Abstract
The bacterial respiratory pathogens Bordetella pertussis and Bordetella bronchiseptica employ multiple alternative iron acquisition pathways to adapt to changes in the mammalian host environment during infection. The alcaligin, enterobactin, and heme utilization pathways are differentially expressed in response to the cognate iron source availability by a mechanism involving substrate-inducible positive regulators. As inducers, the iron sources function as chemical signals termed ferrimones. Ferrimone-sensing allows the pathogen to adapt and exploit early and late events in the infection process.
Keywords: Bordetella, Iron, Heme, Siderophore, Regulation, Ferrimone
Introduction
Bordetella species
Members of the genus Bordetella are Gram-negative β-proteobacteria of the family Alcaligenaceae. Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica, the so-called classical Bordetella species, are highly genetically related respiratory pathogens of mammals (Mattoo and Cherry 2005; Parkhill et al. 2003). B. pertussis is a human-adapted species and is the agent of whooping cough or pertussis. B. parapertussis infects humans and other animals including sheep (Mattoo and Cherry 2005) and B. bronchiseptica causes respiratory diseases in a variety of nonhuman mammalian hosts. Based on genomic sequence analyses, B. bronchiseptica or a B. bronchiseptica-like organism is the hypothesized progenitor of B. pertussis and B. parapertussis (Parkhill et al. 2003).
B. pertussis, B. parapertussis, and B. bronchiseptica colonize the cilia of the ciliated cells of the host respiratory epithelium where they multiply and elaborate a variety of factors, resulting in disease symptoms. Their virulence factors include adhesins such as filamentous hemagglutinin, a type III secretion system, dermonecrotic toxin and an adenylate cyclase/hemolysin (Confer and Eaton 1982; Fennelly et al. 2008; Hewlett and Wolff 1976; Katada and Ui 1982; Kozak et al. 2007). Tracheal cytotoxin is also released by the bacteria. This toxin is a peptidoglycan-derived disaccharide tetrapeptide that triggers host nitric oxide production that leads to extrusion of the ciliated cells from the epithelium (Flak and Goldman 1999). An additional virulence factor produced only by B. pertussis is the ADP-ribosylating pertussis toxin. Production of most of the known Bordetella virulence factors is transcriptionally controlled by a two-component phosphorelay system consisting of the BvgS transmembrane sensor kinase and the BvgA DNA-binding response regulator (Uhl and Miller 1994; Weiss et al. 1983).
Bordetella iron retrieval
During infection, host cells and Bordetella cells coordinate their activities in response to one another in a dynamic environment. As both host and bacterial cells are damaged and cellular products are liberated, the nutrients that become available to Bordetella, including iron, are likely to fluctuate over the course of the infection. Sensitive iron source-sensing mechanisms allow Bordetella to adapt to these fluctuations.
B. pertussis, B. parapertussis and B. bronchiseptica have 14–19 genes encoding TonB-dependent outer membrane receptors that are known or predicted to be involved in iron uptake. To obtain iron, these species produce and utilize the siderophore alcaligin (Brickman et al. 1996; Moore et al. 1995). B. pertussis and B. bronchiseptica are also capable of using the xenosiderophores enterobactin (Beall and Sanden 1995a), ferrichrome and desferrioxamine B (Beall and Hoenes 1997). B. bronchiseptica has also been reported to use ferrichrysin, ferrirubin, aerobactin, protochelin, schizokinen, ferricrocin, vicibactin, and pyoverdin (Pradel and Locht 2001). In addition, these Bordetella species can also utilize hemin as an iron source (Vanderpool and Armstrong 2001). The Bordetella genetic systems for only three known iron sources have been characterized to date: alcaligin, enterobactin and heme. Under iron-replete growth conditions the genes of these three iron systems are repressed by Fur (Beall and Sanden 1995b; Brickman and Armstrong 1995; Brickman et al. 2004). Importantly, each is also under positive transcriptional regulation that involves induction by the cognate iron source under derepressing conditions.
