Abstract
Kinetochores are the elaborate protein assemblies that attach chromosomes to spindle microtubules in mitosis and meiosis. The kinetochores of point-centromere yeast appear to represent an elementary module, which repeats a number of times in kinetochores assembled on regional centromeres. Structural analyses of the discrete protein subcomplexes that make up the budding-yeast kinetochore have begun to reveal principles of kinetochore architecture and to uncover molecular mechanisms underlying functions such as transmission of tension and establishment and maintenance of bipolar attachment. The centromeric DNA is probably wrapped into a compact organization, not only by a conserved, centromeric nucleosome, but also by interactions among various other DNA-bound kinetochore components. The rod-like, heterotetrameric Ndc80 complex, roughly 600 Å long, appears to extend from the DNA-proximal assembly to the plus end of a microtubule, to which one end of the complex is known to bind. Ongoing structural studies will clarify the roles of a number of other well-defined complexes.
Kinetochores are DNA–protein complexes that assemble on chromosomes, generally at a single location, and direct equal partitioning of replicated DNA into daughter cells in mitosis and meiosis. They do so by performing three key functions: (1) connecting chromosomes to microtubules (MTs) of the spindle; (2) transmitting forces that move paired sister chromosomes back and forth along the spindle, establishing and maintaining bipolar MT attachments; and (3) originating the spindle checkpoint signals that prevent cells from progressing into anaphase until all chromatid pairs have achieved bipolarity (Cheeseman and Desai 2008; Santaguida and Musacchio 2009).
Centromeres—the chromosomal assembly sites for kinetochores—vary in length from the “point centromeres” of budding yeast (~125 bp in Saccharomyces cerevisiae) to the large, “regional centromeres” of mammalian cells (several million base pairs). The former attach to a single MT; the latter, to as many as several dozen. Conservation of sequences in many of the kinetochore proteins, and conservation of structures in the several subcomplexes studied to date nonetheless imply that fundamental architectural features of all kinetochores are similar (Meraldi et al. 2006; Fukagawa and De Wulf 2009). Moreover, the copy number of conserved proteins does not scale with the length of centromeric DNA but more closely with the number of MT attachments (Joglekar et al. 2008). That is, a large centromere appears to comprise a set of MT attachment structures (kinetochore modules), each of which is likely to resemble a kinetochore built on a point centromere. These modules appear to cluster into a single large complex on the surface of the extensively coiled, centromeric heterochromatin (McEwen and Dong 2010).
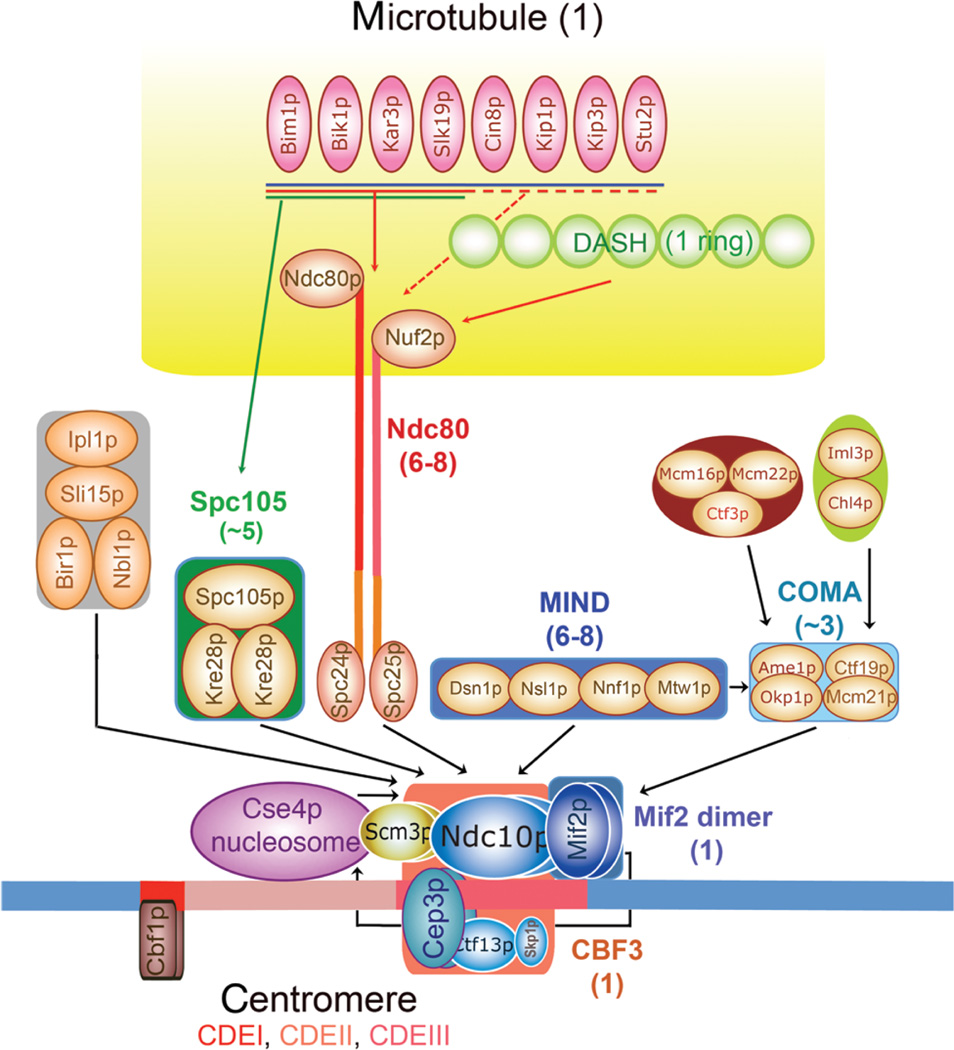
Even the kinetochores of point-centromere yeasts turn out to have a remarkably large complement of components (Fukagawa and De Wulf 2009). Roughly 70 distinct proteins can be designated as “kinetochore-associated.” Most of these proteins are present in multiple copies, and many are probably added or removed at different stages in kinetochore assembly. Developing a structural description is somewhat simplified by a clear organizational hierarchy (Fig. 1) (De Wulf et al. 2003; Fukagawa and De Wulf 2009). Many of the critical proteins are components of stable subassemblies, even in the absence of DNA or other kinetochore proteins. Roughly a dozen of these subassemblies have been identified, each having between two and 10 unique subunits. Structural analyses of the molecular architecture of kinetochores can thus focus initially on a description of these subcomplexes (each a relatively elaborate molecular entity) and move up the hierarchy stepwise toward their longer-range organization. We outline here the status of this project at the time of writing the present paper (July 2010).
