merit (Merit) (p = variable) | Yes | Top.Merit.PA |
| No | Top.Merit | |
| None (p = 0) | Yes | PA |
RESULTS AND DISCUSSION
Predictive Ability of HDP Models and PA
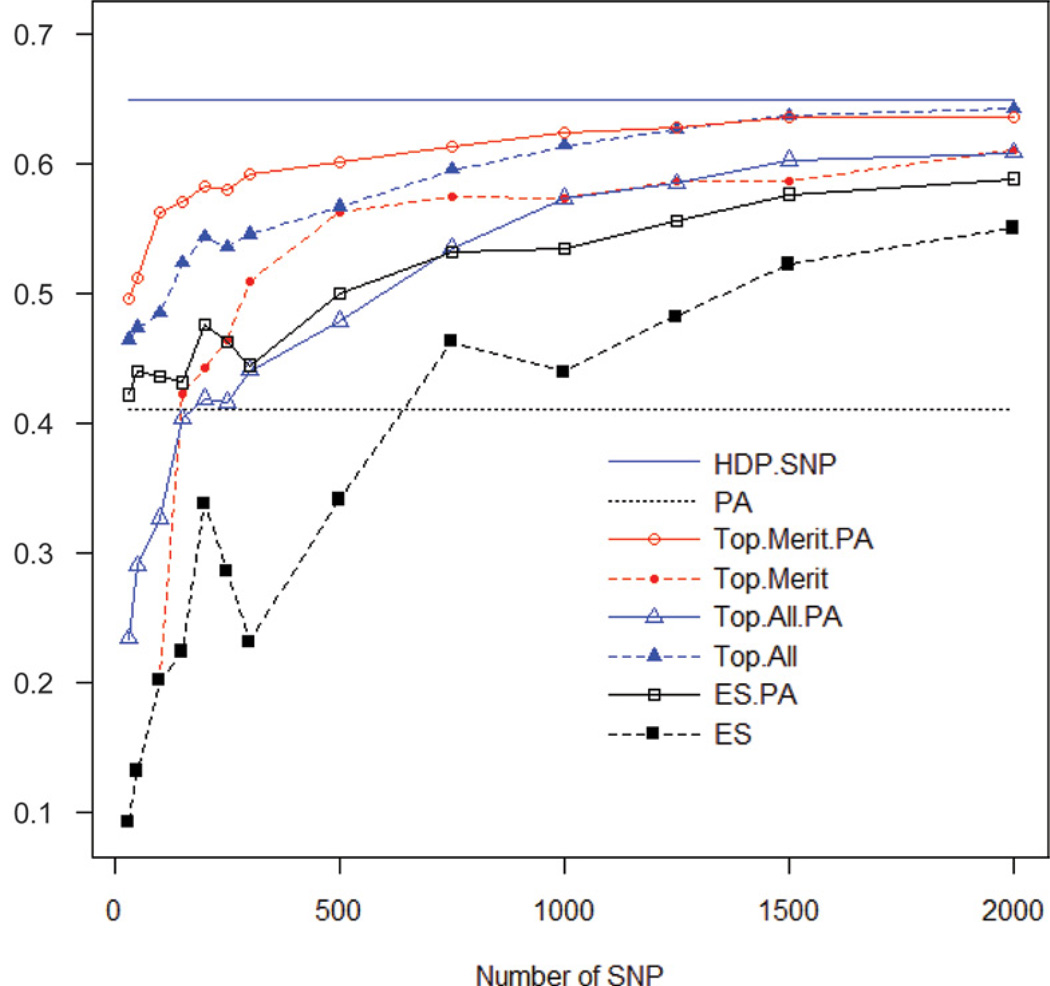
Genome-enabled prediction of PTA with HDP panels was highly correlated with the 2008 PTA for all traits, including milk yield (0.70), fat yield (0.68), protein yield (0.71), productive life (0.68), SCS (0.67), and DPR (0.68) (Figure 1 and Table 2). These results are in agreement with previous reports by VanRaden et al. (2009) and Weigel et al. (2009), who indicated that genome-enabled predictions with HDP could attain high levels of predictive ability for economically relevant traits in dairy cattle. Genome-based predictions for net merit were also highly correlated with the PTA in models HDP.SNP (0.65) and HDP.SNP.PA (0.65), but the regression on PA had a lower correlation (0.41) (Figure 2). Genomic predictions with PA as a covariate had correlations with PTA that were 158% larger than those from predictions using only PA. VanRaden et al. (2009) reported a similar difference between PA and genome-based predictions for net merit in Holsteins (0.33 and 0.53, respectively). In an infinitesimal additive model in the absence of inbreeding, PA can account for up to half of the additive variance of the offspring, the other half being accounted for by Mendelian segregation. Molecular markers allow tracking Mendelian segregation at several points in the genome, and this can be used to refine early predictions. Finally, HDP alone performed almost as well as HDP panels including PA; for those models, the correlation was almost the same (0.65 for both models).