Alcaligin siderophore system
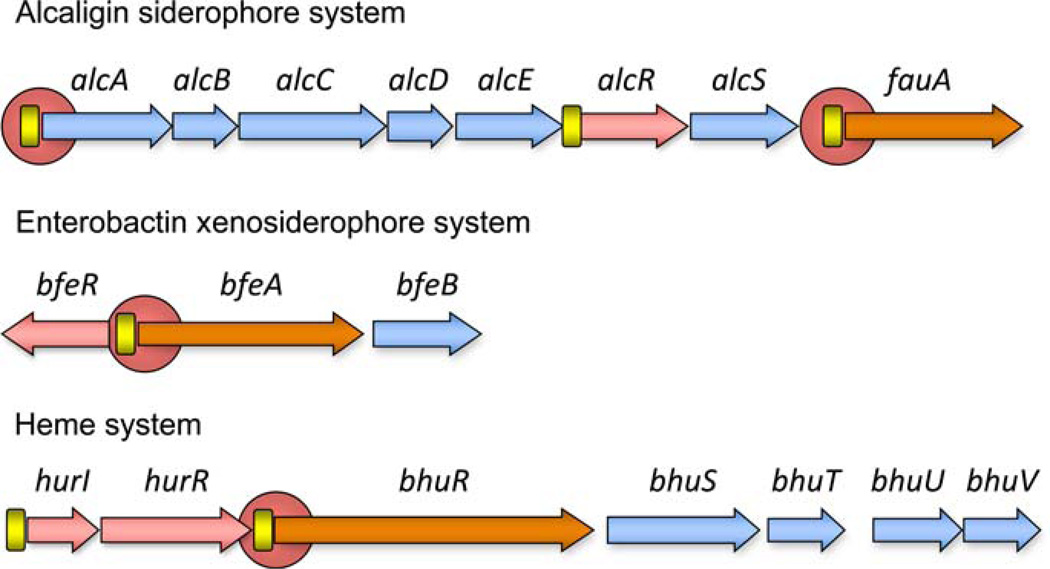
Alcaligin (1,8(S),11,18(S)-tetrahydroxy-1,6,11,16-tetraazacycloeicosane-2,5,12,15-tetrone) is a 20-membered macrocyclic dihydroxamate siderophore (Nishio et al. 1988; Nishio and Ishida 1990). The alcaligin system gene cluster spans a 10.6-kb contiguous region of the Bordetella chromosome (Fig. 1), and encodes alcaligin biosynthesis enzymes, the AlcR substrate-inducible transcriptional regulator (Beaumont et al. 1998; Pradel et al. 1998; Brickman et al. 2001), the alcaligin exporter AlcS (Brickman and Armstrong 2005), and the outer membrane FauA receptor protein for transport of ferric alcaligin (Brickman and Armstrong 1999). The organization of the alcaligin system gene cluster is conserved among the classical Bordetella species (Parkhill et al. 2003). The genetic system has four transcriptional units: a polycistronic alcABCDER transcript, and monocistronic transcripts for alcR, alcS and fauA (Beaumont et al. 1998; Kang and Armstrong 1998; Pradel et al. 1998; Brickman and Armstrong 1999; Brickman and Armstrong 2005).
Fig. 1.
Genetic organization of the alcaligin, enterobactin, and heme utilization systems in Bordetella species. Arrows denote the limits and transcriptional orientations of genes. Locations of Fur-binding sites (rectangles) are shown, and circles represent control regions that are responsive to the cognate positive regulator and iron source inducer
The alcABCDER operon encodes alcaligin biosynthesis and regulatory functions (Armstrong and Clements 1993; Giardina et al. 1995; Kang et al. 1996; Giardina et al. 1997; Kang and Armstrong 1998; Beaumont et al. 1998; Pradel et al. 1998). The pathway for alcaligin biosynthesis has not been fully elucidated and mechanistic details remain mostly hypothetical (Brickman and Armstrong 1996; Giardina et al. 1995; Kang et al. 1996; Pradel et al. 1998; Challis 2005); however, the initial step in alcaligin biosynthesis in Bordetellae is known to involve decarboxylation of ornithine to produce putrescine, catalyzed by the pyridoxal phosphate-dependent decarboxylase Odc (Brickman and Armstrong 1996). It is believed that putrescine is then converted to N-hydroxyputrescine by AlcA, a putative FAD-dependent monooxygenase, by a reaction similar to that catalyzed by IucD in aerobactin biosynthesis. AlcB is then thought to mediate the N-acylation of the hydroxylamine group with succinyl CoA by a process similar to the IucB-dependent acylation of N6-hydroxylysine with acetyl CoA in the aerobactin pathway. The product could then undergo C-hydroxylation mediated by AlcE, which has similarity to the ring-hydroxylating dioxygenase family of iron–sulfur cluster proteins. Finally, AlcC, which belongs to a siderophore synthetase family that includes IucC, could catalyze nucleotide triphosphate-dependent dimerization, followed by nucleotide triphosphate-dependent macrocyclization to yield alcaligin. The role for AlcD in alcaligin biosynthesis is unclear. Alcaligin binds ferric iron at a 3:2 molar ratio (Fe2Alc3) at physiological pH, and the complex has a stability constant estimated at 1037 M−1 (Nishio et al. 1988).
AlcR is a 36-kDal protein of the AraC/XylS family of transcriptional regulators (Gallegos et al. 1997) that has high similarity to the Bordetella BfeR regulator that activates enterobactin utilization genes in response to enterobactin and certain other catechol compounds (Anderson and Armstrong 2004). During iron starvation, transcription of fauA and alcABCDER is activated by AlcR by a mechanism requiring induction by alcaligin (Brickman et al. 2001; Brickman and Armstrong 2002). Thus, AlcR functions as a substrate-inducible regulator of alcaligin biosynthesis and transport genes. Expression of alcR itself is subject to positive autoregulation. AlcR-mediated transcriptional activation is ultrasensitive, allowing the bacterium to sense and respond to extremely low concentrations of inducer in its environment (Brickman et al. 2001).