Figure 1.
Hierarchical molecular organization of the S. cerevisiae kinetochore. Between the proteins that assemble directly on centromeric DNA (bottom, conserved subsites CDEI [red], CDEII [pink], and CDEIII [rose]) and those that interact with the spindle MT (top, in yellow) is a “layer” of intermediate linker complexes. Defined protein complexes (CBF3, MIND, etc.) are in colored boxes or ovals. The heterotetrameric Ndc80 complex is illustrated with an explicit representation of its extended structure, to emphasize that it is the one linker known to bind specifically to the MT. The numerals in parentheses after the name of each complex are the current best estimates of the number of the respective complexes in one kinetochore (Joglekar et al. 2006). The arrows indicate recruitment dependencies, as determined by genetic and imaging methods. Thus, the Bir1, Spc105, MIND, and Ndc80 complexes all require CBF3 for kinetochore localization, as does the Cse4p nucleosome. (The figure is adapted from Pagliuca et al. 2009 and reprinted with express permission from the Public Library of Science © 2009.)
THE CENTROMERIC PLATFORM
The budding yeast point centromere has three conserved centromere-determining elements, designated CDEI, II, and III (Fig. 2A), each of which binds different protein complexes (Fitzgerald-Hayes et al. 1982). CDEI binds a “borrowed” bHLHZ (basic-region/helix–loop–helix/leucine-zipper) transcription factor, Cbf1, which recognizes an 8-bp sequence similar to the consensus for this class of DNA-binding proteins (Mellor et al. 1991). CDEII incorporates the “specialized nucleosome” found in all kinetochores, in which a centromere-specific, H3-like histone (Cse4p in budding yeast; CENP-A or CenH3 in higher eukaryotes) replaces H3 (Meluh et al. 1998). The amount and identity of the remaining histones in the specialized nucleosome core remain, at this writing, matters of debate. CDEII is generally AT-rich, but its nucleotide sequence is less important for function than its length: CDEII is ~80 bp long in nearly all point-centromere yeast for which genome sequences are known, and just twice that length in two of them (Kluyveromyces lactis and Aphis gossypii) (Meraldi et al. 2006). Lengths shorter than ~75 bp are incompatible with centromere function. Thus, the specialized nucleosome wraps only 80 bp rather than the ~150 bp coiled on a canonical nucleosome, suggesting that CDEII may coil only once around the specialized nucleosome rather than twice. Finally, CDEIII binds a four-component complex known as CBF3, which contains a dimer of Cep3p, a dimer of Ndc10p, and a Skp1p:Ctf13p heterodimer (Lechner and Carbon 1991; Doheny et al. 1993; Russell et al. 1999). DNA contacts of the CBF3 components have been mapped by cross-linking and DNA modification methods (Fig. 2A) (Espelin et al. 1997). Results from high-resolution chromatin immunoprecipitation (ChIP) from yeast cells are consistent with the locations determined by chemical modification of in vitro–reconstituted complexes (Cohen et al. 2008). They also show that Mif2p (the yeast ortholog of higher-eukaryotic CENP-C) binds over CBF3, probably to the 3′ side of the Ndc10p site.