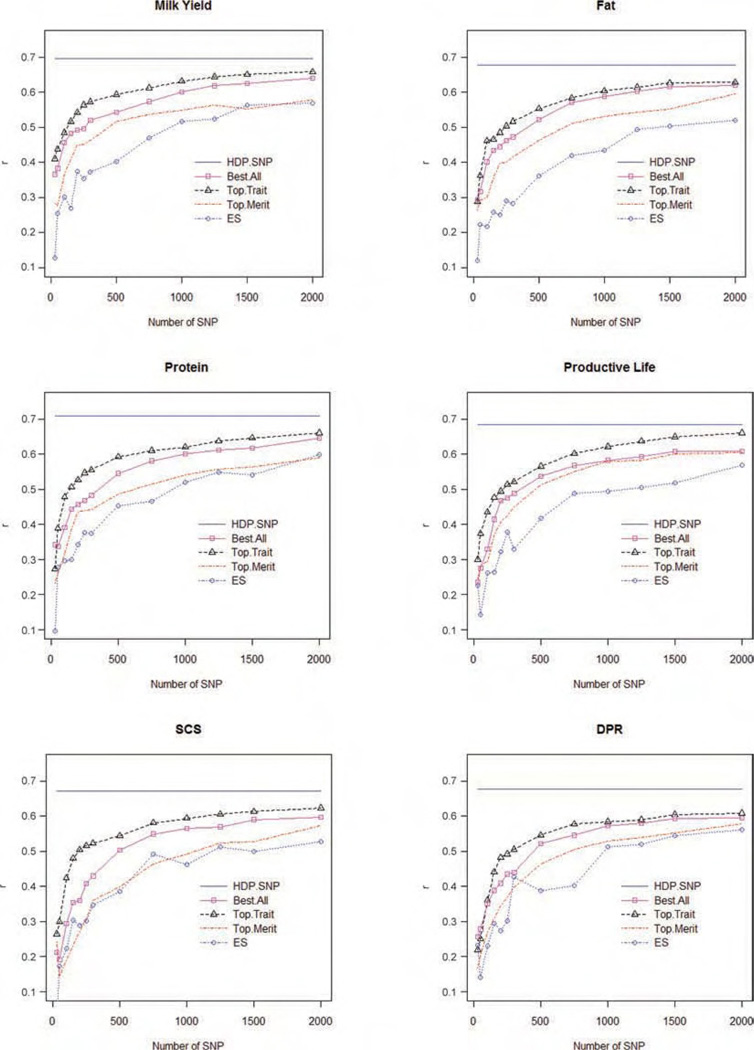
Figure 1.
Correlations between progeny-test PTA and genome-enabled predictions from the high-density panel (HDP), the top SNP for HDP of each trait (Top.Trait), the top SNP for the 6 traits (Top.All), the top SNP for HDP of net merit (Top.Merit), and evenly spaced SNP (ES) for milk, fat, and protein yields, productive life, SCS, and daughter pregnancy rate (DPR). Color version available in the online PDF.
Table 2.
