Abstract
The interferon-stimulated gene viperin has been shown to have antiviral activity against hepatitis C virus (HCV) in the context of the HCV replicon, although the molecular mechanisms responsible are not well understood. Here we demonstrate that viperin plays an integral part in the ability of interferon to limit replication of cell culture derived HCV (JFH-1) that accurately reflects the complete viral life cycle. Using confocal microscopy and Fluorescence Resonance Energy Transfer (FRET) analysis we demonstrate that viperin localizes and interacts with HCV NS5A at the lipid droplet interface. In addition viperin also associates with NS5A and the pro-viral cellular factor, VAP-A at the HCV replication complex. The ability of viperin to limit HCV replication was dependent on residues within the C-terminus as well as an N-terminal amphipathic helix. Removal of the amphipathic helix redirected viperin from the cytosolic face of the ER and the lipid droplet to a homogenous cytoplasmic distribution, coinciding with a loss of antiviral effect. C-terminal viperin mutants still localized to the lipid droplet interface and replication complexes but did not interact with NS5A proteins as determined by FRET analysis. In conclusion we propose that viperin interacts with NS5A and the host factor VAP-A to limit HCV replication at the replication complex. This highlights the complexity of host control of viral replication by interferon stimulated gene expression.
Keywords: Hepatitis C Virus, Viperin, Replication Complex, Antiviral, VAP-A
Introduction
Hepatitis C virus (HCV) is a leading cause of chronic hepatitis and liver-related morbidity worldwide. A significant proportion of infected individuals fail to develop an effective host antiviral response and develop a chronic infection, often resulting in a progressive liver disease including cirrhosis and hepatocellular carcinoma (1). The current standard-of-care therapy for chronic hepatitis C (CHC) is a combination of pegylated interferon alpha (IFN-α) and ribavirin that results in sustained viral clearance in at best 50% of patients.
Viral infection of mammalian cells results in activation of a number of viral recognition pathways triggered by replication intermediates and/or viral proteins, that ultimately induce innate defences to limit viral replication (2-4). Pivitol to this antiviral response is the induction of IFN. The type I IFN's (IFN-α and β) are essential for immune defences against viruses, and following binding to the type I IFN receptor induce the expression of hundreds of interferon-stimulated genes (ISGs), many of which act to limit viral replication. While a number of these ISGs have well characterised antiviral activity (i.e., MxA, PKR and 2,5-OAS), the complete spectrum of antiviral ISGs and their mechanisms of action remain to be elucidated (3).
The ISGs responsible for controlling HCV replication in response to IFN (either endogenously induced or therapeutically given) remain ill defined, although a picture of the ISG's capable of controlling HCV replication is emerging. The ISG 2,5-OAS has been shown to inhibit HCV replication through the RNAse L pathway (5), while IFN-α mediated suppression of HCV replication in vitro is independent of MxA (6). A number of less well characterised ISGs have also been demonstrated to inhibit HCV replication; studies have demonstrated that ISG6-16 can enhance the anti-HCV activity of IFN-α (7), while ISG56 has direct anti-HCV activity through its ability to suppress HCV IRES translation (8). More recently, PKR and the 3′-to-5′ exonuclease ISG20 have been demonstrated to inhibit HCV replication (9, 10). Clearly anti-HCV ISG effectors remain to be discovered and characterised.
Viperin is an evolutionarily conserved type I ISG, previously demonstrated by our laboratory and others to have antiviral properties against HCV in vitro (9, 11), and a number of other viruses including human cytomegalovirus (HCMV), influenza, alphaviruses, HIV and dengue (reviewed in 12). However, the mechanism by which viperin exerts its anti-HCV effect is unknown. Viperin localizes to both the ER and lipid droplets (LD) and considering the LD is central to the HCV life cycle it has been hypothesised that viperin inhibits HCV replication at this location (12, 13). In this study, we show that viperin suppresses replication of cell culture derived infectious HCV, and demonstrate for the first time that viperin interacts with the NS5A protein at the LD interface and within the replication complex (RC). Furthermore we also show that viperin co-localizes with the known proviral cellular factor, VAP-A, within the HCV RC, strongly suggesting viperin exerts its effect at the level of HCV RNA replication.
Experimental Procedures
Cell Lines
The human hepatoma cell lines Huh-7, Huh-7.5 (Charles Rice, Rockefeller University, NY, USA), NNeoC5B and NNeo3-5B (14) were maintained as previously described (15). Huh-7 cells stably expressing viperin shRNA were generated using a 5 clone shRNA set in pLKO.1 purchased from Open Biosystems (Thermo Scientific, AL, USA). These constructs, including a shRNA control were co-transfected with the packaging vectors psPAX2 and pMD2.G into 293T cells to generate VSV-G pseudotyped lentiviral particles. Supernatants containing virus were pooled 48 and 72 hrs after transfection, 0.45μm filtered and placed on Huh-7 cells at a ratio of 1:5 with standard culture media and 8μg/ml polybrene. Polyclonal cell populations were selected with 3μg/ml puromycin. Knockdown of viperin expression was confirmed by treatment of selected polyclonal cell lines with 10 and 50 U/ml of IFN-α, and real-time PCR utilized to assess the upregulation of viperin compared to the control shRNA cell line.
Viruses and antibodies
Infectious genotype 2a JFH-1 HCV was prepared as previously described (16, 17). The HCV monoclonal NS5A antibody (9E10) was a kind gift from Charles Rice. The mouse monoclonal HCV core (C7-50) antibody was purchased form Abcam (Cambridge, MA, USA). Mouse monoclonal anti-FLAG, rabbit polyclonal anti-FLAG and goat anti-GFP biotinylated antibodies were respectively obtained form Sigma (St Louis, MO, USA) and Rockland (Gilbertsville, PA, USA). Rabbit polyclonal viperin antibodies were generated as previously described (18). Bodipy 493/503 (Invitrogen, Carlsbad, CA, USA) was prepared as a stock solution of 1 mg/ml in ethanol.
Plasmids and transfections
Human FDPS was amplified from human liver cDNA, and cloned into pLNCX2 between Not I and Xba I using the following primers: 5′- attcgcggccgcatgcccctgtcccgctggttgagatc-3; and 5′-aacctctagatcaagcgtagtctgggacgtcgtatgggtactttctccgcttgtagattttgcgcgcaag-3′, engineering it to contain a 3′-HA tag. pLenti6-mCherry was generated by cloning mCherry cDNA (lacking a stop codon) into BamHI and XhoI sites of pLenti6/V5-D-TOPO (Invitrogen). Human VAP-A (transcript variant 2) and Rab5A cDNA were PCR-amplified from Huh-7.5 cell cDNA using the following oligonucleotides (restriction sites are italicized): VAP-A (5′- catctcgagctatggcgtccgcctcaggg-3′ and 5′-ggtacgcgttgcatgcttcactctacaagatgaatttc-3′) and Rab5A (5′-catctcgagcttcaaccatggctagtcgaggcgcaa-3′ and 5′- ggtacgcgtttagttactacaacactgattcct-3′) and cloned, in-frame, into XhoI and MluI sites of pLenti6-mCherry. The expression plasmid pHalo-PI4K-IIIα was purchased from Promega (Kazusa DNA Research Institute clone pFN21AB1434). pEGFPC1-ALDI, pEGFPC1-ADRP were generously provided by Albert Pol (University of Barcelone, Barcelona, Spain), John McLauchlan (MRC Virology Unit, Glasgow, UK) respectively. The human viperin plasmid has previously been described (11), and mutant versions of the plasmid were constructed in pLNCX2 either via mutagenesis PCR utilising a QuickChange mutagenesis II system (Stratagene, CA, USA) or via PCR cloning using the HindIII and NotI sites and 5′FLAG tagging the constructs, using the primers listed in table 1. Transfection of all plasmids was performed using Fugene6 (Roche, NJ, USA) according to the manufacturers' recommendations.