Export of monomeric alcaligin from the bacterial cell requires AlcS, a permease of the major facilitator superfamily class of proton motive force-dependent membrane efflux pumps (Brickman and Armstrong 2005). The exporter activity of AlcS is also critical for maintenance of appropriate intracellular alcaligin substrate levels for normal AlcR-dependent induction of alcaligin system gene expression. The alcS gene is expressed constitutively from a monocistronic transcript that is produced independent of cellular iron status (Kang and Armstrong 1998; Brickman and Armstrong 2005).
Bordetella alcaligin system genes are repressible by Fur and iron (Beall and Sanden 1995b; Brickman and Armstrong 1995) (Fig. 1) acting at three control regions. The alcABCDER operon is transcribed from a Fur and iron-repressible promoter upstream of alcA (Brickman and Armstrong 1995; Kang et al. 1996; Kang and Armstrong 1998). The alcR gene is also expressed as a monocistronic transcript originating at another Fur-regulated promoter immediately upstream of the alcR gene; the activity of this secondary alcR promoter is independent of the autoregulatory influence of AlcR acting at the alcABCDER control region (Beaumont et al. 1998). A third Fur-controlled promoter-operator region resides upstream of the fauA outer membrane receptor gene (Brickman and Armstrong 1999).
Enterobactin xenosiderophore system
Enterobactin has an extremely high ferric ion stability constant of 1049 M−1 (Loomis and Raymond 1991) and is commonly produced by members of the Enterobacteriaceae. As obligate mucosal pathogens, B. pertussis and B. bronchiseptica have no known external environmental reservoirs; however, they can use enterobactin to obtain iron and their expression of enterobactin transport systems implies that they naturally encounter enterobactin. The ability to use this high-affinity siderophore may confer to them a competitive advantage in the host.
The Bordetella BfeA outer membrane receptor is required for transport and utilization of ferric enterobactin (Beall and Sanden 1995a). Downstream of bfeA is the predicted cotranscribed and translationally coupled bfeB gene that encodes a member of the α/β superfamily of hydrolases (Fig. 1). BfeB exhibits significant amino acid sequence similarity to the periplasmic Salmonella enterica IroE protein involved in hydrolytic cleavage of salmochelin, the glucosylated form of enterobactin (Zhu et al. 2005). Divergently transcribed from bfeA is the bfeR gene that encodes an AraC/XylS-like regulator that is highly similar to Bordetella AlcR involved in alcaligin gene regulation, with the greatest similarity residing in the putative C-terminal DNA binding domain. Three Fur repressor DNA-binding sites had been predicted upstream of bfeA in B. pertussis (Beall and Sanden 1995a) and functional binding of Fur to the 0.4 kb bfeA-bfeR intergenic region in E. coli was demonstrated using the Fur titration assay (Anderson and Armstrong 2004).
During iron starvation, expression of bfeA, and likely bfeB, is stimulated in the presence of the transport system substrate, enterobactin, by a BfeR-dependent mechanism (Anderson and Armstrong 2004). Since BfeR is a predicted cytoplasmic AraC family regulator, its activation likely requires direct contact with enterobactin. Interestingly, although growth stimulation by ferric enterobactin requires the BfeA receptor, transcriptional activation of the bfeA gene by BfeR and enterobactin requires neither BfeA (Anderson and Armstrong 2004) nor TonB (Anderson and Armstrong 2006). These observations indicate that, as with the FauA- and TonB-independent induction of the alcaligin system by alcaligin, the enterobactin inducer also enters the cell by a route that does not rely on the ferric siderophore’s outer membrane receptor or any other TonB dependent receptor.
Since the upper respiratory tract is the natural colonization site for B. pertussis and B. bronchiseptica, yet enterobactin is produced primarily by inhabitants of the intestinal tract, the utilization of enterobactin by Bordetella is obscure. It is possible that certain respiratory tract commensals produce enterobactin or a structurally similar siderophore, or that members of the Enterobacteriaceae can transiently colonize the host upper respiratory tract and release enterobactin. Alternatively, the authentic ligand for BfeA may be a host molecule that bears structural similarity to enterobactin.
The ability of other catechol siderophores to be transported into Bordetella cells via BfeA was examined. The B. bronchiseptica BfeA receptor protein could transport the ferric catechol siderophores salmochelin and corynebactin to promote bacterial growth in iron-restricted medium. When these siderophores were tested for the ability to activate BfeR-mediated transcription of bfeA, salmochelin was found to have inducing activity but there was little, if any, induction by corynebactin. In comparison, enterobactin was the most potent inducer and stimulator of Bordetella growth (Anderson and Armstrong 2006).
Host neuroendocrine catecholamines such as epinephrine, norepinephrine, and dopamine bear structural similarity to the iron-binding ligands of enterobactin and also have the ability to bind iron (Jewett et al. 1997; Raymond et al. 1976). Norepinephrine was reported to induce E. coli FepA enterobactin receptor production (Burton et al. 2002). Previous studies have examined the role of norepinephrine in removing the iron from transferrin and lactoferrin, making it available for growth of E. coli (Burton et al. 2002; Freestone et al. 2003) and Salmonella enteriditis (Methner et al. 2008). Epinephrine, norepinephrine, and dopamine were found to induce bfeA expression levels in Bordetella by more than 28-fold (Anderson and Armstrong 2006). Induction by norepinephrine is iron-regulated and dependent on the BfeR regulator, similar to induction by enterobactin. Since catecholamine-induced bfeA transcription may be solely due to structural similarity to enterobactin and not necessarily a biologically relevant observation, the ability of norepinephrine to provide iron for Bordetella growth was examined.