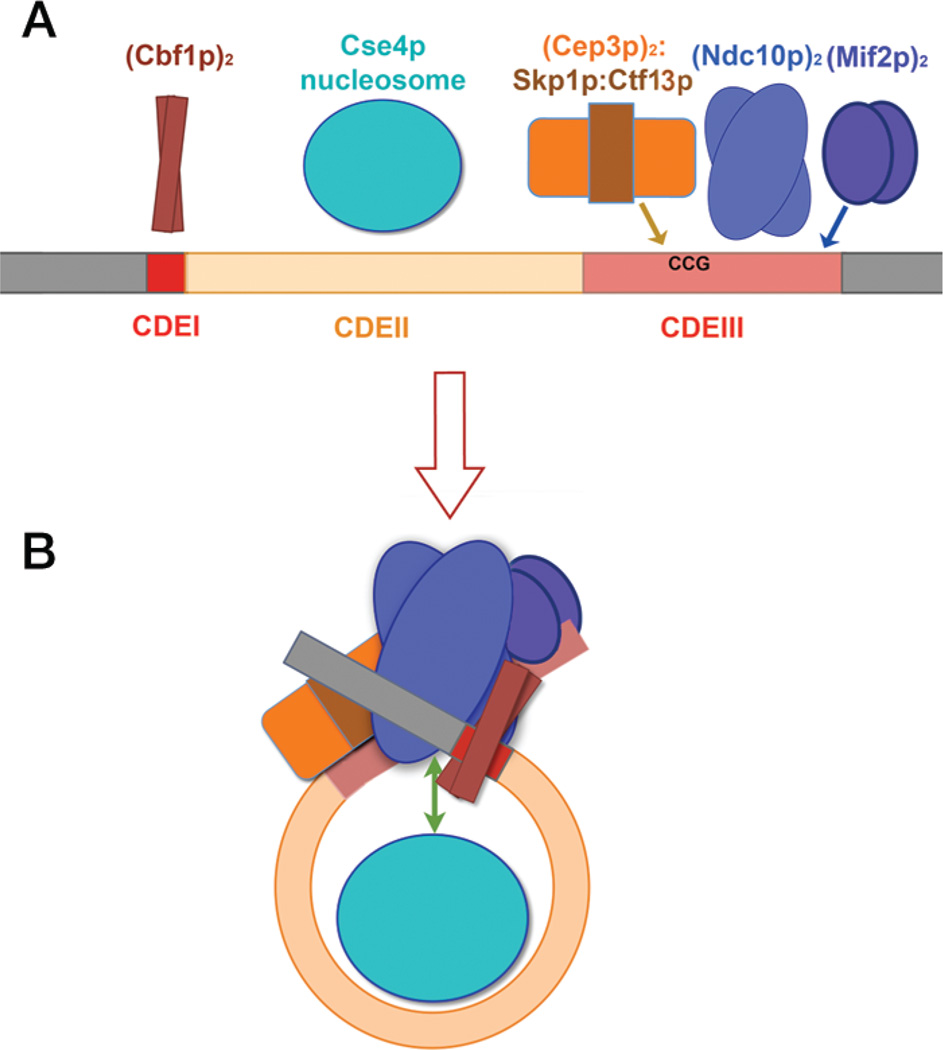
Figure 2.
Diagram of the budding yeast point centromere and its potential compaction by centromere-binding factors. (A) Conserved elements CDEI (red), CDEII (beige), and CDEIII (rose) and proteins known to bind them. The approximate lengths of these elements in S. cerevisiae are 8, 80, and 40 bp, respectively; CDEII in K. lactis is ~160 bp in length. There may be contacts with flanking, pericentromeric DNA (gray). (B) Proposed condensation of the centromere into a compact structure, established by interactions among the various binding components. Scm3p interacts with Ndc10p and the Cse4p nucleosome core (double-headed green arrow).
The Cep3p homodimer appears to be the principal contributor to sequence-specific positioning, as neither Ndc10p nor Mif2p exhibits strong sequence preferences beyond a requirement (in the absence of other factors) for relatively AT-rich regions (Espelin et al. 1997, 2003). Cep3p has an amino-terminal “Zn cluster,” similar to the DNA-binding module of Gal4p, connected to a large, block-like dimerization domain (Bellizzi et al. 2007). The latter resembles a set of five helical zigzags (e.g., HEAT repeats) wound together. One Zn cluster domain evidently recognizes a conserved CCG element at about position 20 in CDEIII; there is no corresponding element on the opposite side of the approximate twofold symmetry axis defined by the pattern of protection and cross-linking, although there is a conserved G:C base pair (Espelin et al. 1997). The consequences of this asymmetry for (Cep3p)2 DNA binding are not yet clear.
The Skp1p component of the Skp1p:Ctf13p heterodimer is not an exclusively kinetochore-dedicated protein; it is the F-box-binding subunit that is a component of the SCF3 ubiquitin ligase complex (Russell et al. 1999). Ctf13p has an F-box sequence at its amino-terminus, and examination of the sequence shows that a set of leucine-rich repeats (LRRs) makes up the bulk of the molecule. It may therefore be related to Skp2, an SCF3 ubiquitin ligase component, which has a similar organization and for which a yeast homolog has not been designated (Schulman et al. 2000). In addition to the LRR domain, there is a carboxy-terminal extension, which in the Skp1–Skp2 crystal structure folds across the LRR concavity and contacts proteins directed for ubiquitylation and degradation. Skp1p:Ctf13p associates tightly with (Cep3p)2 into a stable and self-contained complex we have called the “CBF3 core.” Unlike its role as part of ubiquitin ligases, Skp1p activates the CBF3 core rather than directing its degradation (Kaplan et al. 1997).