Table 1.
| Plasmid # | Sense primer (5′-3′) | Antisense primer (5′-3′) |
|---|---|---|
| Mutagenesis primers | ||
| L142835A | L2835A | |
| gtggaggagcgcggtcccgctgttctgctgggcgagggcaaccttct | agaaggttgccctcgcccagcagaacagcgggaccgcgctcctccac | |
| Followed by L14 | ||
| ctgcttttgctgggaagctcgcgagtgtgttcaggcaacctgcgagc | gctcgcaggttgcctgaacacactcgcgagcttcccagcaaagcag | |
| SAM1 | cactcgccaggccaactacaaagccggcttcgctttccacacagc | gctgtgtggaaagcgaagccggctttgtagttggcctggcgagtg |
| SAM2 | gcttaaggaagctggtatggagaagaacaaccaatcacaacaaaagccatttcttcaagaccggggagaatacctgg | ccaggtattctccccggtcttgaagaaatggcttttgttgtgattggttgttcttctccataccagcttccttaagc |
| SAM3 | ccagcgtgagcatcgtggcccttgcaagcctgatccggg | cccggatcaggcttgcaagggccacgatgctcacgctgg |
| SAM4 | ggacattctcgctatcgcctgtctcgcctttgacgaggaagtcaatgtcc | ggacattgacttcctcgtcaaaggcgagacaggcgatagcgagaatgtcc |
| Cloning primers | ||
| W361A | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | caatgcggccgcccgctctacgcatccagcttc |
| 5′Δ17 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | attagcggccgcctaccaatccagcttcagatcagcctta |
| 5′Δ33 | aataagcttatggactacaaggacgacgatgacaagatgctgagggcaacc | attagcggccgcctaccaatccagcttcagatcagcctta |
| 5′Δ50 | tgagcttatggactacaaggacgacgatgacaagatggtcctgagagggccagatg | attagcggccgcctaccaatccagcttcagatcagcctta |
| 3′Δ17 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | attgcggccgcctacttcagaaacatcttttc |
| 3′Δ33 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | attgcggccgcctaaccaacatccaggatgg |
| 3′Δ50 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | attgcggccgcctacagaaagcgcatatattc |
| 3′Δ4 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | ttatctcgagctacagatcagccttactcc |
| 3′Δ6 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | ttatctcgagctaagccttactccatatgtattt |
| 3′Δ10 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | ttatctcgagctatatgtattttcctcctcgcttc |
| 3′Δ14 | ctcaagcttatggactacaaggacgacgatgacaagatgtgggtgcttacacctgc | ttatctcgagctatcctcgcttcagaaacatcttttc |
plncx2 backbone
Real-time polymerase chain reaction
All experiments involving real-time PCR were performed in 12 well plates with Huh-7 cells seeded at 8 × 104/well, 24 hours prior to transfection/infection, and performed at least in triplicate. RNA was extracted from cells using Trizol reagent (Invitrogen). First strand cDNA was synthesized from total RNA and real-time PCR analysis was utilised to quantitate the relative levels of HCV RNA and viperin mRNA in comparison to the house keeping gene RPLPO. Reaction conditions and primers are as described previously (11).
Immunofluorescence staining
Huh-7 cells were seeded on 0.2% gelatin coated cover slips in 24 well trays (4 × 104 cells/well) 24 hrs prior to transfection/infection. Cells were either fixed using methanol/acetone (1:1) for 5 minutes on ice for standard microscopy, or with 4% paraformaldehyde for 10 minutes on ice followed by a 10 minute incubation in 0.1% Triton-X in PBS for confocal microscopy; prior to incubation with primary antibodies for 1 hr at RT. Cells were washed in PBS and incubated with secondary antibodies for 1 hr at RT before being mounted with Prolong Gold reagent (Invitrogen). Images were acquired with a Bio-Rad Radiance 2100 Confocal or a Nikon TiE inverted microscope.
Fluorescence Energy Resonance Transfer (FRET) analysis
Acceptor photobleaching was carried out essentially as described in (19) with the use of Alexa555 (Invitrogen) and Cy5 (Jackson laboratories, Westgrove, PA, USA) conjugated secondary antibodies or GFP and mCherry tagged protein constructs. Images of the acceptor and donor flurophores were acquired using a Zeiss Axioplan2 upright microscope, using a 63X PlanApo objective. Acceptor photobleaching was performed at maximum light intensity for 30-180 sec, followed by re-imaging of the donor and acceptor fluorophores (this was an automated process ensuring identical imaging conditions). The FRET signal (increase in signal postbleach) was determined by the subtraction of the pre- from postbleach donor image using Image J software (20). Lipid droplets and putative replication complexes positive for both proteins being examined were selected and the average intensity in that region was compared on the aligned pre- and postbleach images (the plug-in StackReg was utilized to control for lateral image displacement, and the FRET signal was displayed using the ‘fire’ lookup table). Multiple lipid droplets and /or cytoplasmic foci that were positive for both proteins being measured were examined from at least 10 different cells in each of at least two independent experiments to ensure reproducibility. Negative slides were prepared by either omitting the primary antibody for the acceptor molecule or in the case of the GFP/mCherry FRET imaging cells with only the donor molecule present.
Luciferase assays
Luciferase assays were performed as previously mentioned (10, 21). Briefly, Huh-7 cells were seeded at 8 × 104 in 12 well plates 24 hours prior to transient transfection using Fugene, with either pLNCX2-viperin, pLNCX2-viperin3′Δ17 or empty vector. 24 hours following transfection, cells were transfected using 2μg of in vitro transcribed RNA (DMRIE-C) representing SGRm-JFH1BlaRL (10). Input renilla luciferase was measured at 3 hrs post RNA transfection to obtain a background reading, with further measurements being taken at 24 and 48 hrs. All time points were performed in quadruplicate. Luciferase assays involving the dicistronic reporter plasmid, pRLHL (21) were performed in a similar manner, with firefly and renilla luciferase measured at 24 hrs post vector transfection. RLuc is translated via cap-dependent translation, whereas the translation of FLuc is directed by the HCV IRES.
Statistical analysis
Student t-tests were utilised to analyse the distributions of 2 normally distributed data sets. All statistical analysis was performed using SPSS 10.