Growth of B. bronchiseptica in iron-depleted and serum-supplemented medium was enhanced by addition of norepinephrine (Anderson and Armstrong 2006). This growth promotion was siderophore-independent. When iron-depleted culture medium was supplemented with partially iron-saturated transferrin instead of serum, growth of B. bronchiseptica was dramatically enhanced by norepinephrine (Anderson and Armstrong 2008). Most importantly, B. bronchiseptica alcA alcaligin biosynthesis mutants that do not produce siderophore can obtain transferrin-bound iron via norepinephrine. These observations contrast with those showing that norepinephrine-mediated growth of E. coli or S. enterica requires either enterobactin or breakdown products of enterobactin or salmochelin (Burton et al. 2002; Methner et al. 2008; Freestone et al. 2003). In Bordetella, norepinephrine-mediated transferrin-iron uptake requires neither the BfeA enterobactin receptor nor the alcaligin siderophore or its FauA receptor (Anderson and Armstrong 2008). Preliminary evidence indicates involvement of theTonB system in norepinephrine-mediated iron acquisition (Anderson and Armstrong 2008). Since Bordetella cells inhabit the respiratory mucosa, one may reason that lactoferrin would be the host glycoprotein most commonly encountered. During infection, host catecholamines as well as transferrin from the bloodstream may access epithelial surfaces damaged by Bordetella toxins; thus it is conceivable that the Bordetella catecholamine iron retrieval mechanism may be biologically relevant to growth in the host.
Heme system
B. pertussis and B. bronchiseptica can use hemin and hemoglobin as sole sources of iron, and this utilization requires the TonB system (Nicholson and Beall 1999; Pradel et al. 2000). The B. pertussis and B. bronchiseptica hurIR bhuRSTUV gene cluster (Fig. 1) associated with heme utilization and regulatory functions has been identified and characterized (Vanderpool and Armstrong 2001). The bird pathogen Bordetella avium is a more genetically distant species and has an orthologous heme utilization gene cluster, rhuIR bhuRSTUV (Murphy et al. 2002). BhuR is the outer membrane receptor for hemin; it has a predicted TonB box C motif and the amino acid sequence motif (FRAP/NPNL) that is conserved among members of the bacterial heme receptor family (Vanderpool and Armstrong 2001). For the transport of heme, BhuT, BhuU and BhuV are the predicted periplasmic heme-binding protein, cytoplasmic membrane permease and ATP binding proteins, respectively. The possible function of BhuS is not clear. The homologous ShuS protein of Shigella dysenteriae can bind heme and is thought to function as a chaperone to sequester internalized heme, thus preventing heme toxicity (Wyckoff et al. 2005).
The BhuR outer membrane heme receptor has an N-terminal extension of 70 amino acids that is similar to the specialized N-terminal regions of members of the E. coli ferric citrate receptor FecA superfamily. These proteins are involved in iron source-inducible extracytoplasmic function (ECF) sigma factor-dependent regulation of iron genes. The receptor N-terminal extension is essential for the signaling cascade that leads to transcription initiation via a cognate ECF σ factor (Kim et al. 1997; Braun and Mahren 2005). In Bordetella cells, the other participants in the signaling cascade are the homologs of E. coli FecI and FecR proteins, HurI (ECF sigma factor) and the HurR anti-sigma membrane sensor (Vanderpool and Armstrong 2001). Initial studies demonstrated that in iron-starved Bordetella cells, production of the BhuR heme receptor was significantly elevated in response to addition of hemin to the culture (Vanderpool and Armstrong 2001). Increased BhuR production was transcriptionally controlled and required a functional hurI gene (Vanderpool and Armstrong 2003). As predicted, hurI mutants are unable to use heme effectively but can be complemented by hurI. Expression of hurI from a high copy number plasmid suppressed the heme inducer requirement, resulting in heme-independent bhuR expression.
Transcriptional activation of the bhu genes in response to heme is dose-dependent and specific for heme; other heme structural analogs such as chlorophyll a, protoporphyrin IX and zinc-protoporphyrin IX did not activate bhu gene expression (Vanderpool and Armstrong 2004). The promoters controlling expression of the hurIR bhuRSTUV genes were mapped (Vanderpool and Armstrong 2004). The hurI promoter has similarity to σ70-like promoters and its transcription is Fur- and iron-repressible and heme independent. Results from those studies indicate that transcription originating at the hurI promoter reads through the hurR–bhuR region, contributing to bhuR expression under iron-starvation conditions in the absence of heme. Heme-responsive transcription originating at bhuR is HurI-dependent. A recent report by the Connell group showed that HurP, a predicted RseP protease homolog encoded outside of the hur–bhu gene cluster, is required for heme utilization and heme-inducible bhuR expression and is proposed to function by modifying the HurR protein (King-Lyons et al. 2007).