Ndc10p from S. cerevisiae is a dimeric, 100-kDa protein (Russell et al. 1999). It has a relatively high affinity for DNA, but binding is promiscuous and nonspecific. For example, in the absence of other factors, it can bind CDEII as well as CDEIII (Espelin et al. 2003). When coupled with the CBF3 core, however, it has CDEIII as its defined location, as detected initially by DNase I footprinting (Espelin et al. 1997). Limited proteolytic cleavage of Ndc10p yields three distinct fragments: a large, amino-terminal fragment bearing the DNA-binding activity, a central fragment that appears to mediate dimer formation, and a carboxy-terminal fragment of unknown function (K Simons, unpubl.). High-resolution ChIP analysis shows a distribution with a nearly bimodal character—a peak corresponding roughly to what one would expect from the DNase I protection pattern and a potential upstream contact (Cohen et al. 2008). Because the specialized nucleosome wraps the CDEII DNA into a coil, it is reasonable to suppose that CDEIII and CDEI will be spatially contiguous, consistent with genetic data showing interactions between CDEI and CDEIII and between the factors that bind them (Meluh and Koshland 1995). An additional Ndc10p contact—for example, just upstream of Cbf1p—could be part of a generally compact structure (Fig. 2B).
CBF3 is required for deposition of the Cse4p nucleosome. There is good evidence that the assembly factor, Scm3p, is the bridge between Ndc10p and Cse4p (Fig. 2C) (Camahort et al. 2007; Mizuguchi et al. 2007). Because Scm3p is a relatively small protein, its role as a connector between CBF3 and the specialized nucleosome suggests that the centromere and its associated proteins form a tightly organized assembly. Mif2p also participates: It binds the centromere and recruits “middle-layer” components, including COMA (Fig. 1). We propose that this compact assembly is the platform on which the rest of the kinetochore is constructed.
MT ATTACHMENT: REGULATED AND PROCESSIVE
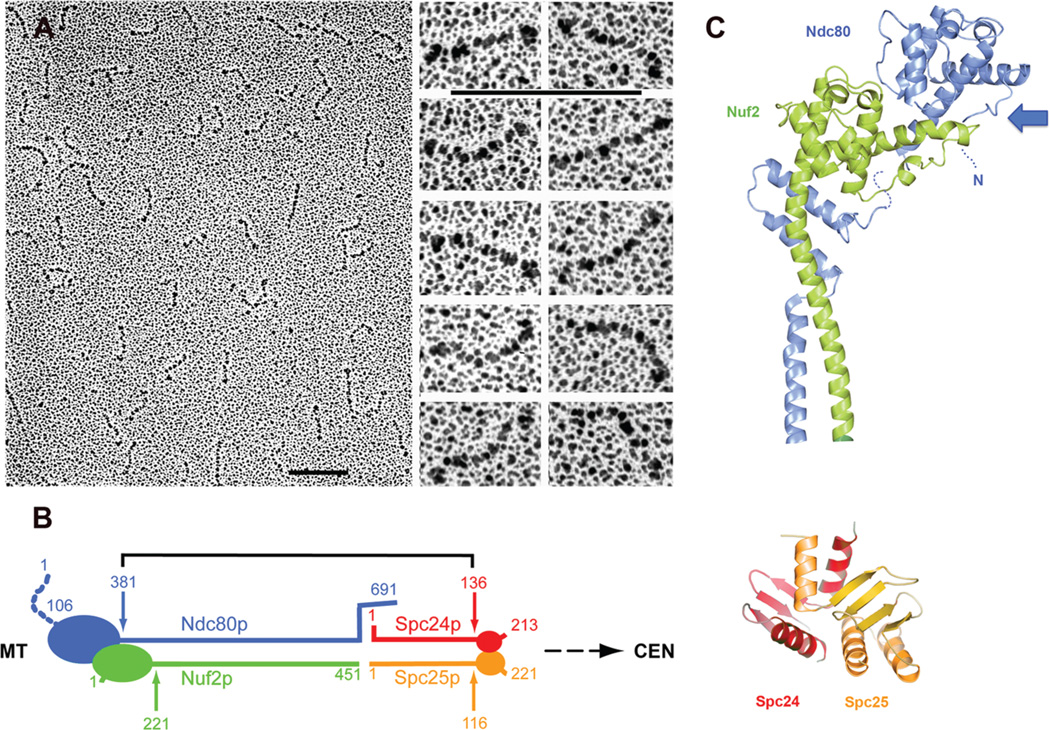
The central element in the connection between MTs and the centromeric platform is the Ndc80 complex, the most conserved of the kinetochore subassemblies, essential for MT attachment in all organisms examined. It is a 550 Å–long heterotetrameric rod, with an α-helical, coiled-coil shaft and globular ends (Fig. 3) (Wei et al. 2005). Two heterodimers, Ndc80p:Nuf2p and Spc24p:Spc25p, join end-to-end to form the full rod. The connection preserves amino-to-carboxyl polarity. That is, the globular domains of Ndc80p and Nuf2p are at the amino termini of their respective chains; the globular domains of Spc24p and Spc25p, at the carboxyl termini (Fig. 3A,B). The globular “heads” of both Ndc80p and Nuf2p are calponin homology (CH) domains, a roughly 100-residue module found in proteins that bind MTs or actin (Fig. 3C) (Wei et al. 2007; Ciferri et al. 2008). The two CH domains are staggered along the axis of the complex, suggesting that they might form a composite MT-binding interface (Fig. 3C). A short, two-headed heterodimer composed of the amino-terminal parts of Ndc80p and Nuf2p binds MTs (Wei et al. 2007), as does the complete Ndc80 complex from Caenorhabditis elegans (Cheeseman et al. 2006). In Ndc80, ~100 residues precede the CH domain, and reasonably tight binding to MTs requires this region, as well as the CH and the residues following it that associate with the Nuf2p head (Wei et al. 2007; Guimaraes et al. 2008).