Results
Viperin is induced during HCV infection of Huh-7 cells and inhibits replication
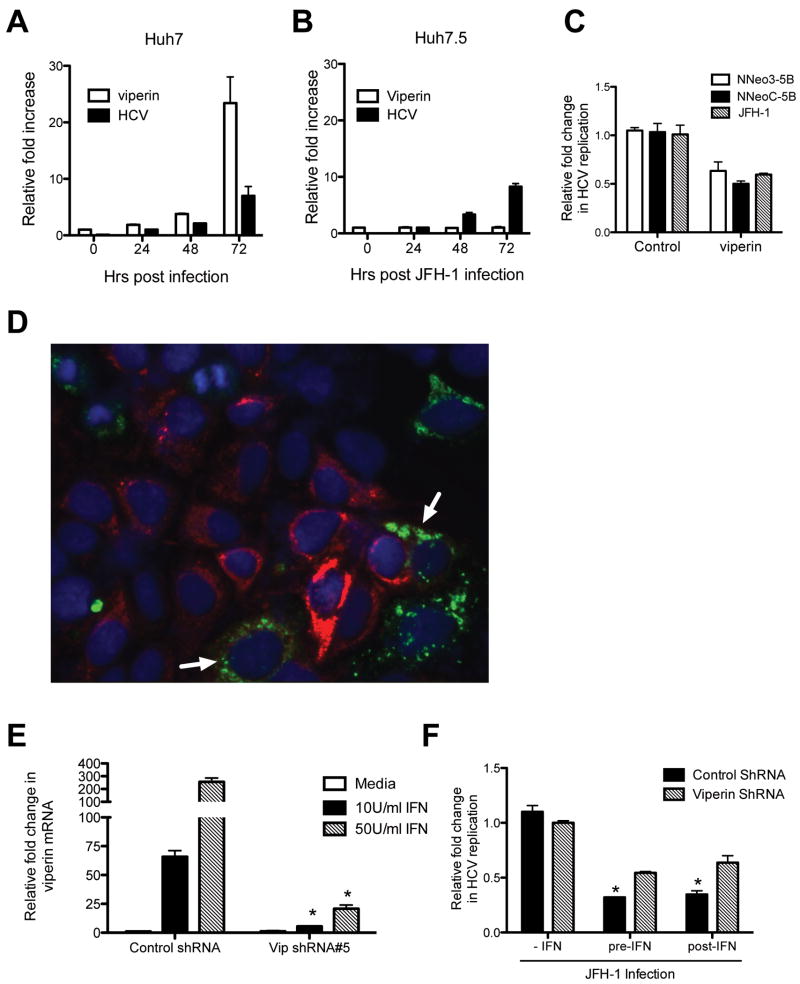
We have previously demonstrated that viperin mRNA expression in Huh-7 cells is responsive to either the dsRNA analogue poly I:C or in vitro transcribed HCV RNA (11). To extend these observations in the context of the complete HCV life-cycle, we infected Huh-7 cells with HCV JFH-1 and monitored viperin mRNA expression. Viperin mRNA expression was significantly increased (∼25 fold) at 72 hrs post infection which coincided with an increase in HCV RNA (Fig 1A). Interestingly similar experiments performed in the Huh7.5 cell line which is defective in dsRNA signaling via a mutation in the pathogen-recognition receptor RIG-I (22), showed only a slight increase in viperin mRNA expression, even though greater than 95% of cells were infected (Fig 1B and Fig S1), implying that its expression in the Huh-7's was RIG-I mediated. We also extended our previous results using the HCV replicon system to show that following transient expression of viperin, HCV JFH-1 replication is inhibited by approximately 45% (Fig 1C). Interestingly, dual immunostaining for both HCV antigen (NS5A) and viperin revealed few cells expressing both antigens, even though control cells were approximately 90% positive for HCV (Figure 1D). In those cells expressing both NS5A and viperin a much lower level of HCV NS5A expression was noted (Fig 1D arrows). These results confirm the anti-HCV activity of viperin using the more physiologically relevant JFH-1 infectious virus system.
Figure 1. Viperin is antiviral and important in the anti-HCV properties of IFN.
(A and B) Expression of viperin in HCV infected Huh-7 and Huh-7.5 cells. Cells were infected with JFH-1 at an MOI of 0.03 and the RNA harvested for Real-time PCR at the indicated time points. (Data are represented as mean + SEM) (C) Viperin expression limits HCV replication. Huh-7 cells were transfected with pLNCX2-viperin 24 hrs prior to JFH-1 infection (MOI =0.03), and RNA harvested 24 hrs post infection. HCV replicon cells were transfected with pLNCX2-viperin and cell sorted for positive expression 48 hrs post transfection. (Data are represented as mean +/- SEM with a significance of p < 0.05 calculated using a student t test for all three samples) (D) Viperin limits HCV replication. Huh-7 cells were transfected with pLNCX2-viperin 24 hrs pre-JFH-1 infection (MOI=0.03) and immunofluorescence staining performed 48 hrs later using a rabbit polyclonal anti-viperin antibody and a mouse monoclonal anti-NS5A antibody, followed by an Alexa488-conjugated goat anti-rabbit Ig and a Alexa555 conjugated goat anti-mouse Ig secondary antisera. Red cells are stained for NS5A antigen, and green cells are expressing viperin, with dual labeled cells indicated by arrow heads. (E) shRNA-mediated inhibition of IFN induced viperin expression in Huh-7 cells. Polyclonal Vip-shRNA cell line #5 and its control were infected with JFH-1 (MOI=0.03) and then treated with IFN-α 24 hrs post infection for 16 hours before real-time PCR was performed on extracted RNA. (Data are represented as mean + SEM, *p < 0.001 for Vip-shRNA cell line #5 vs the control at the indicated treatments) (F) Viperin knockdown reduces the anti-HCV activity of IFN. The polyclonal Vip-shRNA cell line #5 and its control were either pre-treated (24 hrs) prior to JFH-1 infection (MOI=0.03) or post-treated 24 hrs following infection for 16 hrs with 50 U/ml IFN-α. RNA was analysed using Real-time PCR. (Data are represented as mean + SEM, with a significance of p < 0.05 for control cells vs viperin shRNA in the pre- and post-IFN calculated using a student t test)
shRNA knockdown of viperin reduces the antiviral effect of IFN-α
To determine if endogenous viperin has anti-HCV activity following IFN-α stimulation we created a number of polyclonal Huh-7 cell lines stably expressing shRNA targeting viperin mRNA (Fig S2). The cell line Vip shRNA#5 compared to a control cell line expressing non-specific shRNA produced significantly less viperin mRNA following IFN-α stimulation (Fig 1E, *p < 0.001). Vip shRNA#5 cells stimulated for 24 hrs with varying concentrations of IFN-α had approximately 90-95% less viperin mRNA than their control counterparts. The Vip shRNA#5 cells were then either pre-treated with IFN-α for 24 hours before JFH-1 infection, or treated with IFN-α for 24 hours following JFH-1 infection. As can be seen in Figure 1F, the control cell line was able to reduce HCV replication levels by 69 and 66% respectively following either pre- or post- IFN-α treatment, whereas the Vip shRNA#5 cell line was only able to reduce HCV replication by 45 and 37% respectively under the same conditions. These results demonstrate that viperin plays an important, but not exclusive role in the antiviral effects of IFN-α against HCV in vitro.