Based on results from our studies (Vanderpool and Armstrong 2001, 2003, 2004), those from Connell and colleagues (King et al. 2005; King-Lyons et al. 2007; Kirby et al. 2001; Murphy et al. 2002), and knowledge from the E. coli Fec system (Harle et al. 1995; Ochs et al. 1995, 1996), a model of Bordetella bhu gene regulation has emerged. Upon iron-depletion, Fur derepression at the hurI promoter leads to hurIR transcription and read-through transcription of bhuRSTUV, resulting in basal production levels of the heme transport proteins. Under these conditions, the HurR sensor is predicted to bind HurI. When BhuR binds heme, this signal is transmitted across the periplasm via contact of its specialized N-terminus with HurR. BhuR–HurR interaction allows release and/or activation of HurI, enabling it to associate with RNA polymerase to promote transcription at the bhuR promoter. HurP may serve to proteolytically alter HurR to modulate the signaling cascade. This Bordetella mechanism of cell surface signaling by heme ensures that bhuRSTUV are up-regulated only when the cognate heme source is present.
Temporal signaling and the infection process
On the upper respiratory tract epithelium, the interaction of B. pertussis with its human host characteristically results in inflammation, activation of immune responses, and injury to host tissues. Over the course of infection, changes in host niche conditions alter the array and abundance of iron sources available to the organism. The involvement of the substrate-inducible positive regulators AlcR (Beaumont et al. 1998; Pradel et al. 1998), BfeR (Anderson and Armstrong 2004), and HurI (Vanderpool and Armstrong 2001) ensures that the iron systems can be differentially expressed, allowing for effective adaption to changes in iron source availability during infection.
The importance of the alcaligin (Brickman and Armstrong 2007), heme (Brickman et al. 2006) and enterobactin (Brickman et al. 2008) systems for in vivo growth of B. pertussis was established in mixed infection competition experiments using a wild-type strain together with an isogenic fauA, bhuR or bfeA receptor mutant in a mouse respiratory infection model system. Alcaligin, enterobactin and heme receptor mutants were each attenuated for virulence, indicating that each iron utilization system makes a distinct and essential contribution to in vivo multiplication and survival of B. pertussis, albeit at different stages of infection. The alcaligin system had a crucial role in initial colonization and over the entire course of infection. The enterobactin system contributed significantly to initial growth but to a lesser extent than the alcaligin system, and its role diminished after peak bacterial multiplication. Interestingly, the heme system was not needed for growth until after the peak of B. pertussis growth, at which time it was essential. Notably, the phasing of alcaligin, enterobactin and heme system involvement implies that host iron sources indeed change as infection progresses and that B. pertussis can adapt to those changes by deploying different iron acquisition systems.
Temporal expression patterns of the alcaligin, enterobactin and heme systems were examined using recombinase-based in vivo expression technology (RIVET) in a mouse respiratory infection model (Brickman et al. 2008). The three systems were found to be differentially expressed in vivo, showing early induction of the alcaligin and enterobactin siderophore systems and delayed induction of the heme system. These results correlated with those from our competition infection experiments indicating the importance of siderophores early in infection and the heme system at later stages of infection. These RIVET transcription studies confirmed that B. pertussis expresses these three iron acquisition systems in a host and the relevant iron sources are present, while the competition infection experiments demonstrated that B. pertussis requires these iron acquisition systems in vivo. Overall, the results render a pattern of Bordetella iron acquisition in the host that is characterized by early reliance on siderophores and subsequent dependence on heme.
Pertussis patients mount an antibody response to Bordetella iron receptors. A collection of human sera obtained from normal uninfected adults and from B. pertussis culture-positive patients was screened for antibody reactivity with iron-regulated Bordetella membrane proteins (Brickman et al. 2008). Eight of ten sera from infected donors reacted with multiple iron-repressible proteins, including the receptors for alcaligin (FauA), enterobactin (BfeA) and heme (BhuR), whereas 3 of 10 samples from uninfected donors reacted strongly with these receptor proteins. This survey supplied evidence that B. pertussis produces these receptors in the human host during natural infection, confirms that B. pertussis experiences iron starvation in vivo, and implies that the cognate iron source inducers are present in the human host environment.