Figure 3.
The Ndc80 complex. (A) Electron microscopy of rotary-shadowed, recombinant Ndc80 complexes. (Left) Field of molecules; (right) higher magnification of individual complexes (Wei et al. 2005). Bar, 1000 Å. (B) Diagram of molecular organization of the complex (Wei et al. 2005). (C) Ribbon representations of the Ndc80p/Nuf2p and Spc24p/Spc25p globular ends of the complex. The former is from the structure of the human homologs (Ciferri et al. 2008). The latter is from Wei et al. (2007). Colors as in B. The location of the amino-terminal extension of Ndc80p (the human homolog is Hec1), an unstructured region with Ipl1p/Aurora B phosphorylation sites, is indicated by a dotted line and labeled “N.” (Arrow) Likely surface for MT binding. Note that this surface is probably a composite of Ndc80p and Nuf2p and that the amino-terminal extension of Ndc80p projects from it. (A and B, Reprinted from Wei et al. 2005 with permission from the National Academy of Sciences © 2005; C [top], modified from Ciferri et al. 2008 and reprinted with permission from Cell Press © 2008; C [bottom], reprinted from Wei et al. [2007] with permission from Nature Publishing Group © 2007.)
The amino-terminal arm of Ndc80p, which precedes the CH domain in the polypeptide chain, is present in Ndc80p orthologs, but it varies in length and in amino-acid sequence. One conserved feature appears to be the presence of phosphorylation sites for the Ipl1/Aurora B kinase (DeLuca et al. 2006). Aurora B is thought to cause detachment of chromosomes from MTs (Biggins et al. 1999; Pinsky et al. 2006). In mammalian cells, altering the Aurora B sites in the Ndc80 amino-terminal arm to prevent their phosphorylation appears to cause enhanced association (DeLuca et al. 2006). This observation is consistent with our inference from the CH-domain homology, that Ndc80p attachment at MTs is a conserved, critical part of the kinetochore–MT interaction. It further indicates that changing the equilibrium between MT attachment and detachment of Ndc80p is an important mode of regulation and also a step in activating the spindle assembly checkpoint response.
A kinetochore must remain attached to its MT through multiple rounds of local polymerization and depolymerization—that is, it must follow local excursions of the MT plus end. There are six to eight Ndc80 complexes per kinetochore in budding yeast, and probably at least that number per MT in higher eukaryotes (Joglekar et al. 2006, 2008). The observed Kd for the (unphosphorylated) Ndc80p:Nuf2p is ~5 µm (Wei et al. 2007). If attachment of the complex to centromere-proximal components at the Spc24p:Spc25p end is firm, then the effective concentration of the Ndc80p:Nuf2p head in the vicinity of an MT end is between 10 and 100 mm (depending on assumptions regarding the volume within which the unattached head can swing), and each head will spend most of its time attached. The probability that all six to eight complexes will dissociate from the MT at once, causing the chromosome to detach, is thus extremely low. The probability of dissociation of a given Ndc80 complex is nonetheless appreciable, and the protofilament(s) to which it binds can polymerize or depolymerize during the dissociation period. Rebinding of the Ndc80 complex can then occur at a new, longitudinally displaced, position along the MT. Such an asynchronous dissociation/polymerization or depolymerization/reassociation will allow the kinetochore to track the MT end, provided that the Ndc80p:Nuf2p part of the complex has some flexibility—either an internal hinge or a flexible attachment at one end. Electron microscopy has provided some evidence for an internal hinge, but its significance remains to be determined (Wang et al. 2008). MT polymerases and rescue factors, such as Stu1p and Stu2p (Brouhard et al. 2008; Al-Bassam et al. 2010), which are present at kinetochore-attached MT plus ends, could in principle coordinate Ndc80-complex fluctuations and tubulin polymerization.