Viperin interacts with HCV core and NS5A
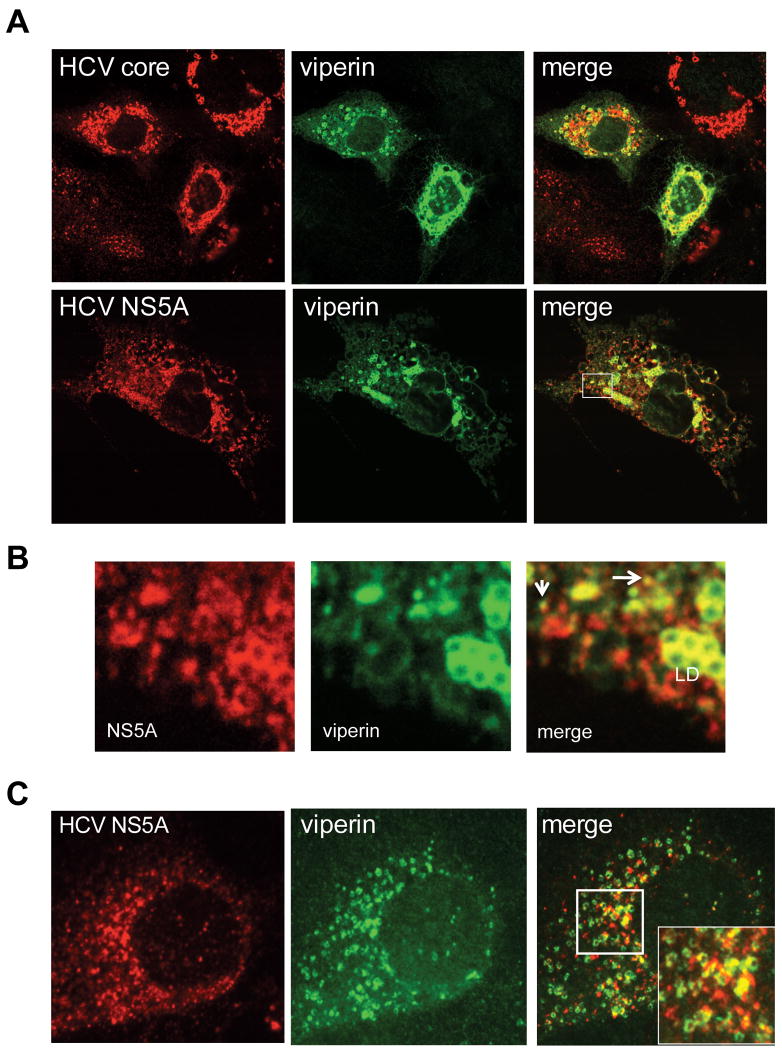
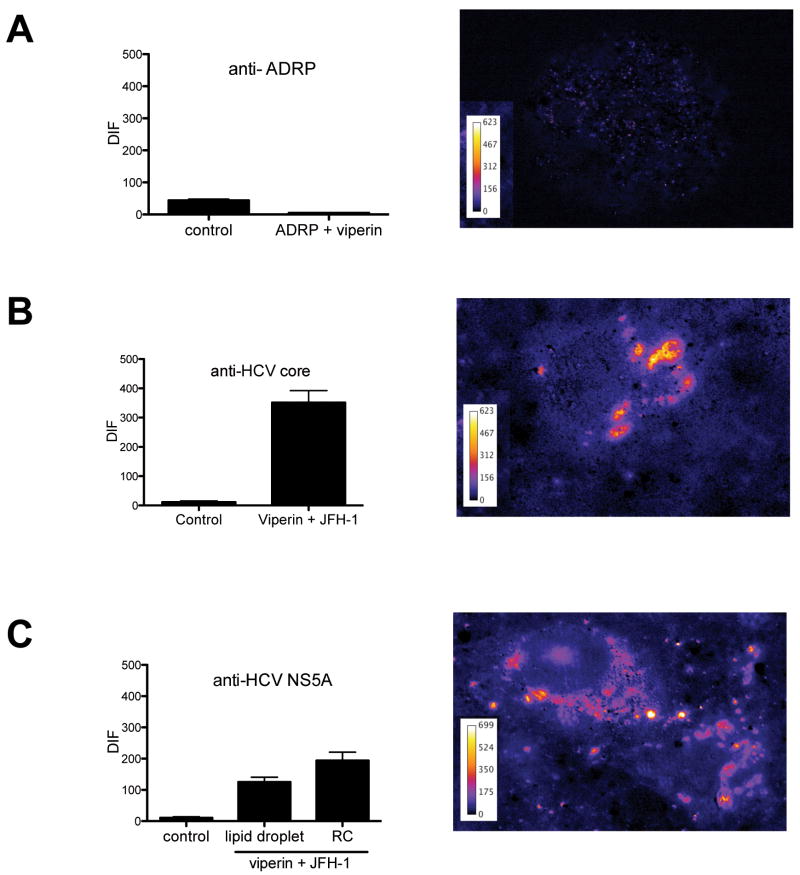
Previous reports suggest that viperin localises to the ER (18, 23), however we and others have observed that viperin also localises to lipid droplets (LD) in Huh-7 cells (Fig S3 and S4)(24). The LD plays an important role in the HCV life cycle as both core and NS5A localise to the LD surface (25). With this in mind we investigated the distribution of viperin, HCV core and NS5A in Huh-7 cells productively infected with JFH-1 using confocal microscopy. These studies revealed considerable but not absolute co-localisation between viperin and both core and NS5A proteins, surrounding LDs (Fig 2A). In addition viperin also co-localised with NS5A in a proportion of small cytoplasmic foci that have been well characterised as part of the HCV replication complex (RC) (26) (arrow heads in Figure 2B). Next we investigated localisation of viperin and NS5A in cells harbouring a HCV subgenomic replicon that are devoid of the HCV structural proteins. In these cells viperin co-localised with NS5A in a similar manner to that observed for JFH-1 infected cells with co-localisation of viperin and NS5A at the RC and LD surface (Fig 2C inset), although the latter was not as pronounced as in JFH-1 infection. This suggests that viperin may exert its antiviral effect through a possible interaction with NS5A either within the RC or at the LD surface. To extend our confocal microscopy results and to determine if viperin physically interacts with NS5A and core, fluorescence energy resonance transfer (FRET) was utilised. Even though viperin and ADRP co-localise by confocal analysis no positive FRET was observed (Fig 3A) indicating that it is unlikely these two proteins physically interact. In contrast, Huh-7 cells infected with JFH-1 displayed significant FRET at the LD surface between viperin and either HCV core or NS5A, in addition to positive FRET with NS5A within the HCV RC (Fig 3B and C). Collectively these results strongly suggest that viperin is able to interact with core and NS5A on the LD surface and with NS5A within the RC.
Figure 2. Viperin co-localises with HCV core and NS5A proteins.
(A and B) Huh-7 cells were infected with JFH-1 (MOI=0.03) for 72 hrs, before being transfected with pLNCX2-viperin. Cells were stained 24 hrs following transfection with a rabbit polyclonal anti-viperin antibody,or a mouse monoclonal anti-core or anti-NS5A, followed by an Alexa555-conjugated goat anti-rabbit Ig or an Alexa555 conjugated goat anti-mouse Ig secondary antisera. The boxed region is shown enlarged in panel B, with arrows indicating co-localisation of viperin and NS5A in putative RC (LD = lipid droplet). (C) NNeo3-5B cells were transfected with pLNCX2-viperin and stained as above 48 hours later.
Figure 3. Viperin displays positive FRET with HCV core and NS5A.
Huh-7 cells were infected with JFH-1 (MOI=0.03) for 48 hrs, before being transfected with pLNCX2-viperin or co-transfected with pLNCX2-viperin and pEGFPC1-ADRP. 24 hrs post transfection cells were stained with either a rabbit polyclonal anti-viperin antibody followed by an Alexa555-conjugated goat anti-rabbit Ig and (A) a biotinylated goat anti-GFP antibody and a CY5 conjugated rabbit anti-goat Ig, (B) a mouse monoclonal anti-core antibody followed by a CY5 goat anti-mouse Ig or (C) a mouse monoclonal anti-NS5A followed by a CY5 goat anti-mouse Ig. Slides were analysed on a Zeiss Axioplan microscope for FRET, with a representative cell displaying a positive FRET signal shown for each experiment . (Data are represented as mean +/- SEM with a significance of p < 0.001 for (B) and (C)).