Correlations between progeny-test PTA and genome-enabled predictions obtained from model for net merit1
| Number of SNP | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model2 | 30 | 300 | 500 | 1,000 | 2,000 | 3,000 | 5,000 | 10,000 |
| Correlation between genomic prediction and PTA | ||||||||
| Top.Merit | 0.09 | 0.51 | 0.56 | 0.57 | 0.61 | 0.62 | 0.63 | 0.64 |
| Top.All | 0.23 | 0.44 | 0.48 | 0.57 | 0.61 | 0.62 | 0.65 | 0.65 |
| ES | 0.09 | 0.23 | 0.34 | 0.44 | 0.55 | 0.55 | 0.61 | 0.64 |
| Top.Merit.PA | 0.50 | 0.59 | 0.60 | 0.62 | 0.64 | 0.65 | 0.66 | 0.67 |
| Top.All.PA | 0.46 | 0.54 | 0.57 | 0.61 | 0.64 | 0.65 | 0.67 | 0.67 |
| ES.PA | 0.42 | 0.44 | 0.50 | 0.53 | 0.59 | 0.59 | 0.62 | 0.64 |
| Correlation between genomic prediction and PTA, relative to that obtained with a high-density model (%) | ||||||||
| Top.Merit | 14 | 79 | 86 | 88 | 94 | 96 | 97 | 99 |
| Top.All | 36 | 68 | 74 | 88 | 94 | 96 | 100 | 100 |
| ES | 14 | 35 | 52 | 68 | 85 | 85 | 94 | 99 |
| Top. Merit.PA | 77 | 91 | 92 | 96 | 99 | 100 | 102 | 103 |
| Top.All.PA | 71 | 84 | 87 | 94 | 99 | 100 | 103 | 104 |
| ES.PA | 65 | 68 | 77 | 82 | 91 | 91 | 96 | 99 |
The top half of the table gives the estimated predictive correlation, and the bottom half expresses this correlation relative to that obtained with high-density markers (0.649).
Top.Merit uses markers selected based on the absolute value of the estimated marker effect obtained in the high-density model; Top.All uses markers selected based on the absolute value of the estimated marker effect obtained in the high-density models for milk, fat, protein, productive life, SCS, and daughter pregnancy rate (DPR); ES uses evenly spaced markers; Top.Merit.PA, Top.All.PA, and ES.PA include markers and parent average as predictors.
Figure 2.
Correlations between progeny-test PTA for net merit and genome-enabled predictions obtained with the high-density panel (HDP), the parent average (PA), the top SNP for HDP with (Top.Merit.PA) and without (Top.Merit) parent averages, the top SNP for 6 traits (milk, fat, protein, productive life, SCS, and daughter pregnancy rate) with (Top.All.PA) and without (Top.All) parent averages, and evenly spaced SNP with (ES.PA) and without (ES) parent averages, by set sizes of SNP. HDP.SNP (—) and PA (….) have a fixed number of SNP. Color version available in the online PDF.
Predictions Based on LDP
Figures 1 and 2 give the estimated predictive correlation between genome-based predictions and the 2008 PTA for net merit (Figure 2) and all other traits (Figure 1) versus subset size. Table 2 gives the predictive correlations obtained for net merit for each subset of markers, relative to that obtained with a HDP. The HDP models set an upper bound for the predictive ability of LDP and predictive correlations increased with subset size in all situations (Figures 1 and 2). The rate of increase in predictive correlation slowed substantially for LDP larger than 1,500 or 2,000 SNP, depending on the trait and the strategy used to select markers (Figures 1 and 2); these results are in agreement with the literature (Weigel et al., 2009). However, marker-based predictions using 2,000 markers had a predictive correlation that was still substantially smaller than HDP models; that is, the predictive correlations for Top.Trait and ES for a LDP with 2,000 SNP were 94% and 85% relative to the HDP model (100%) for net merit (Table 2). For the other traits, the predictive correlation of panels using 2,000 markers were 98 to 93% for the Top.Trait SNP, 92 to 88% for the Top.Merit SNP, and 89 to 84% for ES SNP relative to that from HDP.
Selection Strategy
The method used to select SNP for the LDP affected predictive ability markedly, especially for small subsets. For a given subset size, Top.Trait always had the highest predictive ability. Any marker associated with a QTL region for that particular trait would be expected to be included in the selected subset. These results provide a reasonable upper bound for other LDP for multiple traits (Figure 1). However, a disadvantage of this method is that it builds an optimal LDP specific for each trait of interest, which is not practical when the selection goal involves several traits.