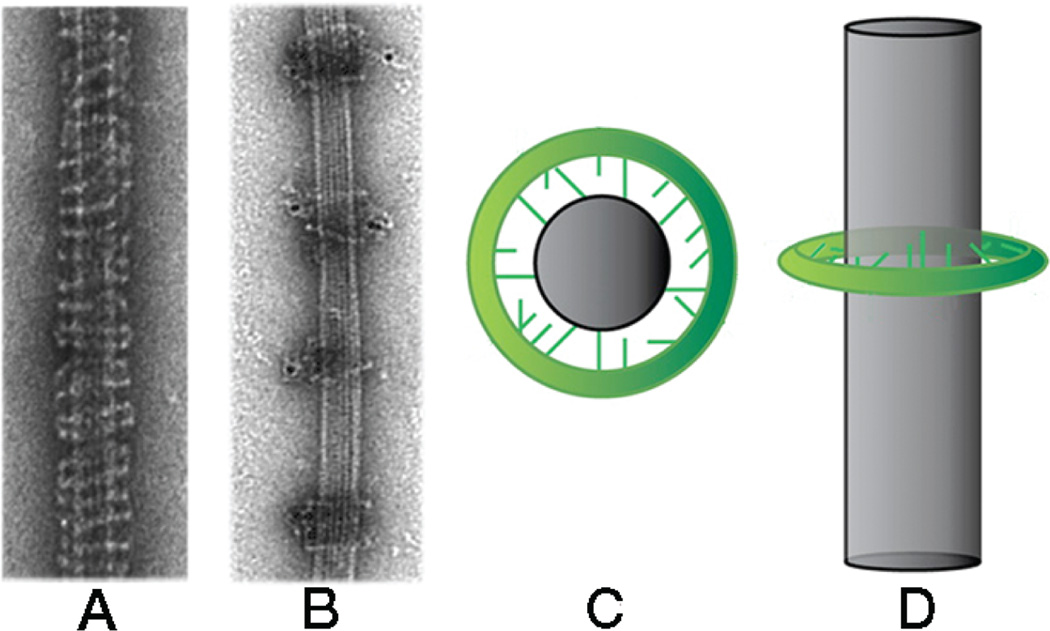
When multiple MTs terminate at a single kinetochore, the mechanism just described may be adequate to ensure nearly error-free processivity. When a kinetochore captures only one MT, as in point-centromere yeasts, additional processivity factors may be necessary. A candidate for such a role is the DASH/Dam1 complex, a heterodecamer that polymerizes into an MT-encircling ring (Fig. 4) (Miranda et al. 2005; Westermann et al. 2005). The rings, which contain ~20–25 heterodecamers (estimates by different methods vary slightly), are ~500 Å in diameter. They do not form in free solution; contacts with the MT lattice are probably required for their integrity. These contacts are through flexibly extended “arms” from one or more of the subunits; the arms probably interact electrostatically with the negatively charged MT surface (Miranda et al. 2007; Wang et al. 2007). Pairwise clustering of the rings by antibodies (against an affinity tag) showed originally that the rings could slide along an MT (Miranda et al. 2005); other groups have observed translocation directly by single-molecule methods (Westermann et al. 2006). Two recent reports provide evidence that DASH/Dam1 is an Ndc80-complex processivity factor in vitro and that phosphorylation by Ipl1p of one or more components of the DASH complex eliminates this functional interaction (Lampert et al. 2010; Tien et al. 2010).
Figure 4.
DASH/Dam1 complex: formation of rings on microtubules (MTs). (A) Image of negatively stained MT “decorated” with DASH at high concentration. (B) Image of negatively stained MT decorated with DASH at lower concentration, followed by addition of gold-tagged antibody (against the His tag on the DASH components). The antibody clusters the rings in pairs; with no antibody added, the rings are randomly spaced (not shown). (C,D) Diagrams showing flexible connections between a DASH ring and an MT. The connections can adapt to different registers of the MT protofilaments. (A and B, Reprinted from Miranda et al. 2005 with permission from Nature Publishing Group © 2005; C and D, reprinted from Miranda et al. 2007 with permission from American Society for Cell Biology © 2007.)