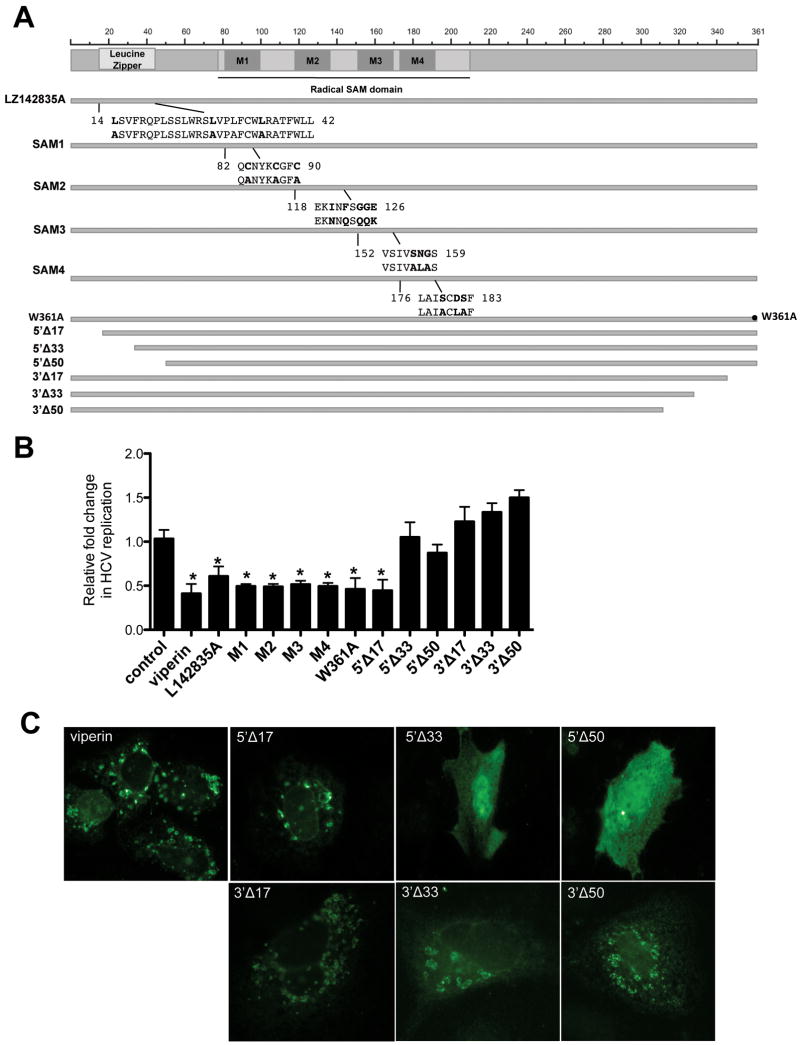
Mutagenic analysis reveals important regions of viperin for anti-HCV activity
Little is known about the functional aspects of viperin that contribute to its antiviral activity. Recent work has demonstrated that viperin is able to bind the enzyme FDPS and interrupt the mevalonate pathway, causing a restriction in influenza budding from lipid rafts (23). Restoration of this pathway did not rescue HCV replication in viperin expressing cells, thereby indicating an alternative anti-viral mechanism for the protein in the context of HCV (Fig S6).
Viperin is a member of the radical SAM family of enzymes (27), and contains 4 radical SAM motifs in addition to a putative leucine zipper domain, which may be of importance in protein-protein interactions. In order to further understand the anti-HCV mechanism of viperin, mutations were made to destabilise the leucine and SAM1-4 domains (Figure 4A). In contrast to previous reports (9), all viperin mutations retained anti-HCV activity in JFH-1 infected Huh-7 cells (Fig 5B, M1-4). Next we created a panel of deletion mutants from the N- and C-termini of viperin (Figure 4A). Deletion of 33 aa or 17 aa from the N and C termini respectively, abrogated viperin's anti-HCV function, (Fig 4B). Interestingly, coincident with loss of anti-HCV activity for the N-terminal deletions was a redistribution of viperin from the LD's and ER to a homogeneous cytoplasmic pattern (Fig 4C). This is not entirely unexpected given the presence of a N terminally located amphipathic-alpha helix (13), which is thought to allow peripheral proteins to anchor into the ER, induce curvature of the ER and bind lipid droplet surfaces (28). In contrast to previous reports (9), the 6 terminal aa were not required for anti-viral activity (Fig 4B and Fig S5). However, deletion of 10aa abrogated the anti-HCV action of viperin.
Figure 4. Viperin N'- and C'- terminal mutants are not anti-viral.
(A) Schematic diagram of viperin and mutant derivatives. (B) Analysis of the anti-HCV activity of the viperin mutants. Huh-7 cells were transfected with pLNCX2-viperin or the indicated mutant viperin plasmid and 24 hrs later infected with JFH-1 (MOI=0.03) for 24 hrs before RNA harvest and real-time PCR analysis. (Data are represented as mean +/- SEM, *p < 0.05) (C) N-terminal mutants of viperin have altered localisation. Huh-7 cells were transfected with pLNCX2-viperin or one of the 6 pLNCX2-viperin truncation mutants. Cells were stained 24 hrs following transfection with either a rabbit polyclonal anti-viperin antibody followed by an Alexa488-conjugated goat anti-rabbit Ig or in the case of the C-terminal mutants, a mouse monoclonal anti-FLAG antibody followed by an Alexa488-conjugated goat anti-mouse Ig antisera.
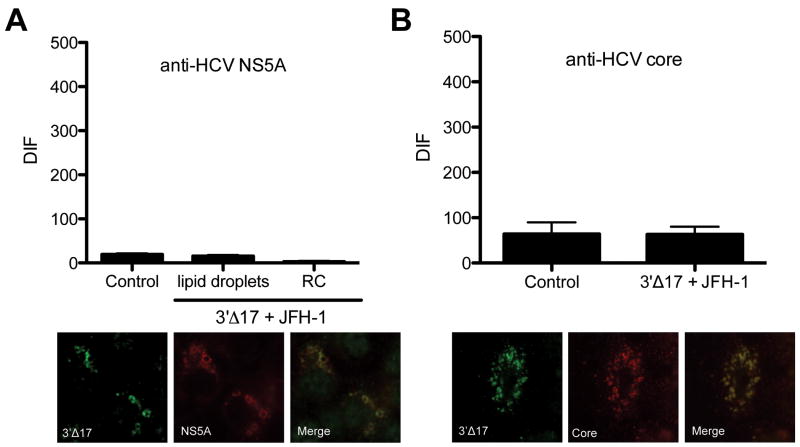
Figure 5. The C terminus of viperin is important for its interaction with HCV core and NS5A.
Huh-7 cells were infected with JFH-1 (MOI=0.03) for 48 hrs, before being transfected with viperin 3′Δ17. 24 hours post transfection cells were stained with a rabbit polyclonal anti-FLAG antibody followed by an Alexa555-conjugated goat anti-rabbit Ig and (A) a mouse monoclonal anti-NS5A antibody followed by a CY5 goat anti-mouse Ig or (B) a mouse monoclonal anti-core antibody followed by a CY5 goat anti-mouse Ig. Slides were analysed on a Zeiss Axioplan microscope for FRET. (Data are represented as mean +/- SEM).
In contrast to N-terminal deletions, C-terminal truncations of viperin localised to the ER and LD (Figure 4C) and co-localised with HCV core and NS5A (Figures 5A and 5B), even though its antiviral activity had been abrogated (Fig 4B). FRET analysis of JFH-1 infected Huh-7 cells expressing the 3′Δ17 viperin mutant revealed that viperin was no longer associated with either HCV core or NS5A (Fig 5A and B).
Collectively these results demonstrate that the final 10aa of the C-terminal region of viperin are essential for its ability to limit intracellular HCV RNA levels through interaction with HCV NS5A and/or core.