Iron source ‘ferrimones’ integrate environmental and intracellular signals to prioritize expression of the iron transport systems
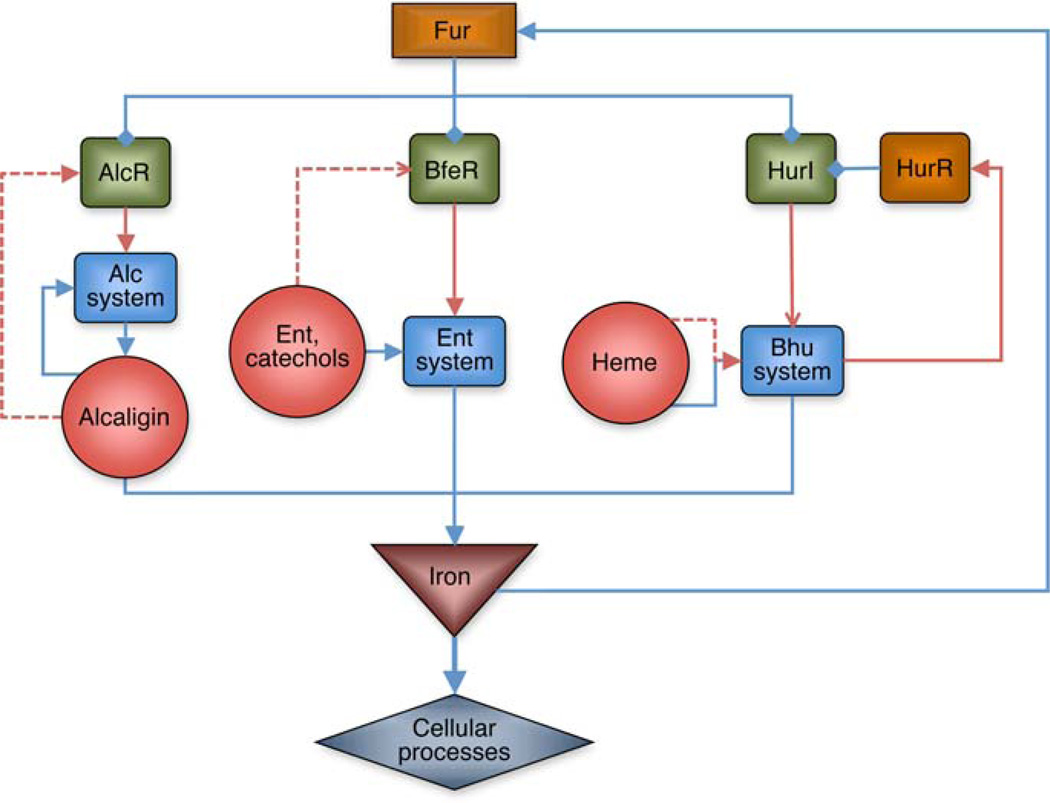
Intercellular chemical signaling enables a population of bacteria to regulate gene expression collectively in response to environmental signals produced by the same or different species, and in response to host signals and environmental cues. Bordetella species possess multiple ultrasensitive iron source-sensing circuits organized in parallel (Fig. 2). In their role as inducers, the alcaligin, heme and enterobactin iron sources function as signaling molecules we have termed ferrimones that activate the cognate iron transport system through the action of the specific substrate-inducible positive regulators, AlcR, BfeR and HurIR. Ferrimone-sensing underlies the bacterium’s ability to discriminate and integrate multiple iron source availability cues to coordinate expression of the iron transport systems. As the iron acquisition systems are activated independently, they are not subject to interference or signal antagonism, and all circuits converge to supply the requisite nutrient iron. When the bacterium’s iron demands are satisfied, iron activates the DNA-binding activity of the Fur repressor and iron transport system expression is globally repressed. Ferrimone-sensing thus acts to support, promote, and exploit early and late events in the infection process to assure growth and survival of the pathogen.
Fig. 2.
Regulatory circuits controlling expression of the alcaligin, enterobactin, and heme utilization systems in Bordetella species. Fur and iron control the expression of three parallel regulatory circuits that respond to the cognate iron source via distinct substrate-inducible positive regulators (AlcR, BfeR, and HurI). The circuits are activated independently, and are not subject to interference or signal antagonism. All three circuits converge to supply the requisite nutrient iron. When the bacterium’s iron demands are satisfied, iron activates the DNA-binding activity of the Fur repressor and iron transport system expression is globally repressed
Acknowledgements
The authors’ research described herein was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
References
- Anderson MT, Armstrong SK. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J Bacteriol. 2004;186:7302–7311. doi: 10.1128/JB.186.21.7302-7311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MT, Armstrong SK. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J Bacteriol. 2006;188:5731–5740. doi: 10.1128/JB.00495-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MT, Armstrong SK. Norepinephrine mediates acquisition of transferrin-iron in Bordetella bronchiseptica. J Bacteriol. 2008;190:3940–3947. doi: 10.1128/JB.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SK, Clements MO. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J Bacteriol. 1993;175:1144–1152. doi: 10.1128/jb.175.4.1144-1152.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Hoenes T. An iron-regulated outermembrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology. 1997;143(Pt 1):135–145. doi: 10.1099/00221287-143-1-135. [DOI] [PubMed] [Google Scholar]
- Beall B, Sanden GN. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology. 1995a;141(Pt 12):3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- Beall BW, Sanden GN. Cloning and initial characterization of the Bordetella pertussis fur gene. Curr Microbiol. 1995b;30:223–226. doi: 10.1007/BF00293637. [DOI] [PubMed] [Google Scholar]
- Beaumont FC, Kang HY, Brickman TJ, Armstrong SK. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Mahren S. Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol Rev. 2005;29:673–684. doi: 10.1016/j.femsre.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J Bacteriol. 1995;177:268–270. doi: 10.1128/jb.177.1.268-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J Bacteriol. 1996;178:54–60. doi: 10.1128/jb.178.1.54-60.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Essential role of the ironregulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J Bacteriol. 1999;181:5958–5966. doi: 10.1128/jb.181.19.5958-5966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Bordetella interspecies allelic variation in AlcR inducer requirements: identification of a critical determinant of AlcR inducer responsiveness and construction of an alcR(Con) mutant allele. J Bacteriol. 2002;184:1530–1539. doi: 10.1128/JB.184.6.1530-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J Bacteriol. 2005;187:3650–3661. doi: 10.1128/JB.187.11.3650-3661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Impact of alcaligin siderophore utilization on in vivo growth of Bordetella pertussis. Infect Immun. 2007;75:5305–5312. doi: 10.1128/IAI.00849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Hansel JG, Miller MJ, Armstrong SK. Purification, spectroscopic analysis and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. Biometals. 1996;9:191–203. doi: 10.1007/BF00144625. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, Kang HY, Armstrong SK. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J Bacteriol. 2001;183:483–489. doi: 10.1128/JB.183.2.483-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Vanderpool CK, Armstrong SK. Bordetella. In: Crosa JH, Mey AR, Payne SM, editors. Iron transport in bacteria. Washington, DC: ASM Press; 2004. pp. 311–328. [Google Scholar]