INTERMEDIATE CONNECTIONS
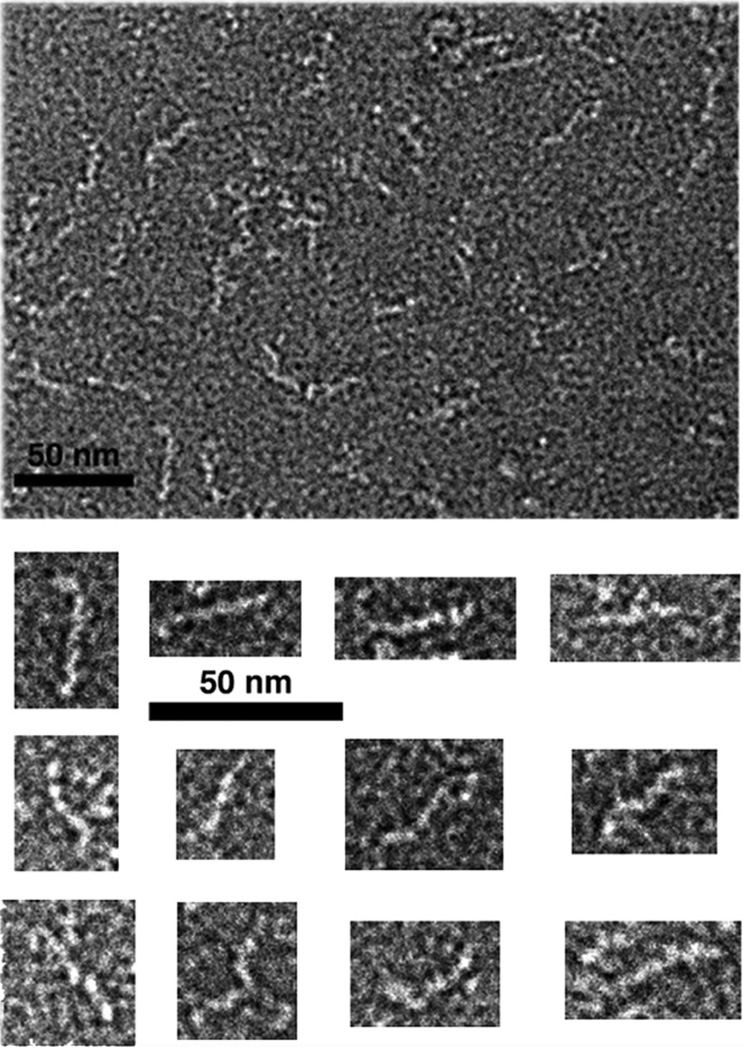
At the centromere-proximal end of the Ndc80 complex, the Spc24p:Spc25p globular domain probably attaches to the MIND complex (a heterotetramer of Mtw1, Nsl1, Nnf1, and Dsn1—known as the Mis12 complex in Schizosaccharomyces pombe and other eukaryotes) (De Wulf et al. 2003; Obuse et al. 2004). The mammalian forms of the two heterotetramers associate with each other and with a third protein (KNL-1; Spc105 in S. cerevisiae) to form a “KMN (KNL-1/Mis12/Ndc80) supercomplex” (Cheeseman and Desai 2008). The MIND complex is an ~250 Å–long rod, with a relatively uniform diameter at the resolution of negative-stain electron microscopy (Fig. 5). Its mass per unit length is substantially greater than that of a coiled-coil; the predicted α-helix content would be consistent, for example, with an array of helical zigzags. One adaptor-like function of the MIND complex may be to mediate the transition between a single-copy centromeric platform and the six to eight Ndc80 complexes that bind the MT.
Figure 5.
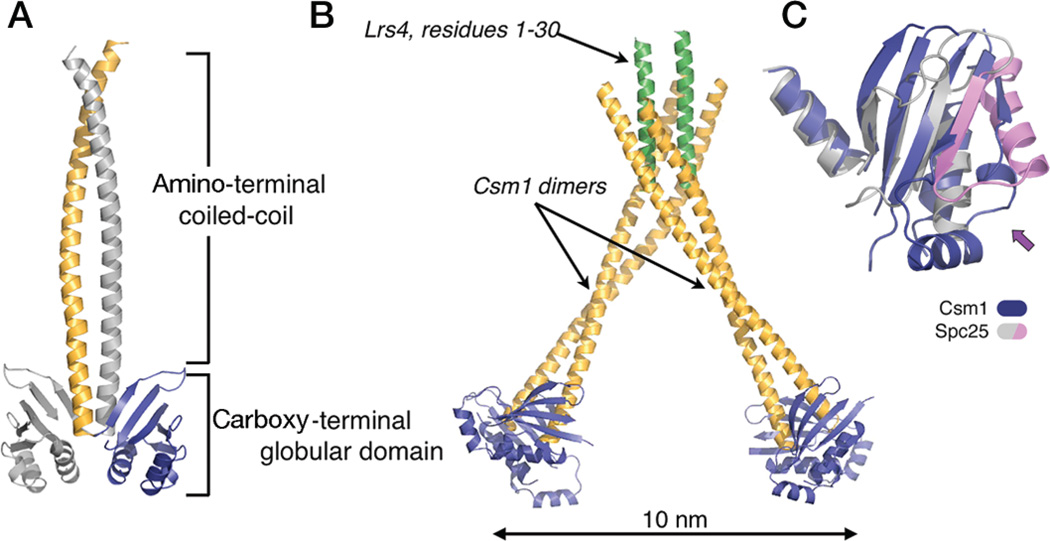
Structure of the Csm1p/Lrs4p subcomplex of yeast monopolin. (A) Molecular organization of Csm1p. (B) Complex of Csm1p with Lrs4p. The amino-terminal 30 residues of Lrs4p form a dimerizing, α-helical segment; the dimeric segment, in turn, binds two Csm1p dimers, creating a V-shaped heterohexamer. The remaining residues of Lrs4p are disordered. (C) Superposition of the globular domains of Csm1p (blue) and Spc25p (magenta/gray). The arrow points to a lateral surface with somewhat divergent structures on the two proteins, as well as in Spc25p with respect to Spc24p (see Fig. 2C); this surface contains the hydrophobic patch, conserved among Csm1p orthologs, that binds Dsn1p (a component of the MIND complex) and Mif2p. (Figure from Corbett et al. 2010 and reprinted with permission from Cell Press © 2010.)