Viperin interrupts HCV replication
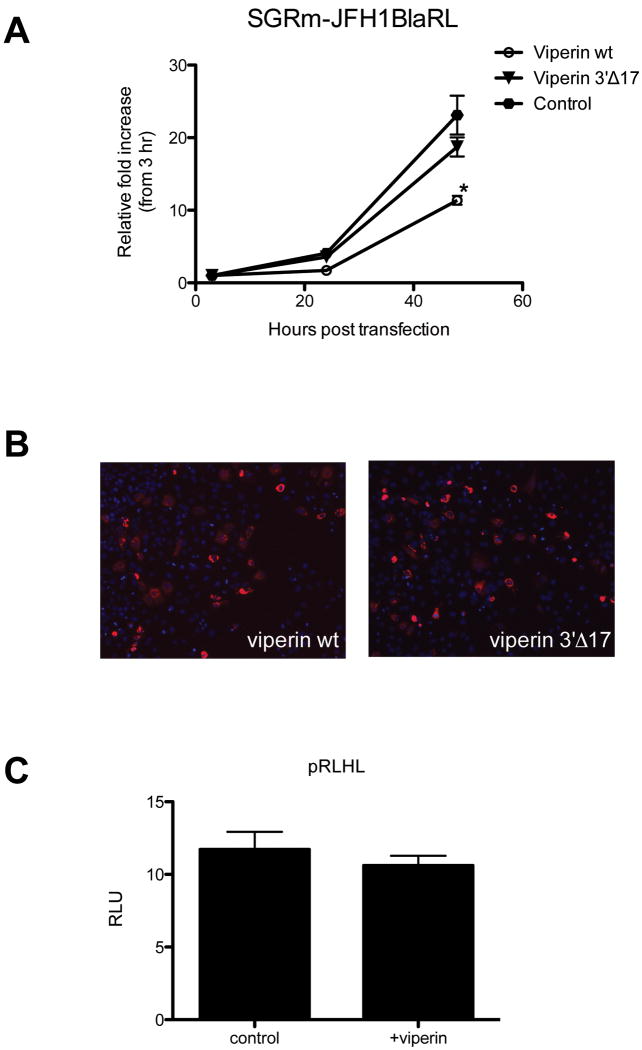
Given the ability of viperin to limit the HCV subgenomic replicon (Fig 1C), and its interaction with NS5A within the RC (Fig 3B) we hypothesised that viperin was acting at the level of HCV RNA replication. To investigate the mechanism of viperin action in more detail we used a subgenomic HCV replicon that encodes the renilla luciferase reporter gene (SGRm-JFH1BlaRL) that allows us to investigate the kinetics of HCV RNA replication uncoupled from virion assembly (10). Compared to control (empty plasmid) or a viperin 3′ deletion mutant that has no antiviral activity (pLNCX2-viperin3′Δ17), cells transfected with WT viperin expressing plasmid revealed a significant decrease in Renilla output (p = 0.005) at 48 hours post transfection (Fig 6A). We also discounted any effect of viperin on HCV IRES directed translation (Fig 6B). Collectively these results suggest that viperin acts at the level of HCV RNA replication via a direct interaction of viperin with NS5A at the RC.
Figure 6. Viperin inhibits HCV RNA replication but not IRES activity.
Huh-7 cells were transiently transfected with pLNCX2-viperin, pLNCX2-viperin3′Δ17 or empty vector 24 hours prior to RNA transfection with (A) SGRm-JFH1BlaRL. Representative transfections efficiencies of the viperin plasmids are demonstrated in (Data are represented as mean +/- SEM, *p < 0.05) (B). The effects of viperin on HCV IRES activity were ascertained via transient transfection of the HCV-IRES plasmid pRLHL with either pLNCX2 or pLNCX2-viperin into Huh-7 cells. Luciferase readouts were taken 24 hours after transfection and were controlled by renilla expression driven from the . (Data are represented as mean +/- SEM)
Viperin co-localizes with VAP-A, a known NS5A interacting cellular factor
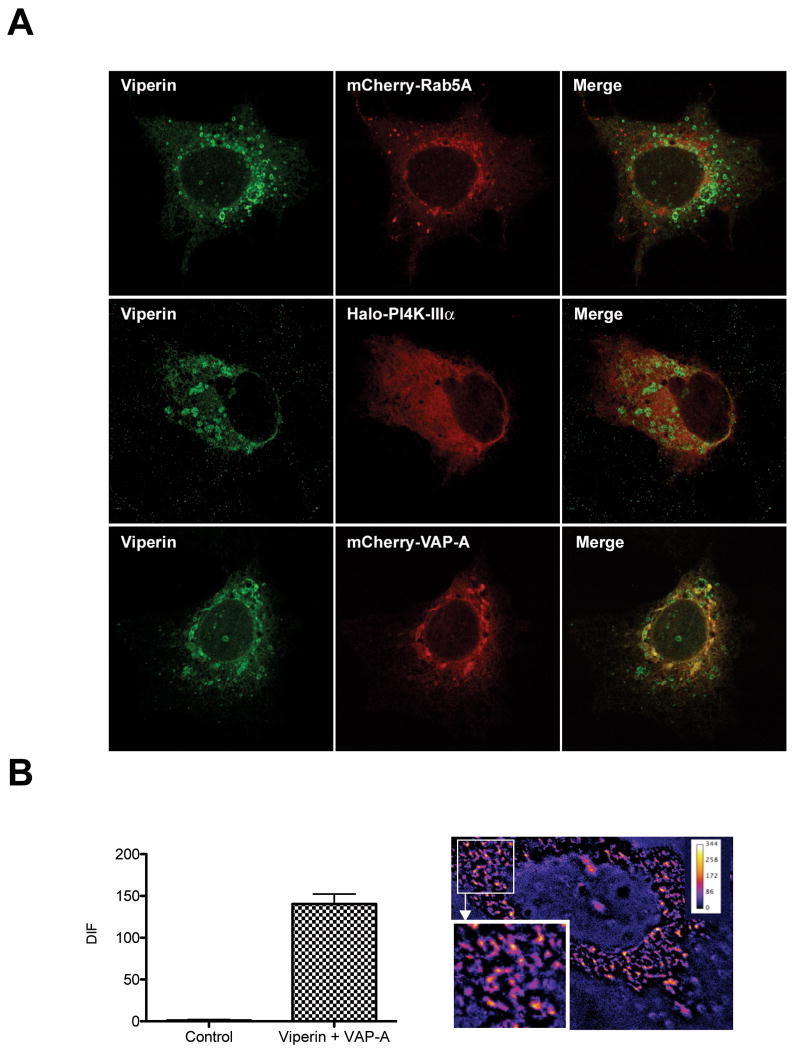
A number of cellular factors, including Rab5a, VAP-A and the ER lipid kinase PI4K-IIIa are essential for HCV replication and co-localize with NS5A at the RC (29, 30). We therefore investigated the potential for interaction between the above mentioned factors and viperin using confocal microscopy. In the absence of productive HCV infection, viperin co-localized with VAP-A, but not Rab5a or PI4K-IIIa (Fig 7A). To investigate the significance of the viperin/VAP-co-localization in the context of HCV replication, FRET analysis between viperin and VAP-A was performed in Huh-7 cells harbouring the HCV full length genomic replicon. Viperin was found to positively interact with VAP-A in small cytoplasmic foci representing RC's (Fig 7B). These results confirm that viperin/NS5A cytoplasmic structures are RC's and suggest that the interaction of viperin with VAP-A at the RC may destabalise HCV RNA replication.
Figure 7. Viperin interacts with the HCV RC factor VAP-A.
(A) Huh-7 cells were transiently transfected with pLNCX2-viperin and either pLenti6-mCherry-Rab5a, Halo-PI4K-IIIα or pLenti6-mCherry-VAP-A. 24 hrs post transfection, the Halo-TMR ligand (Promega) was mixed with the cells expressing PI4K-IIIα for 15 min before being washed out and all cells fixed. Cells were stained for viperin using a mouse monoclonal anti-FLAG antibody followed by an Alexa488-conjugated goat anti-mouse Ig antisera.
(B) NNeoC-5B cells were co-transfected with pEGFP-viperin and pLenti6-mCherry-VAP-A. 24 hrs post transfection cells were fixed and the slides were analysed on a Zeiss Axioplan microscope for FRET, with a representative cell displaying a positive FRET signal shown. (Data are represented as mean +/- SEM with a significance of p < 0.001).