- Brickman TJ, Vanderpool CK, Armstrong SK. Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect Immun. 2006;74:1741–1744. doi: 10.1128/IAI.74.3.1741-1744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Hanawa T, Anderson MT, Suhadolc RJ, Armstrong SK. Differential expression of Bordetella pertussis iron transport system genes during infection. Mol Microbiol. 2008;70:3–14. doi: 10.1111/j.1365-2958.2008.06333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton CL, et al. The growth response of Escherichia coli to neurotransmitters and related catecholamine drugs requires a functional enterobactin biosynthesis and uptake system. Infect Immun. 2002;70:5913–5923. doi: 10.1128/IAI.70.11.5913-5923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem : a European journal of chemical biology. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- Confer DL, Eaton JW. Phagocyte impotence caused by an invasive bacterial adenylate cyclase. Science. 1982;217:948–950. doi: 10.1126/science.6287574. [DOI] [PubMed] [Google Scholar]
- Fennelly NK, et al. Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses. Infect Immun. 2008;76:1257–1266. doi: 10.1128/IAI.00836-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flak TA, Goldman WE. Signalling and cellular specificity of airway nitric oxide production in pertussis. Cell Microbiol. 1999;1:51–60. doi: 10.1046/j.1462-5822.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- Freestone PP, Haigh RD, Williams PH, Lyte M. Involvement of enterobactin in norepinephrine-mediated iron supply from transferrin to enterohaemorrhagic Escherichia coli. FEMS Microbiol Lett. 2003;222:39–43. doi: 10.1016/S0378-1097(03)00243-X. [DOI] [PubMed] [Google Scholar]
- Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina PC, Foster L-A, Toth SI, Roe BA, Dyer DW. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- Giardina PC, Foster LA, Toth SI, Roe BA, Dyer DW. Analysis of the alcABC operon encoding alcaligin biosynthesis enzymes in Bordetella bronchiseptica. Gene. 1997;194:19–24. doi: 10.1016/s0378-1119(97)00094-2. [DOI] [PubMed] [Google Scholar]
- Harle C, Kim I, Angerer A, Braun V. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 1995;14:1430–1438. doi: 10.1002/j.1460-2075.1995.tb07129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlett E, Wolff J. Soluble adenylate cyclase from the culture medium of Bordetella pertussis: purification and characterization. J Bacteriol. 1976;127:890–898. doi: 10.1128/jb.127.2.890-898.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett SL, Eggling S, Geller L. Novel method to examine the formation of unstable 2:1 and 3:1 complexes of catecholamines and iron(III) J Inorg Biochem. 1997;66:165–173. [Google Scholar]
- Kang HY, Armstrong SK. Transcriptional analysis of the Bordetella alcaligin siderophore biosynthesis operon. J Bacteriol. 1998;180:855–861. doi: 10.1128/jb.180.4.855-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HY, Brickman TJ, Beaumont FC, Armstrong SK. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J Bacteriol. 1996;178:4877–4884. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada T, Ui M. ADP ribosylation of the specific membrane protein of C6 cells by islet-activating protein associated with modification of adenylate cyclase activity. J Biol Chem. 1982;257:7210–7216. [PubMed] [Google Scholar]
- Kim I, Stiefel A, Plantor S, Angerer A, Braun V. Transcription induction of the ferric citrate transport Biometals genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- King ND, Kirby AE, Connell TD. Transcriptional control of the rhuIR-bhuRSTUV heme acquisition locus in Bordetella avium. Infect Immun. 2005;73:1613–1624. doi: 10.1128/IAI.73.3.1613-1624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Lyons ND, Smith KF, Connell TD. Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica. J Bacteriol. 2007;189:6266–6275. doi: 10.1128/JB.00629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby AE, Metzger DJ, Murphy ER, Connell TD. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect Immun. 2001;69:6951–6961. doi: 10.1128/IAI.69.11.6951-6961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak NA, Panina EM, Miller JF. Type III secretion in Bordetella subspecies. In: Locht C, editor. Bordetella molecular microbiology. Norfolk: Horizon Bioscience; 2007. pp. 119–139. [Google Scholar]