For multiple-trait selection purposes with LDP, selection of markers using Top.All, Top.Merit, and ES yields a single LDP for all traits. Top.Trait subsets, on the other hand, lead to trait-specific panels. The Top.All was the best selection strategy in all the situations. The Top.Merit subsets of SNP outperformed ES for most traits (Figure 2). For example, with 300 SNP, genome-based predictions using Top.All, Top.Merit, and ES panels had predictive correlations of 0.52, 0.46, and 0.37, respectively, for milk yield; 0.47, 0.42, and 0.28 for fat yield; 0.48, 0.44, and 0.38 for protein yield; 0.49, 0.45, and 0.33 for productive life; 0.43, 0.36, and 0.35 for SCS; and 0.44, 0.40, and 0.43 for DPR. Net merit is a linear combination of economically important traits, and it is very likely that favorable markers for net merit are in regions of the chromosome associated with genetic variation for relevant traits, but it was not as effective as directly selecting the best SNP for each trait (Top.All). The difference in predictive ability of Top.Merit and ES was more important for traits with high weight in net merit or traits that are correlated with traits weighted heavily in the index (e.g., milk, fat, protein yield, and productive life) and was less important for traits with relatively low weight (SCS and DPR). Note that in this study, an ES set does not include all SNP of a smaller ES set. For this reason a smaller subset, including markers closer to regions that contribute highly to the genetic variance, might predict better that the following larger subset besides the panel is denser.
Table 3 shows the correlation between the order of importance of the SNP effects for each of the 6 individual traits and net merit. Net merit has correlations that range from 0.28 to 0.71 with these traits. This would lead to an expectation of a good response in all traits when selecting markers based on net merit, and the LDP Top.Merit strategy was indeed better than ES. However, selecting SNP for the component traits of net merit (Top.All) was superior in every situation. One could expect a decline in DPR when using the Top.All LDP, because the order of the SNP effects for DPR was negatively correlated with most of the other traits (in a range of −0.17 to −0.20 for SCS and the production traits), but this did not happen (Figure 1, last panel).
Table 3.
Correlation between the estimated marker effects obtained in the high-density model for each trait
| Protein | Fat | Net merit |
Productive life |
SCS | DPR1 | |
|---|---|---|---|---|---|---|
| Milk | 0.82 | 0.37 | 0.49 | 0.07 | 0.08 | −0.20 |
| Protein | — | 0.58 | 0.65 | 0.10 | 0.07 | −0.18 |
| Fat | — | — | 0.61 | 0.08 | −0.04 | −0.18 |
| Net merit | — | — | — | 0.71 | −0.39 | 0.28 |
| Productive life | — | — | — | — | −0.45 | 0.56 |
| SCS | — | — | — | — | — | −0.17 |
Daughter pregnancy rate.
Inclusion of Parent Average in Models for LDP
Genome-enabled predictions based on small LDP subsets substantially improved, when PA was included in the regression (Figure 1 and Table 2). The smaller the set, the larger the improvement, and this was observed regardless of the strategy used to select the markers. For example, for 300 SNP, adding PA in the model increased predictive correlations from 0.51 to 0.59 for Top.Trait versus Top.Trait.PA, 0.44 to 0.54 for Top.All versus Top.All.PA, and from 0.23 to 0.44 for ES versus ES.PA. Note that the predictions based on 300 ES SNP yielded a correlation that was almost half of that attained by a simple regression on PA (0.23 vs. 0.41).
The LDP with selected SNP and PA as a covariate (i.e., Top.Merit.PA) can yield high predictive correlations. For example, with only 300 SNP, the Top.Merit. PA model had a predictive correlation that was 91% of that attained with HDP-SNP, and that correlation improved to more than 95% of the HDP when the predictions were based on 1,000 SNP. Other sets of SNP needed to be larger to achieve 95% of the HDP; for example, Top.Trait sets needed at least 2,500 SNP, Top.All.PA needed 2,500, Top.All needed 1,000, ES.PA needed 5,000 SNP, and the ES regression needed at least 7,500 SNP (Table 2). The increase in predictive ability obtained by including PA in models with LDP decreases as the number of markers in the panel increases (Table 2); for example, at 10,000 SNP all model predictions achieve 99% of the predictions based on the HDP.SNP model.