In meiosis I, an additional protein complex termed “monopolin” cross-links and “co-orients” sister kinetochores during the reductional division of meiosis I (Toth et al. 2000; Rabitsch et al. 2003; Monje-Casas et al. 2007). The conserved core of this complex, composed of Csm1p and Lrs4p, forms an elongated V-shaped complex with two dimers of Csm1p linked by two copies of Lrs4p (Corbett et al. 2010). The tips of this V, spaced ~100 Å apart, are dimers of the Csm1 carboxy-terminal domain, which closely resemble the paired carboxy-terminal domains of Spc24p and Spc25p (Fig. 6). Csm1p has a conserved, relatively hydrophobic patch on its globular domain, which we have found binds two kinetochore proteins: Dsn1p of the MIND complex, and Mif2p. Mutations in this patch disrupt binding to both of these kinetochore proteins in vitro and cause severe chromosome mis-segregation in vivo. Each (Csm1p)4(Lrs4p)2 complex has four such conserved surface patches, two closely spaced and two substantially more distant. Thus, the complex could cross-link MIND and Mif2p on a single kinetochore or, as seems more likely given its function in directing sister kinetochores to a single pole, one or both partners on one sister kinetochore to one or both partners on the other.
Figure 6.
Electron microscopy of negatively stained, recombinant MIND complex (heterotetramer) from S. cerevisiae. Selected images from the field at the top are shown in the lower panels. Bars, 1000 Å. (Figure from Corbett et al. 2010 and reprinted with permission from Cell Press ©2010.)
CONCLUSIONS
The relatively modest number of kinetochore-related molecular structures now in hand suggests one or two basic architectural principles. A point-centromere kinetochore (and probably each modular element of a regional-centromere kinetochore) connects a centromeric platform to a single MT. In budding yeasts, the kinetochore is an asymmetric assembly of 1–2 MDa, which, if it were a compact structure, would probably have outer dimensions of ~150–200 Å. A single MT is a 250 Å–diameter cylinder, probably made much larger by “fraying” of disassembling MT protofilaments and by the presence of plus-end directed MT polymerases such as Stu2p. In addition, the bridge that kinetochores form between the centromere and the MT must also be able to track growth and shrinkage of the MT tip. A relatively open, adaptable connecting structure composed of rod-like elements, joined by hinged links, appears to be a logical way to satisfy both the geometrical and functional constraints. The Ndc80 complex is clearly one such rod-like connector; the MIND complex and the rod-like Spc105 complex (Pagliuca et al. 2009) are probably others. Finally, the subtle regulation of the Ndc80:MT interface by phosphorylation suggests that control of kinetochore function can be achieved by modulating the affinity of interfaces within and between kinetochore subcomplexes.
ACKNOWLEDGMENTS
This summary is a report on an extended, ongoing collaboration, and we thank other members of the Harrison and Sorger laboratories for critical input and discussion. We acknowledge the following sources of support: NIH Grants R01-GM51464 (to P.K.S.) and K99-GM84292 (to J.A.-B.), the Leukemia and Lymphoma Society (Postdoctoral fellowships to U.-S.C. and J.J.B.), the Helen Hay Whitney Foundation (Postdoctoral fellowship to K.D.C.), the Charles King Trust (Fellowship to K.S.), the Italian Association for Cancer Research (AIRC: grant to P.D.W.), and the National Science Foundation (Predoctoral fellowship to J.M.). S.C.H. is an Investigator in the Howard Hughes Medical Institute.
REFERENCES
- Al-Bassam J, Kim H, Brouhard GJ, van Oijen A, Harrison SC, Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi JJ, III, Sorger PK, Harrison SC. Crystal structure of the yeast inner kinetochore subunit Cep3p. Structure. 2007;15:1422–1430. doi: 10.1016/j.str.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol Biol Cell. 2008;19:4480–4491. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee L-S, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA–protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, De Wulf P. Kinetochore composition, formation and organization. In: De Wulf P, Earnshaw WC, editors. The kinetochore: From molecular discoveries to cancer therapy. New York: Springer; 2009. pp. 133–191. [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore–microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore–microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- McEwen BF, Dong Y. Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell Mol Life Sci. 2010;67:2163–2172. doi: 10.1007/s00018-010-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J, Rathjen J, Jiang W, Barnes CA, Dowell SJ. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 1991;19:2961–2969. doi: 10.1093/nar/19.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Miranda JJ, King DS, Harrison SC. Protein arms in the kinetochore–microtubule interface of the yeast DASH complex. Mol Biol Cell. 2007;18:2503–2510. doi: 10.1091/mbc.E07-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol. 2004;6:1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P. Roles for the conserved spc105p/kre28p complex in kinetochore–microtubule binding and the spindle assembly checkpoint. PLoS One. 2009;4:e7640. doi: 10.1371/journal.pone.0007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Russell ID, Grancell AS, Sorger PK. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: A kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Wang HW, Ramey VH, Westermann S, Leschziner AE, Welburn JP, Nakajima Y, Drubin DG, Barnes G, Nogales E. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore–microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niderstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore–microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]