- Loomis LD, Raymond KN. Solution equilibria of enterobactin and metal-enterobactin complexes. Inorg Chem. 1991;30:906–911. [Google Scholar]
- Mattoo S, Cherry JD. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev. 2005;18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methner U, Rabsch W, Reissbrodt R, Williams PH. Effect of norepinephrine on colonisation and systemic spread of Salmonella enterica in infected animals: role of catecholate siderophore precursors and degradation products. Int J Med Microbiol. 2008;298:429–439. doi: 10.1016/j.ijmm.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Moore CH, Foster LA, Gerbig DG, Jr, Dyer DW, Gibson BW. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J Bacteriol. 1995;177:1116–1118. doi: 10.1128/jb.177.4.1116-1118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ER, et al. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect Immun. 2002;70:5390–5403. doi: 10.1128/IAI.70.10.5390-5403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson ML, Beall B. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, hemin and hemoglobin as iron sources. Microbiol Read U K. 1999;145:2453–2461. doi: 10.1099/00221287-145-9-2453. [DOI] [PubMed] [Google Scholar]
- Nishio T, Ishida Y. Production of dihydroxamate siderophore alcaligin by Alcaligenes xylosoxidans subsp. xylosoxidans. Agric Biol Chem. 1990;54:1837–1839. [Google Scholar]
- Nishio T, Tanaka N, Hiratake J, Katsube Y, Ishida Y, Oda J. Isolation and structure of the novel dihydroxamate siderophore alcaligin. J Am Chem Soc. 1988;110:8733–8734. [Google Scholar]
- Ochs M, Veitinger S, Kim I, Welz D, Angerer A, Braun V. Regulation of citrate-dependent iron transport of Escherichia coli: fecR is required for transcription activation by FecI. Mol Microbiol. 1995;15:119–132. doi: 10.1111/j.1365-2958.1995.tb02226.x. [DOI] [PubMed] [Google Scholar]
- Ochs M, Angerer A, Enz S, Braun V. Surface signaling in transcriptional regulation of the ferric citrate transport system of Escherichia coli: mutational analysis of the alternative sigma factor FecI supports its essential role in fec transport gene transcription. Mol Gen Genet. 1996;250:455–465. doi: 10.1007/BF02174034. [DOI] [PubMed] [Google Scholar]
- Parkhill J, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Pradel E, Locht C. Expression of the putative siderophore receptor gene bfrZ is controlled by the extracytoplasmic-function sigma factor BupI in Bordetella bronchiseptica. J Bacteriol. 2001;183:2910–2917. doi: 10.1128/JB.183.9.2910-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Guiso N, Locht C. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J Bacteriol. 1998;180:871–880. doi: 10.1128/jb.180.4.871-880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Guiso N, Menozzi FD, Locht C. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect Immun. 2000;68:1919–1927. doi: 10.1128/iai.68.4.1919-1927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond KN, Isied SS, Brown LD, Fronczek FR, Nibert JH. Coordination isomers of biological iron transport compounds. VI. Models of the enterobactin coordination site. A crystal field effect in the structure of potassium tris(catecholato)chromate(III) and -ferrate(III) sesquihydrates, K3[M(O2C6H4)3] × 1.5H2O, M = chromium, iron. J Am Chem Soc. 1976;98:1767–1774. doi: 10.1021/ja00423a022. [DOI] [PubMed] [Google Scholar]
- Uhl MA, Miller JF. Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc Natl Acad Sci USA. 1994;91:1163–1167. doi: 10.1073/pnas.91.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. The Bordetella bhu locus is required for heme iron utilization. J Bacteriol. 2001;183:4278–4287. doi: 10.1128/JB.183.14.4278-4287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. Heme-responsive transcriptional activation of Bordetella bhu genes. J Bacteriol. 2003;185:909–917. doi: 10.1128/JB.185.3.909-917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. Integration of environmental signals controls expression of Bordetella heme utilization genes. J Bacteriol. 2004;186:938–948. doi: 10.1128/JB.186.4.938-948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss AA, Hewlett EL, Myers GA, Falkow S. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect Immun. 1983;42:33–41. doi: 10.1128/iai.42.1.33-41.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff EE, Lopreato GF, Tipton KA, Payne SM. Shigella dysenteriae ShuS promotes utilization of heme as an iron source and protects against heme toxicity. J Bacteriol. 2005;187:5658–5664. doi: 10.1128/JB.187.16.5658-5664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Valdebenito M, Winkelmann G, Hantke K. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology. 2005;151:2363–2372. doi: 10.1099/mic.0.27888-0. [DOI] [PubMed] [Google Scholar]