Discussion
The standard treatment for chronic hepatitis C is IFN-α2/ribavirin combination therapy, however at the molecular level its mode of action is not well understood. In individuals that clear HCV following IFN therapy there is a rapid first phase of decline lasting approximately 1–2 days, followed by a slower second phase of decline (31). This early decline suggests that IFN has a direct effect on HCV replication via expression of antiviral ISGs in infected hepatocytes. This observation has been validated in vitro with IFN-α treatment of replicon cells resulting in a dose dependant decrease in HCV RNA (32, 33) while long-term treatment with IFN-α can completely cure these cells of HCV replication (34). However the ISGs responsible for this decrease in HCV RNA are not well characterised. Considering that hundreds of ISGs are induced following IFN stimulation, a systematic approach is required to identify novel ISGs with antiviral activity.
We previously identified the ISG viperin as being significantly expressed in the HCV infected liver and subsequently demonstrated viperin to have anti-HCV activity (11). In this study we now show that viperin is antiviral in the context of the complete HCV life cycle. Consistent with viperin being an ISG we also observed that in Huh-7 cells there was a significant increase in viperin mRNA following infection with HCVcc (JFH-1). Interestingly this increase in viperin mRNA was not seen in Huh-7.5 cells that are highly permissive for HCV infection and are defective in the dsRNA sensing molecule RIG-I. This strongly suggests that viperin expression is induced through the cellular innate dsRNA response and subsequent interferon production that feeds back to induce viperin expression through IRF3 and ISGF3 dependent mechanisms (35, 36). To confirm viperin's anti-HCV activity we knocked down viperin expression using an RNAi approach and were able to demonstrate for the first time that viperin plays an important but not exclusive role in the anti-HCV activity of IFN-α. Considering that many genes are differentially regulated in Huh-7 cells following IFN-α stimulation it is highly likely that a coordinated ISG response is responsible for control of HCV replication.
A number of studies have suggested that viperin has an ER distribution (18, 23), however, we and others have observed that viperin localises to both LDs and in our studies the HCV NS5A positive RCs (24). LDs have recently been shown to be an essential component of the HCV life cycle (25) and it is thought that the close association of the LD and ER membranes provides a microenvironment that is essential for HCV RNA replication and virion production. It has been hypothesized that the interaction of viperin with NS5A at the LD surface is the possible mechanism whereby viperin exerts its antiviral effect through disruption of virion assembly (12, 13). However, a number of lines of evidence suggest that this is unlikely. Firstly, viperin exerts its anti-HCV effect against the HCV subgenomic replicon, that lacks the HCV structural proteins and is defective in virion assembly. This would also suggest that the viperin-core interaction we observed is not fundamental to viperins antiviral activity, and that the interaction with NS5A is critical. It is plausible that the observed interaction between viperin and core at the surface of the LD is mediated by the ability of core to recruit and interact with NS5A at the LD surface to initiate virion assembly. Secondly, viperin is antiviral against a genotype 2a HCV subgenomic replicon (SGRm-JFH1BlaRL) in which the HCV IRES drives expression of the luciferase reporter gene to allow for quantitative measurement of HCV RNA replication kinetics uncoupled from virion assembly, following transfection of in vitro transcribed HCV RNA (10). Expression of viperin significantly suppressed luciferase output from this HCV subgenomic replicon suggesting that the anti-HCV effect of viperin was at the level of HCV replication and not virion assembly. Finally, through confocal miscroscopy and FRET analysis we have conclusively shown that viperin interacts with both NS5A and the pro-viral host factor VAP-A within the HCV RC. VAP-A (also known as hVAP-33) is a known interacting partner with NS5A (and NS5B) and required for the efficient replication of HCV genomic RNA (30). While the exact mechanism of how VAP-A enhances HCV replication is unknown, VAP-A is involved in the regulation, biosynthesis and trafficking of sterols and lipids and is thought to be involved in the formation of the HCV RC (30). Thus based on our observations it is conceivable to envisage that viperin interacts with VAP-A at the HCV RC and that this interaction perturbs the association of NS5A with VAP-A that is essential for efficient viral RNA replication.
In this study we have shown that the ISG viperin is physically associated with the HCV NS5A and core proteins at LD interface while interacting with the pro-viral host factor VAP-A at the HCV RC. Through mutational analysis of viperin we have also demonstrated that the presence of the N-terminal amphipathic helix of viperin is important to localize the protein to the LD and RC, and that the C-terminal region of viperin is essential for its ability to interact with NS5A and exert its anti-HCV action. Furthermore, we have demonstrated that viperin is antiviral against HCV JFH-1 and the HCV subgenomic replicon (genotype 1b and 2a) suggesting that the interaction with core protein is not essential for its antiviral activity. This strongly implicates the association of viperin with NS5A and VAP-A at the RC as the site where viperin exerts its novel antiviral activity through altering the stability and/or functionality of the HCV RC. Interestingly, while the list of viruses towards which viperin exerts its antiviral effect on is growing, its mode of action is unique in many cases highlighting the complexity of this multi-functional protein. This work adds to our understanding of the viral host interaction and the hepatocyte response to overcome viral infection. Moreover, defining the mechanism of action of these ISGs will add to our understanding of HCV replication and may present novel therapeutic strategies for chronic hepatitis C.
Supplementary Material
Huh-7 or Huh7.5 cells were infected with JFH-1 HCVcc (MOI= 0.03) and fixed at 24, 48 and 72 hrs post infection. Cells were stained for HCV NS5A expression using a mouse monoclonal anti-NS5A antibody followed by detection with an Alexa488-conjugated goat anti-rabbit Ig.
Huh-7 cells were infected with lentivirus expressing either Control shRNA or Vip shRNA#1 - #5. Polyclonal cell lines were established, excepting Vip shRNA#3 which was determined to have a cellular growth defect. Stable cell lines were treated with Media alone or 100 U/ml of IFN-α for 16 hrs before RNA extraction and subsequent real-time PCR to establish the ability of Vip shRNA clones to limit viperin induction.
Viperin co-localises with resident lipid droplet proteins. Huh-7 cells were co-transfected with pLNCX2-viperin and either pEGFPC1-ADRP or pEGFPC1-ALDI. Cells were stained 24 hrs following transfection with a rabbit polyclonal anti-viperin antibody followed by an Alexa555-conjugated goat anti-rabbit Ig.
(A) The lipid content of Huh-7 cells effects viperin distribution. Huh-7 cells were either cultured with a standard 10% FCS medium or starved of serum for 48 hrs prior to transfection (to reduce the LD content) with pLNCX2-viperin and subsequent staining using a rabbit polyclonal anti-viperin antibody followed by an Alexa488-conjugated goat anti-rabbit Ig. (B) Viperin co-localises with BODIPY. Huh-7 cells were transfected with pLNCX2-viperin, and stained 24 hrs following transfection with a rabbit polyclonal anti-viperin antibody followed by an Alexa555-conjugated goat anti-rabbit Ig. BODIPY staining was performed in conjunction with the secondary antibody.
Huh-7 cells were transfected with pLNCX2-viperin or the indicated mutant viperin plasmid and 24 hrs later infected with JFH-1 (MOI=0.03) for 24 hrs before RNA harvest and real-time PCR analysis.
(A) Viperin does not inhibit FDPS activity. Huh-7 cells were transfected with either pLNCX2-viperin, pLNCX2-FDPS or both plasmids. 24 hrs following transfection cells were infected with JFH-1 (MOI=0.03) and RNA harvested for real-time PCR 24 hrs following infection. (B) Farnesol and geranylgeraniol supplementation does not abrogate the anti-HCV activity of viperin. Huh-7 cells were transfected with either pLNCX2-viperin and/or pLNCX2-FDPS, and 8 hours later treated with 10μM of either farnesol or geranylgeraniol for 16 hrs in an attempt to restore the mevalonate pathway for which farnesyl diphosphate synthetase is a key enzyme. Cells were then infected with JFH-1 (MOI=0.03) for 4 hours before the replacement of the farnesol and geranylgeraniol. RNA was harvested for real-time PCR 24 hrs following infection.