CONCLUSIONS
The overall improvement resulting from the use of molecular genomic information instead of PA for prediction was approximately 58%. Low-density panels with targeted SNP can provide reasonable predictive ability of genetic merit of animals (e.g., a platform with 300 SNP yielded correlations of 0.51 and 0.23 for Top.Trait and ES SNP for net merit, not considering PA). The ES subsets should include a larger number of SNP (about 5,000) or consider imputation of missing genotypes to improve predictive ability. Additionally, if PA information is available, a platform of 300 SNP combined with these averages could attain predictions that were only 9% below those from a HDP (0.59). Top.All LDP is a single platform for all traits, and the strategy provided predictions that outperformed both Top.Merit and ES in all situations, even though some traits had negative correlations between their estimated marker effects. Over generations, the effectiveness of the LDP would be reduced, linkage disequilibrium would change, and other forces such as selection and mutation would act. The LDP panel is breed-specific, because linkage disequilibrium and marker effects are expected to change between breeds. These results are specific to the Holstein population considered and might not extend to other situations. For other species, the number of markers would change depending on the level of linkage disequilibrium.
ACKNOWLEDGMENTS
The authors gratefully acknowledge scientists at the USDA-ARS Beltsville Agricultural Research Center (Beltsville, MD) for providing genotypic and phenotypic data for the present study. G. J. M. Rosa and K. A. Weigel acknowledge financial support from the Wisconsin Agriculture Experiment Station and from the National Association of Animal Breeders (Columbia, MO), respectively. D. B. Allison acknowledges NIH grant number R01GM077490. Suggestions by the anonymous reviewers of this manuscript are gratefully acknowledged.
REFERENCES
- Cole JB, VanRaden PM Multi-State Project S-1040. Net merit as a measure of lifetime profit: 2010 revision. [Accessed May 2010];2010 http://aipl.arsusda.gov/reference/nmcalc.htm.
- de los Campos G, Naya H, Gianola D, Crossa J, Legarra A, Manfredi E, Weigel K, Cotes JM. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics. 2009;182:375–385. doi: 10.1534/genetics.109.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D, Fernando RL, Dekkers JCM. Genomic selection using low-density marker panels. Genetics. 2009;182:343–353. doi: 10.1534/genetics.108.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Thompson R. Efficiency of marker assisted selection in the improvement of quantitative traits. Genetics. 1990;124:743–756. doi: 10.1093/genetics/124.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Casella G. The Bayesian LASSO. J. Am. Stat. Assoc. 2008;103:681–686. [Google Scholar]
- R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2009. R: A language and environment for statistical computing. http://www.R-project.org. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J. Royal Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- VanRaden PM, Van Tassell CP, Wiggans GR, Sonstegard TS, Schnabel RD, Taylor RD, Schenkel FS. Reliability of genomic predictions for North American dairy bulls. J. Dairy Sci. 2009;92:16–24. doi: 10.3168/jds.2008-1514. [DOI] [PubMed] [Google Scholar]
- Weigel KA, de los Campos G, Gonzáez-Recio O, Naya H, Wu XL, Long N, Rosa GJM, Gianola D. Predictive ability of direct genomic values for lifetime net merit of Holstein sires using selected subsets of single nucleotide polymorphism markers. J. Dairy Sci. 2009;92:5248–5257. doi: 10.3168/jds.2009-2092. [DOI] [PubMed] [Google Scholar]
- Weigel KA, de los Campos G, Vazquez AI, Rosa GJM, Gianola D, Van Tassell CP. Accuracy of direct genomic values derived from imputed single nucleotide polymorphism genotypes in Jersey cattle. J. Dairy Sci. 2010;93:5423–5435. doi: 10.3168/jds.2010-3149. [DOI] [PubMed] [Google Scholar]