Acknowledgments
The authors wish to thank Takaji Wakita for the use of JFH-1, and Charles Rice for the use of the Huh7.5 cells and the kind gift of the HCV monoclonal NS5A antibody (9E10). We would also like to thank John McLauchlan and Albert Pol for supplying us with the plasmids, pEGFPC1-ADRP and pEGFPC1-ALDI respectively.
Financial support: This work was supported by the NH&MRC of Australia (M.R.B and K.J.H) and the NIH (U19-AI40035-14, R21-AI081058-1, S.M.L. and R01-AI069285 K.L)
Abbreviations
- IFN
interferon
- HCV
hepatitis C virus
- ISG
interferon stimulated gene
- CHC
chronic hepatitis C
- LD
lipid droplet
- RC
replication complex
References
- 1.Theodore D, Fried MW. Natural history and disease manifestations of hepatitis C infection. Curr Top Microbiol Immunol. 2000;242:43–54. doi: 10.1007/978-3-642-59605-6_3. [DOI] [PubMed] [Google Scholar]
- 2.Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 3.Sen GC. Viruses and interferons. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 4.Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem. 2010;285:22741–22747. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han JQ, Barton DJ. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA. 2002;8:512–525. doi: 10.1017/s1355838202020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frese M, Pietschmann T, Moradpour D, Haller O, Bartenschlager R. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J Gen Virol. 2001;82:723–733. doi: 10.1099/0022-1317-82-4-723. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Zhao H, Collins CD, Eckenrode SE, Run Q, McIndoe RA, Crawford JM, et al. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology. 2003;37:1180–1188. doi: 10.1053/jhep.2003.50184. [DOI] [PubMed] [Google Scholar]
- 8.Wang C, Pflugheber J, Sumpter R, Jr, Sodora DL, Hui D, Sen GC, Gale M., Jr Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77:3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, et al. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, Wang N, Woodson SE, Dong Q, Wang J, Liang Y, Rijnbrand R, et al. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology. 2010;409:175–188. doi: 10.1016/j.virol.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helbig KJ, Lau DT, Semendric L, Harley HA, Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42:702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald KA. The interferon inducible gene: Viperin. J Interferon Cytokine Res. 2010;31:131–135. doi: 10.1089/jir.2010.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinson ER, Cresswell P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci U S A. 2009;106:20452–20457. doi: 10.1073/pnas.0911679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda M, Yi M, Li K, Lemon SM. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helbig KJ, Ruszkiewicz A, Semendric L, Harley HA, McColl SR, Beard MR. Expression of the CXCR3 ligand I-TAC by hepatocytes in chronic hepatitis C and its correlation with hepatic inflammation. Hepatology. 2004;39:1220–1229. doi: 10.1002/hep.20167. [DOI] [PubMed] [Google Scholar]
- 16.Helbig KJ, Ruszkiewicz A, Lanford RE, Berzsenyi MD, Harley HA, McColl SR, Beard MR. Differential expression of the CXCR3 ligands in chronic hepatitis C virus (HCV) infection and their modulation by HCV in vitro. J Virol. 2009;83:836–846. doi: 10.1128/JVI.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- 20.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 21.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell GA, Lemon SM. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 22.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Hinson ER, Cresswell P. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem. 2009;284:4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 26.Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, et al. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol. 2004;78:7400–7409. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duschene KS, Broderick JB. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 2010;584:1263–1267. doi: 10.1016/j.febslet.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Cordes KR, Farese RV, Jr, Walther TC. Lipid droplets at a glance. J Cell Sci. 2009;122:749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perelson AS, Herrmann E, Micol F, Zeuzem S. New kinetic models for the hepatitis C virus. Hepatology. 2005;42:749–754. doi: 10.1002/hep.20882. [DOI] [PubMed] [Google Scholar]
- 32.Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001;75:8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanford RE, Guerra B, Lee H, Averett DR, Pfeiffer B, Chavez D, Notvall L, et al. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J Virol. 2003;77:1092–1104. doi: 10.1128/JVI.77.2.1092-1104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frese M, Schwarzle V, Barth K, Krieger N, Lohmann V, Mihm S, Haller O, et al. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology. 2002;35:694–703. doi: 10.1053/jhep.2002.31770. [DOI] [PubMed] [Google Scholar]
- 35.Fredericksen B, Akkaraju GR, Foy E, Wang C, Pflugheber J, Chen ZJ, Gale M., Jr Activation of the interferon-beta promoter during hepatitis C virus RNA replication. Viral Immunol. 2002;15:29–40. doi: 10.1089/088282402317340215. [DOI] [PubMed] [Google Scholar]
- 36.Severa M, Coccia EM, Fitzgerald KA. Toll-like receptor-dependent and - independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J Biol Chem. 2006;281:26188–26195. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Huh-7 or Huh7.5 cells were infected with JFH-1 HCVcc (MOI= 0.03) and fixed at 24, 48 and 72 hrs post infection. Cells were stained for HCV NS5A expression using a mouse monoclonal anti-NS5A antibody followed by detection with an Alexa488-conjugated goat anti-rabbit Ig.
Huh-7 cells were infected with lentivirus expressing either Control shRNA or Vip shRNA#1 - #5. Polyclonal cell lines were established, excepting Vip shRNA#3 which was determined to have a cellular growth defect. Stable cell lines were treated with Media alone or 100 U/ml of IFN-α for 16 hrs before RNA extraction and subsequent real-time PCR to establish the ability of Vip shRNA clones to limit viperin induction.
Viperin co-localises with resident lipid droplet proteins. Huh-7 cells were co-transfected with pLNCX2-viperin and either pEGFPC1-ADRP or pEGFPC1-ALDI. Cells were stained 24 hrs following transfection with a rabbit polyclonal anti-viperin antibody followed by an Alexa555-conjugated goat anti-rabbit Ig.
(A) The lipid content of Huh-7 cells effects viperin distribution. Huh-7 cells were either cultured with a standard 10% FCS medium or starved of serum for 48 hrs prior to transfection (to reduce the LD content) with pLNCX2-viperin and subsequent staining using a rabbit polyclonal anti-viperin antibody followed by an Alexa488-conjugated goat anti-rabbit Ig. (B) Viperin co-localises with BODIPY. Huh-7 cells were transfected with pLNCX2-viperin, and stained 24 hrs following transfection with a rabbit polyclonal anti-viperin antibody followed by an Alexa555-conjugated goat anti-rabbit Ig. BODIPY staining was performed in conjunction with the secondary antibody.
Huh-7 cells were transfected with pLNCX2-viperin or the indicated mutant viperin plasmid and 24 hrs later infected with JFH-1 (MOI=0.03) for 24 hrs before RNA harvest and real-time PCR analysis.
(A) Viperin does not inhibit FDPS activity. Huh-7 cells were transfected with either pLNCX2-viperin, pLNCX2-FDPS or both plasmids. 24 hrs following transfection cells were infected with JFH-1 (MOI=0.03) and RNA harvested for real-time PCR 24 hrs following infection. (B) Farnesol and geranylgeraniol supplementation does not abrogate the anti-HCV activity of viperin. Huh-7 cells were transfected with either pLNCX2-viperin and/or pLNCX2-FDPS, and 8 hours later treated with 10μM of either farnesol or geranylgeraniol for 16 hrs in an attempt to restore the mevalonate pathway for which farnesyl diphosphate synthetase is a key enzyme. Cells were then infected with JFH-1 (MOI=0.03) for 4 hours before the replacement of the farnesol and geranylgeraniol. RNA was harvested for real-time PCR 24 hrs following infection.